Abstract
The stem-rust-susceptible barley cv. Golden Promise was transformed by Agrobacterium-mediated transformation of immature zygotic embryos with the Rpg1 genomic clone of cv. Morex containing a 520-bp 5′ promoter region, 4,919-bp gene region, and 547-bp 3′ nontranscribed sequence. Representatives of 42 transgenic barley lines obtained were characterized for their seedling infection response to pathotype Pgt-MCC of the stem rust fungus Puccinia graminis f. sp. tritici. Golden Promise was converted from a highly susceptible cultivar into a highly resistant one by transformation with the dominant Rpg1 gene. A single copy of the gene was sufficient to confer resistance against stem rust, and progenies from several transformants segregated in a 3:1 ratio for resistance/susceptibility as expected for Mendelian inheritance. These results unequivocally demonstrate that the DNA segment isolated by map-based cloning is the functional Rpg1 gene for stem rust, resistance. One of the remarkable aspects about the transformants is that they exhibit a higher level of resistance than the original sources of Rpg1 (cvs. Chevron and Peatland). In most cases, the Golden Promise transformants exhibited a highly resistant reaction where no visible sign of infection was evident. Hypersensitive necrotic “fleck” reactions were also observed, but less frequently. With both infection types, pathogen sporulation was prevented. Southern blot and RT-PCR analysis revealed that neither Rpg1 gene copy number nor expression levels could account for the increased resistance observed in Golden Promise transformants. Nevertheless, this research demonstrates that stem-rust-susceptible barley can be made resistant by transformation with the cloned Rpg1 gene.
Resistance of flax (Linum usitatissimum) to the rust fungus Melampsora lini provided the basic genetic framework for exploring interactions between plant disease resistance genes and avirulence genes of pathogens. Based on studies of the genetics of resistance in flax and genetics of virulence in the flax rust pathogen, H. H. Flor (1, 2) proposed that for every dominant resistance gene (R) in the host there was a corresponding gene for avirulence (Avr) in the pathogen. This specific gene-for-gene interaction was later found to be characteristic for many interactions of plants with viral and bacterial, as well as fungal, pathogens (3). Introduction of the maize Ac transposable element by transformation of flax allowed tagging of the L6 gene for flax rust resistance and determination of its structure (4). Similar to other R genes cloned early on (5), the L6 gene encodes a protein with three domains: an N-terminal domain homologous to the cytoplasmic part of the Toll/IL-1 receptor (TIR), a central nucleotide binding site (NBS) domain, and a C-terminal leucine-rich repeat (LRR) domain. The 13 different recognition specificities between the L gene proteins and M. lini avirulence proteins are determined by alleles of a single gene. The alleles differ by single or multiple amino acid changes in the three domains (6, 7). The L6 and L11 proteins differ only in the LRR region, whereas L6 and L7 differ in the TIR domain. By using functional analysis in transgenic flax plants with recombinant alleles constructed in vitro, novel specificities were created, e.g., by combining the LRR of L10 with the two other domains of L2. The L7 specificity could be engineered by combining the TIR domain of L2 with the promoter and NBS+LRR domains specified by the L6 gene. This L7 specificity was also obtained by crossover in progenies from the cross L2/L6 × LH/LH. Flax contains another complex locus (M) that confers resistance to biotrophic pathogens. The M locus is comprised of 15 linked genes with ≈86% amino acid sequence identity to the L genes. These linked genes express seven known resistance specificities (8), but only one member of the cluster is responsible for a given interaction with an avirulence gene.
A dominant gene of the wild rice species Oryza longistaminata (Xa21) was transferred by crossing into the cultivated rice variety IR24 and confers resistance to all known races of the bacterial blight pathogen Xanthomonas oryzae pv. oryzae (9). Xa was isolated by map-based cloning and its identity verified by transformation of susceptible lines and cultivars (10–13). The gene translates into a protein with an LRR domain and a serine–threonine kinase domain. One of the transgenic lines (T103-10) revealed excellent agronomic characteristics in field tests (14).
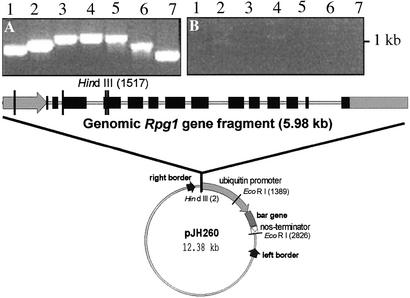
Recently, the Rpg1 gene for resistance to stem rust caused by Puccinia graminis f. sp. tritici in barley was cloned by a map-based approach (15). Rpg1 was identified by high-resolution genetic and physical mapping, comparative sequencing of multiple alleles from resistant and susceptible lines, and analysis of a rare recombinant that combined portions of the gene from resistant and susceptible parents. Other putative candidate genes could be eliminated based on their nucleotide sequences. The in silico translated Rpg1 gene revealed a receptor-like protein of 837 aa with two tandem kinase domains. The Rpg1 gene provides resistance to most pathotypes of P. graminis f. sp. tritici and has been incorporated into North American barley cultivars to protect against stem rust epidemics that plagued the Northern Great Plains production region during the first half of the last century (16). Rpg1 has protected barley cultivars, from significant stem rust losses for >60 years and is remarkable for its durability. Chevron and Peatland are the original sources of Rpg1, but the gene was cloned from one of their derived cultivars, Morex. The nucleotide sequence of Rpg1 from the resistant cultivars Kindred, Chevron, Peatland, Q21861, Leger, Bowman, and 80-TT-29 is identical to that of Morex. The susceptible cultivar Golden Promise lacks the Rpg1 gene as demonstrated by the failure to PCR-amplify the gene with seven primer pairs spanning the length of the 4,919-bp Rpg1 gene in Morex (Fig. 1). The Rpg1 gene in other investigated susceptible barley cultivars contains either multiple mutations or stop codons, or the gene cannot be amplified with appropriate primers (15).
Figure 1.
Absence of the gene Rpg1 in cv. Golden Promise. PCR amplification of seven Rpg1 gene fragments (lanes 1–7) with primer pairs covering the 4,919-bp gene region (primer sequences are given in Table 3, which is published as supporting information on the PNAS web site). Shown is DNA from Morex (A) and Golden Promise (B). Transformation plasmid pNRG040 has been constructed to express the Rpg1 gene in the susceptible cultivar Golden Promise. The genomic Rpg1 gene fragment with its complex exon (black box) and intron (gray line) structure under the control of the Rpg1 gene promoter (gray arrow) and terminator (gray box) was isolated from Morex and ligated into Agrobacterium vector pJH260.
To determine whether the stem-rust-susceptible cultivar Golden Promise can be converted into a resistant cultivar, we transformed it with the Rpg1 genomic clone of Morex. Representatives of 42 transgenic barley lines obtained by Agrobacterium-mediated transformation were then characterized for their infection response to the stem rust fungus.
Materials and Methods
Plant Material.
Hordeum vulgare cvs. Golden Promise, Morex, and transgenic T1 progenies were grown in a greenhouse maintained at 21°C (16 h light) and 16°C (8 h dark). Plants were watered every second day with water supplemented with 100 ppm 20N:20P:20K greenhouse fertilizer. Spikes were harvested about 25 days after anthesis for barley transformation. For RNA isolation, leaves from 15 8-day-old T1 seedlings of each primary transformant were pooled, frozen in liquid nitrogen, and stored at −80°C. Leaf material of Golden Promise and Morex was stored in the same way.
Plasmid Construct.
The genomic Rpg1 gene fragment from cv. Morex (GenBank accession no. AF509748) was isolated after SrfI and HpaI restriction of plasmid pNRG028, which is a 13.4-kb NotI subclone of bacterial artificial chromosome clone 426c16 containing the complete Rpg1 gene sequence (15). The SrfI/HpaI DNA fragment was ligated to HindIII adapters (S1107S and S1140S, New England Biolabs) and cloned into the HindIII-restricted Agrobacterium vector pJH260 (17) without restoring the HindIII site. The resulting plasmid, pNRG040, contains between the left and right border (i) the selectable marker gene bar under the control of the maize ubiquitin promoter and the nos terminator (18) and (ii) the Rpg1 gene with its own promoter and terminator (Fig. 1).
The Rpg1 cDNA was obtained from Morex total RNA by RT-PCR amplification using primers Rpg1_CDS_cw (5′-AAAAAGGATCCGCGTGGACTATTGTTGTG-3′) and Rpg1_Bam_ccw (5′-AAAAAGGATCCGCAGGGTTTATAGCTTCA-3′). The 2,615-bp amplification product was cloned into the intermediate vector pGEM-T Easy (Promega) by using the A/T cloning system. The Rpg1 coding region was excised by using the EagI restriction site located 8 bp upstream of the ATG codon and the PstI site located in the polylinker of the vector and ligated into NotI/PstI digested plasmid pQE-2 (Qiagen, Valencia, CA), resulting in plasmid pNRG072.
Barley Transformation and Selection of Transformants.
Plasmid pNRG040 was transferred into the disarmed Ti plasmid of Agrobacterium-strain AGL-1 by electroporation, resulting in strain H228. Transgenic plants of the cultivar Golden Promise were produced by cocultivation of immature zygotic embryos with Agrobacterium-strain H228 and regenerated as described (19). Genomic DNA was isolated (20), and primary transformants were identified by PCR using primers 228-F1 (5′-GCCGGGGCTGGACGATGAGGAATTC-3′) and 228-R1 (5′-GAACTCGAATGCAAACTCCCTTGTC-3′), amplifying 1,084 bp of the Rpg1 coding region. PCRs of 50 μl contained 100 ng of genomic DNA, 0.2 mM dNTP mix, 25 pmol of each primer, 2.5 μl of REDTaq DNA polymerase (Sigma), and 5 μl of 10× RedTaq reaction buffer. Amplification was performed in a PTC-100 programmable thermal controller (MJ Research, Cambridge, MA) at 95°C for 4 min, followed by 35 cycles of 95°C for 1 min, 62°C for 1 min, and 72°C for 1 min; this was followed by 7 min at 72°C. Thirty-eight transgenic plants were regenerated from an experiment with 53 cocultivated cut embryos and four from another batch of 184 cocultivated cut embryos.
Southern Blot Hybridization.
The methods used for DNA isolation, Southern blotting, and hybridization were as described (21). Five micrograms of HindIII-digested genomic barley DNA was separated by agarose gel electrophoresis, blotted onto nylon membranes, and hybridized to the 2.5-kb cDNA probe of Rpg1. The hybridization probe was obtained by labeling the 2.5-kb NotI restriction fragment of plasmid pNRG072 with [α-32P]dCTP using the All-in-One random labeling system (Sigma). DNA samples from T0 plants for Southern blots were available from transformants H228.1, H228.2c, H228.3, H228.5, and H228.10 T0 DNA for the other analyzed transformants was reconstituted by pooling DNA from T1 seedlings.
Phenotyping for Disease Reaction.
Twenty T1 seeds per transgenic line plus resistant (cvs. Chevron and Morex) and susceptible (cvs. Golden Promise and Steptoe) controls were germinated and sown in 15-cm-diameter pots filled with a 1:1 mix of sandy loam soil and potting mix consisting of peat moss, vermiculite, perlite, and sand. Plants were grown in a growth chamber at 19–21°C with a 14-h photoperiod. Seven-day-old seedlings (first leaf fully expanded) were inoculated with pathotype Pgt-MCC of P. graminis f. sp. tritici using rust inoculators pressured by an air pump. The rate of inoculum applied was ≈0.033 mg per plant. Inoculated plants were placed in mist chambers for 16 h in the dark at 21–22°C (100% relative humidity), exposed to light (120–160 μmol photon⋅m−2⋅s−1), and then allowed to dry slowly for 4 h before being returned to a growth chamber at 26–28°C (80% relative humidity) with a 14-h photoperiod. Nine days after inoculation, the infection types (ITs) were assessed based on a 0–4 rating scale (22). On this scale, IT 0 is characterized by no visible symptom (i.e., an immune reaction); IT 0; is characterized by immune reactions together with hypersensitive “flecks” (small necrotic areas) and no uredinia (infection sites with pathogen sporulation); IT 1 is characterized by minute uredinia surrounded by distinct necrotic areas; IT 2 is characterized by small uredinia surrounded by chlorosis; IT 3 is characterized by medium-sized uredinia often surrounded by chlorosis; and IT 4 is characterized by large uredinia usually without chlorosis. Barley often exhibits mesothetic reactions where two or more ITs may be present on the same leaf (16). The range of ITs observed on individual barley lines were recorded in order of their prevalence. ITs were divided into five general classes: highly resistant with ITs of 0 or 0; resistant with ITs of 0, 1, or 10; moderately resistant with ITs of 12 or 21; intermediate with ITs of 23; and susceptible with ITs of 3 or 4. Selected resistant and susceptible plants were transplanted and grown for Southern blot and RT-PCR analysis.
The surprising result, that the Rpg1 gene could provide an enhanced level of resistance in transformants compared with the source of the cloned gene (cv. Morex), prompted a question as to whether the gene might also confer resistance to other barley rusts. To address this question, transformants with Rpg1 were inoculated with pathotype Pgt-QCC of P. graminis f. sp. tritici (with virulence for Rpg1) and pathotype 8 of Puccinia hordei (barley leaf rust). The T1 progenies tested included those with all resistant seedlings (H228.5 and H228.8) and those segregating for resistance and susceptibility (H228.2a, H228.2c, and H228.12) to pathotype Pgt-MCC. Methods for inoculation, incubation, and disease assessment have been described (23).
RNA Isolation and RT-PCR.
Total RNA from 100-mg leaf tissue was isolated by using the ToTALLY RNA kit followed by DNaseI treatment using the DNA-free kit (both from Ambion, Austin, TX). Integrity and quantity of total RNA was validated by formaldehyde denaturing gel electrophoresis. One microgram of total RNA was used for conversion to cDNA with the Reverse Transcription System (Promega) and oligo(dT)15 primer. Quantification of the Rpg1 and reference gene GAPDH transcripts was performed on the Rotor-Gene 2000 real-time PCR cycler (Corbett Research, Mortlake, New South Wales, Australia) using the QuantiTect SYBR Green PCR system (Qiagen) with gene-specific primers. Primers Rpg1_Ex3_cw2 (5′-GCCGGTGTACTATCCCTTTC-3′) and Rpg1_Ex4_ccw2 (5′-TGTCGGACCCTCATAAGATT-3′) amplified a 250-bp fragment of the Rpg1 coding region. As a standard, 225 bp of the GAPDH gene were amplified by using primers Hv-GAPDH_cw1 (5′-CGTTCATCACCACCGACTAC-3′) and Hv-GAPDH_ccw1 (5′-CAGCCTTGTCCTTGTCAGTG-3′). PCR amplification was performed at 50°C for 2 min; hot start at 95°C for 15 min; 40 cycles of 95°C for 15 s, 60°C for 20 s, 72°C for 30 s, and data collection at 78°C for 15 s; and 1 min at 72°C. Absolute quantification of the mRNAs was obtained with external Rpg1 and GAPDH cDNA standards. Rpg1 mRNA used as a template was normalized with respect to the GAPDH gene mRNA. The amount of Rpg1 transcript in the transgenic lines was calculated as the percentage compared with the Morex mRNA level (100%).
Isolation of T-DNA Flanking Sequences.
DNA sequences adjacent to the left border of the integrated T-DNA from plasmid pNRG040 were isolated and cloned into SmaI restricted vector PUC18 as described (24). The DNA insert was sequenced with the universal M13 forward sequencing primer by using the BigDye Terminator system on an ABI Prism 377 DNA sequencer (Applied Biosystems) at Amplicon (Pullman, WA).
Results
Molecular Analysis of Rpg1 Transformants.
Golden Promise is very susceptible to stem rust pathotype Pgt-MCC and usually exhibits IT 3, similar to the susceptible control Steptoe (Table 1). Golden Promise lacks the Rpg1 DNA sequence, based on the fact that such a sequence could not be amplified by PCR with the seven primer pairs spanning the entire length of the Rpg1 gene of Morex (Fig. 1). For transformation, plasmid pNRG040 was constructed by inserting the genomic 5.98-kb Rpg1 gene fragment with its native exon–intron structure into the barley standard single cassette vector pJH260 (Fig. 1). Transformation with Agrobacterium strain H228 yielded 42 primary transgenic (T0) plants. The transformants were identified by PCR with specific primers amplifying 1,084 bp of the Rpg1 coding region. The amplified fragment indicated the successful transfer of the 9.56-kb T-DNA into the barley genome.
Table 1.
Reaction types in transgenic and control plants 9 days after inoculation with P. graminis f. sp. tritici pathotype MCC
| T0 line | Rpg1 copies | No. of T1 plants
|
χ2 (3:1) | P | ||||
|---|---|---|---|---|---|---|---|---|
| HR | R | MR | I | S | ||||
| H228.3* | 2 | 40 | 0 | 0 | 0 | 0 | ||
| H228.5* | 2 | 39 | 3 | 0 | 0 | 0 | ||
| H228.13 | 14 | 0 | 4 | 0 | 0 | |||
| H228.17* | 44 | 0 | 0 | 0 | 0 | |||
| H228.25 | 7 | 4 | 0 | 0 | 0 | |||
| H228.2a* | 32 | 0 | 0 | 0 | 14 | 0.72 | 0.5 > P > 0.3 | |
| H228.2b | 19 | 0 | 0 | 0 | 1 | 4.27 | 0.05* > P > 0.025* | |
| H228.2c* | 1 | 34 | 0 | 0 | 0 | 11 | 0.01 | 0.95 > P > 0.90 |
| H228.10* | 2 | 32 | 5 | 0 | 0 | 2 | ||
| H228.11 | 21 | 0 | 0 | 0 | 2 | 3.26 | 0.1 > P > 0.05* | |
| H228.12* | 32 | 0 | 0 | 0 | 3 | 5.04 | P ≈ 0.025* | |
| H228.16* | 33 | 0 | 0 | 0 | 12 | 0.07 | P ≈ 0.8 | |
| H228.39 | 8 | 7 | 3 | 0 | 1 | |||
| H228.40 | 7 | 2 | 1 | 0 | 5 | 0.56 | 0.5 > P > 0.3 | |
| H228.55 | 2 | 0 | 0 | 0 | 17 | |||
| H228.66 | 15 | 0 | 0 | 0 | 7 | 0.55 | 0.5 > P > 0.3 | |
| H228.7 | 10 | 0 | 2 | 3 | 0 | |||
| H228.8 | 7 | 6 | 1 | 2 | 0 | |||
| H228.19* | 5 | 19 | 6 | 8 | 4 | 2 | ||
| H228.30 | 8 | 1 | 2 | 4 | 7 | |||
| H228.44 | 3 | 0 | 6 | 4 | 4 | |||
| H228.1 | 9 | 0 | 0 | 0 | 0 | 6 | ||
| Chevron | 0 | 10 | 0 | 0 | 0 | |||
| Morex | 0 | 0 | 10 | 0 | 0 | |||
| Golden Promise | 0 | 0 | 0 | 0 | 10 | |||
| Steptoe | 0 | 0 | 0 | 0 | 10 | |||
HR, highly resistant; R, resistant; MR, moderately resistant; I, intermediate; S, susceptible.
Duplicated experiment.
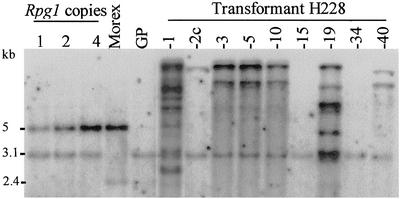
Southern blot analysis was performed to confirm stable integration of the genomic Rpg1 fragment and to estimate the number of inserted copies (Fig. 2). Genomic DNA of eight representative T0 plants was cut with the HindIII restriction enzyme and probed with the 2.5-kb cDNA of Rpg1. Transformation plasmid pNRG040 contains a single HindIII restriction site in the fourth exon (Fig. 1). The cDNA probe hybridizes preferentially to the 2,257 bp of exons 4–14, which results in an undetectable signal from the 461 bp of the first four exons. The HindIII restricted genomic Morex DNA containing one Rpg1 copy per haploid genome displayed the strongly Rpg1 hybridizing band at 5.1 kb and a weakly hybridizing band at 2.4 kb because of a related gene designated ABC 1037 (15). Thus, the number of Rpg1 gene copies inserted into the genome can be estimated from the number of hybridizing bands. For additional quantification, bacterial artificial chromosome clone 244m13 containing Rpg1 was digested with HindIII, and amounts equal to one, two, and four copies were included in the Southern blot. The 3.1-kb band is caused by the ABC 1037 gene present in Golden Promise, which is polymorphic with respect to the Morex band (Fig. 2). Golden Promise genomic DNA was used as a carrier for the copy number lanes. The copy number in the analyzed transformants ranged from one to five. Of the eight tested T0 plants, one line (H228.2c) contained a single copy of the Rpg1 gene, four lines (H228.3, H228.5, H228.10, and H228.40) contained two Rpg1 gene copies, and one line (H228.19) contained five Rpg1 gene copies. Transformants H228.3, H228.5, and H228.10 were obtained from one callus. All three T0 plants were derived from a single transformed cell, resulting in genetically identical plants, as confirmed by their identical hybridization patterns. T0 plant H228.15 was found by PCR to carry the transgene, but the pooled DNA of the T1 seedlings did not hybridize to the Rpg1 cDNA probe. Apparently, Rpg1 was absent from the germline of the T0 plant, indicating a chimeric transformant. The absence of the transgene in line H228.15 was verified in T1 seedlings by their susceptibility to stem rust and the absence of Rpg1 mRNA (Fig. 5A). Transformant H228-1 with approximately nine copies gave rise to 14 progeny plants that died before infection. The remaining six seedlings were inoculated and displayed susceptible reactions.
Figure 2.
Estimate of Rpg1 gene copies in eight transformants. Equivalents of one, two, and four copies (on the left) and HindIII-digested plant DNA were hybridized with 2.5 kb of the Rpg1 cDNA. See text for detailed explanation.
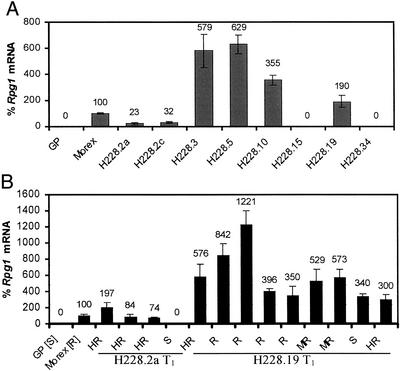
Figure 5.
Rpg1 transcript levels in 15 pooled T1 plants of eight transformants (A) and individual T1 plants after determination of disease resistance from two different transformants (B).
Tests of T1 Plants for Stem Rust Resistance.
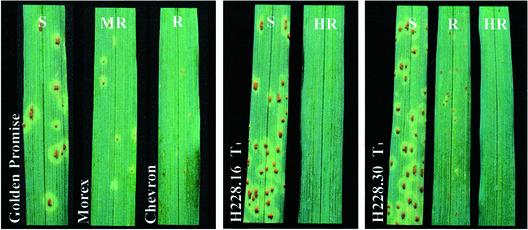
T1 progenies of 23 primary transformants containing Rpg1 were tested for stem rust resistance (Table 1, Fig. 3). Disease resistance tests were repeated for T1 progeny of nine transgenic lines with identical results. The Rpg1(resistant) controls of Chevron and Morex gave resistant (IT = 0;1) and moderately resistant (IT = 210;) reactions, respectively. In contrast, the susceptible controls of Golden Promise and Steptoe exhibited a susceptible reaction (IT = 3). Among the 23 sets of T1 progenies tested, 21 had plants with highly resistant (IT = 0) or resistant (IT = 0;) reactions (Table 1, Fig. 3). This result demonstrates that the Rpg1 gene converted the stem-rust-susceptible Golden Promise wild type into a stem-rust-resistant cultivar. Resistant plants of transformed Golden Promise gave consistently lower ITs (mostly IT 0 to 0;, but occasionally 0;1, and rarely 120;) than Chevron and Morex.
Figure 3.
Genetically engineered stem rust resistance in susceptible (S) cv. Golden Promise with the Rpg1 gene. Highly resistant (HR) T1 seedlings display lower ITs against P. graminis f. sp. tritici pathotype MCC than the moderately resistant (MR) cultivar Morex and the resistant (R) cultivar Chevron. Transgenic line H228.16 segregated into 33 HR and 12 S T1 plants, whereas T1 seedlings of line H228.30 exhibited various degrees of resistance.
Transformant H228.2c, with a single Rpg1 gene copy, segregated 3:1 for resistance/susceptibility (Table 1), indicating that a single copy of Rpg1 is sufficient to confer immunity or near immunity against the stem rust pathogen. Analysis of 10 T1 seedlings from this transformant by Southern blot revealed six seedlings with the expected insert and three seedlings without the transgene (Fig. 4). One seedling showed an aberrant insert but was still highly resistant. Further analysis is required to explain this observation. Transformant H228.5 (and H228.3), containing two copies of Rpg1, gave rise to all resistant progeny in the T1 generation. As seen in Fig. 4, the T1 progeny contained one seedling with a single copy, in addition to five seedlings with two copies, revealing that the two copies represent separate loci. This result agrees with the 15:1 (37 resistant/2 susceptible; χ2 = 0.08; 0.8 > P > 0) segregation ratio obtained for the third identical transformant (H228.10).
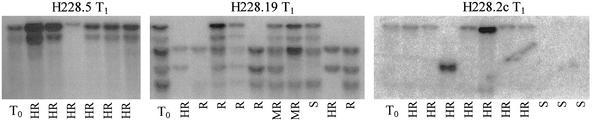
Figure 4.
Southern blot analysis of T0 plants and individual T1 seedlings after determination of disease resistance. Immune T1 plants (HR) are observed in all three transgenic lines. The single Rpg1 gene copy of transformant H228.2c is sufficient to confer immunity against P. graminis f. sp. tritici pathotype MCC, which is also observed in the progeny plants of line H228.5 containing either one or two Rpg1 gene copies. T1 plants of line H228.19 with two to five copies displayed a wide range of ITs, probably because of the complex integration and segregation pattern.
All but four T1 plants of transformants H228.13, H228.17, and H228.25 exhibited highly resistant reactions (IT = 0 to 0;) with no pathogen sporulation. T1 progenies with as yet unknown copy numbers in the clearly segregating category of Table 1 all yielded highly resistant plants (IT = 0 to 0;), several of which segregated with Mendelian ratios. Five T1 progenies segregated for highly resistant (HR), resistant (R), moderately resistant (MR), intermediate (I), and susceptible (S) plants (Fig. 3). Among these is transformant H228.19 with five gene copies in the primary transformant (Fig. 2). Ten progeny seedlings analyzed by Southern blot (Fig. 4) contained between one and five Rpg1 copies. The five gene copies present in the primary transformant have thus inserted into four independent locations. One susceptible seedling appeared to contain four of the copies, which were also present in one moderately resistant plant.
Rpg1 Transcript Levels.
Real-time PCR was used to quantify the Rpg1 mRNA in comparison to Golden Promise (0%) and Morex (100%). Transcript amounts of Rpg1 were determined for 15 pooled seedlings of the T1 progeny from six transgenic lines (Fig. 5A). The amount of transcripts produced by the progenies from H228.2 with a single copy transgene was only 23% and 32% of that of Morex. Four plants of this line were selected for analysis of individual T1 plants after assessment of stem rust resistance. The three immune T1 plants of line H228.2a, which are either homozygous or heterozygous for Rpg1, contain 74%, 84%, and 197% Rpg1 mRNA compared with Morex, whereas the susceptible plant lacks the transcript indicating the absence of the Rpg1 allele in this progeny plant (Fig. 5B). The T1 plant with 197% mRNA may be homozygous, whereas the plants with 84% and 74% may be heterozygous with only half the amount of transcript. The three lines from the transformant with two copies of the transgene (H228.3, H228.5, and H228.10, originating from the same callus) yielded four to six times more Rpg1 transcripts in the resistant and highly resistant pooled progenies than Morex (Fig. 5A).
Nine T1 plants were selected for transcript analysis from line H228.19, segregating for a wide range of ITs (Fig. 5B). The highly resistant, resistant, moderately resistant, and susceptible plants had 3–6, 6–12, 5–6, and 3 times more Rpg1 mRNA than Morex, respectively. There was no correlation between the amounts of Rpg1 transcripts as measured by amplification from total RNA and the level of stem rust resistance in the transgenic plants.
Tests of T1 Plants for Resistance to Pathotype QCC-2 and Leaf Rust Pathotype 8.
All progenies from the transformants exhibited susceptible ITs of 3 to both rusts. These reactions were similar to those observed on cv. Morex, which is susceptible to both rusts. Thus, the Rpg1 transgene appeared to act specifically toward stem rust pathotype Pgt-MCC just as it does in cv. Morex.
Isolation of T-DNA Flanking Sequences.
The site of insertion of the transgene in the genome is of interest for understanding its pattern of transcription, stability, and possible position effects. Relevant sequences isolated adjacent to T-DNA insertions at the left border are presented in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. The 48-bp plant DNA at the T-DNA junction of the single copy transgene in transformant H228.2c originates from repetitive DNA identified by a strong signal with multiple bands and a smear after hybridization of the 32P-labeled flanking sequence to HindIII restricted genomic DNA of Golden Promise. Of the two unlinked copies in the three transgenic plants with identical genotype (H228.3, H228.5, and H228.10), one was linked to 209-bp repetitive DNA, whereas another flanking sequence consisted of 190-bp Rpg1 transgene DNA. Plant H228.40, with two putatively linked transgene copies segregating 3:1 for resistant and susceptible progeny, yielded 74 bp genomic DNA unidentifiable as a specific gene region in the blast search. In plant H228.19 with five transgene copies at four independent locations, a left border in tandem with a right border was isolated, indicating that two of the copies are inserted in a tandem direct repeat configuration, as is the rule in barley (24). Plant H228.1, with nine inserted transgene copies and a seedling lethal phenotype, yielded a 60-bp unidentifiable genomic flanking sequence and three highly scrambled flanking sequences.
Discussion
Golden Promise was converted from a highly susceptible cultivar into a highly resistant one by Agrobacterium-mediated transformation with the Rpg1 genomic clone from cv. Morex. These results unequivocally demonstrate that the DNA segment is indeed the functional Rpg1 gene for stem rust resistance. A single copy of the gene was sufficient to confer resistance against stem rust. One of the most remarkable aspects about the stable Golden Promise transformants is that they exhibit a higher level of resistance than the original sources of Rpg1. The reason for this remains to be discovered, but it shows that stem-rust- susceptible barley cultivars with valuable agronomic or quality traits can be made more resistant by transformation with the functional Rpg1 gene rather than by hybridization with existing resistant cultivars.
Stem rust phenotypes for Rpg1 were determined on plants at the seedling stage in response to pathotype Pgt-MCC under the carefully controlled conditions of the growth chamber. By using these methods, one can usually obtain clear differential phenotypes between lines with and without Rpg1. For barley producers in the Upper Midwest region, the expression of resistance at the adult plant stage in the field is of paramount importance because stem rust inoculum from the southern United States does not reach the crop in the north until after the heading stage (16). Additional studies are required to determine the adult plant reaction of these transformants in the field to pathotype Pgt-MCC and others known to be avirulent for Rpg1.
The specificity of the Rpg1 gene toward pathotype Pgt-MCC conserved in the transgenic plants opens the possibility to further delineate, by site-directed mutagenesis, the amino acids or peptides responsible for the specific interaction with the avirulence protein or components of the signal transduction pathway leading to the incompatible interaction. Technical progress in this direction has been made in identifying two genes that provide race-specific resistance to the barley powdery mildew pathogen Blumeria graminis f. sp. hordei. The barley powdery mildew resistance locus conferring race specific resistance spans 240 kb and contains eight genes of three subfamilies (RGH1, RGH2, and RGH3). The Mla1 gene conferring resistance to AvrMla1 of B. graminis f. sp. hordei is an R gene encoding an N-terminal coiled-coil (CC) protein structure as well as NBS and LRR domains (25). It was pinpointed by isolating a γ-ray induced susceptible mutant with an Mla deletion, identifying a clone corresponding to the deletion in a cosmid library, and carrying out a functional test with the cosmid subclones by using a three-component epidermal single-cell expression system. The functional test used cobombardment of epidermal cells of a generally resistant recessive mlo mutant with a plasmid containing the Mlo gene linked to the GFP gene and a plasmid containing candidate cosmid subclones. Challenge inoculation with conidia of +AvrMla1 vs. −AvrMla1 powdery mildew races identified the cosmid subclone providing resistance to +AvrMla1 spores by a decrease in both fluorescence and mildew colony formation. It was further shown with a rar1 mutant that the Mla1 interaction with Avr1Mla1 does not involve the Rar1 gene-dependent signal transduction pathway. With a similar approach, a RGH1 gene specifying a CC-NBS-LRR protein was identified as the Mla6 gene conferring resistance to AvrMla6 mildew races but using the Rar1 signaling pathway (26). The amino acid sequence of MLA6 is 91.2% identical to that of MLA1, indicating that the specificity is residing in small peptide epitopes.
A rapid gene expression system using plasmids and particle bombardment should be developed for cereal rusts, which unlike powdery mildew infect via the stomates into mesophyll cells. However, the first task is to isolate additional rust-resistance genes from barley and wheat either by map-based cloning or rather by screening of bacterial artificial chromosome and transformation-competent artificial chromosome (27) libraries for R gene homologues and by testing these in transformants after pathogen inoculation. Studies elucidating the basis of race specificity in cereals may lead to the development of novel germ plasm with rust resistance that is potentially more durable.
Supplementary Material
Acknowledgments
This is Scientific Paper No. 021101 from the College of Agriculture and Home Economics Research Center, Washington State University, Project 0196. This work was supported by U.S. Department of Agriculture National Research Initiative Grant 9901325.
Abbreviations
- NBS
nucleotide binding site
- LRR
leucine-rich repeat
- IT
infection type
References
- 1.Flor H H. J Agric Res. 1947;74:241–262. [Google Scholar]
- 2.Flor H H. Adv Genet. 1956;8:29–54. [Google Scholar]
- 3.Keen N T. Annu Rev Genet. 1990;24:447–453. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence G J, Finnegan E J, Ayliffe M A, Ellis J G. Plant Cell. 1995;7:1195–1206. doi: 10.1105/tpc.7.8.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staskawicz B J, Ausubel F M, Baker B, Ellis J G, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 6.Ellis J G, Lawrence G J, Luck J E, Dodds P N. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luck J E, Lawrence G J, Dodds P N, Shepherd K W, Ellis J G. Plant Cell. 2000;12:1367–1377. doi: 10.1105/tpc.12.8.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson P A, Lawrence G J, Morrish B C, Ayliffe M A, Finnegan E J, Ellis J G. Plant Cell. 1997;9:641–651. doi: 10.1105/tpc.9.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khush G S, Bacalangco E, Ogawa T. Rice Genet Newslett. 1990;7:121–122. [Google Scholar]
- 10.Song W-Y, Wang G-L, Chen L-L, Kim H-S, Pi L-Y, Holsten T, Gardner J, Wang B, Zhai W-X, Zhu L-H, Fauquet C, Ronald P. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 11.Wang G-L, Song W-Y, Ruan D L, Sideris S, Ronald P C. Mol Plant–Microbe Interact. 1996;9:850–855. doi: 10.1094/mpmi-9-0850. [DOI] [PubMed] [Google Scholar]
- 12.Tu J, Ona I, Zhang Q, Mew T W, Khush G S, Datta S K. Theor Appl Genet. 1998;97:31–36. [Google Scholar]
- 13.Wang G-L, Ruan D-L, Song W-Y, Sideris S, Chen L-L, Pi L-Y, Zhang S, Zhang Z, Fauquet C, Brandon S G, Whalen M C, Ronald P C. Plant Cell. 1998;10:765–779. doi: 10.1105/tpc.10.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu J, Datta K, Khush G S, Zhang Q, Datta S K. Theor Appl Genet. 2000;101:15–20. [Google Scholar]
- 15.Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, Druka A, Steffenson B, Kleinhofs A. Proc Natl Acad Sci USA. 2002;99:9328–9333. doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steffenson B J. Euphytica. 1992;63:153–167. [Google Scholar]
- 17.Horvath H, Huang J, Wong O T, Kohl E, Okita T, Kannangara C G, von Wettstein D. Proc Natl Acad Sci USA. 2000;97:1914–1919. doi: 10.1073/pnas.030527497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen L G, Politz O, Olsen O, Thomsen K K, von Wettstein D. Hereditas. 1998;129:215–225. [Google Scholar]
- 19.Horvath H, Huang J, Wong O T, von Wettstein D. In: Barley Science. Slafer G A, Molina-Cano J L, Savin R, Araus J L, Romagosa J, editors. New York: Harworth; 2002. pp. 143–176. [Google Scholar]
- 20.Edwards K, Johnstone C, Thompson C. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinhofs A, Kilian A, Maroof M A S, Biyashev R M, Hayes P, Chen F O, Lapitan N, Fenwick A, Blake T K, Kanazin V, et al. Theor Appl Genet. 1993;86:705–712. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
- 22.Stakman E C, Stewart D M, Loegering W O. US Dept Agric Res Serv. 1962;E-617:1–53. [Google Scholar]
- 23.Steffenson B J, Jin Y, Rossnagel B G, Rasmussen J, Kao K N. Plant Breed. 1995;114:50–54. [Google Scholar]
- 24.Stahl R, Horvath H, Van Fleet J, Voetz M, von Wettstein D, Wolf N. Proc Natl Acad Sci USA. 2002;99:2146–2151. doi: 10.1073/pnas.032645299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou F, Kurth J, Wei F, Elliott C, Valè G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P. Plant Cell. 2001;13:337–350. doi: 10.1105/tpc.13.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halterman D, Zhou F, Wei F, Wise R P, Schulze-Lefert P. Plant J. 2001;25:335–348. doi: 10.1046/j.1365-313x.2001.00982.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y-G, Nagaki K, Fujita M, Kawaura K, Uozumi M, Ogihara Y. Plant J. 2000;23:687–695. doi: 10.1046/j.1365-313x.2000.00827.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.