Abstract
Thioredoxins are small multifunctional redox active proteins widely if not universally distributed among living organisms. In chloroplasts, two types of thioredoxins (f and m) coexist and play central roles in regulating enzyme activity. Reduction of thioredoxins in chloroplasts is catalyzed by an iron-sulfur disulfide enzyme, ferredoxin-thioredoxin reductase, that receives photosynthetic electrons from ferredoxin, thereby providing a link between light and enzyme activity. Chloroplast thioredoxins function in the regulation of the Calvin cycle and associated processes. However, the relatively small number of known thioredoxin-linked proteins (about 16) raised the possibility that others remain to be identified. To pursue this opportunity, we have mutated thioredoxins f and m, such that the buried cysteine of the active disulfide has been replaced by serine or alanine, and bound them to affinity columns to trap target proteins of chloroplast stroma. The covalently linked proteins were eluted with DTT, separated on gels, and identified by mass spectrometry. This approach led to the identification of 15 potential targets that function in 10 chloroplast processes not known to be thioredoxin linked. Included are proteins that seem to function in plastid-to-nucleus signaling and in a previously unrecognized type of oxidative regulation. Approximately two-thirds of these targets contained conserved cysteines. We also identified 11 previously unknown and 9 confirmed target proteins that are members of pathways known to be regulated by thioredoxin. In contrast to results with individual enzyme assays, specificity for thioredoxin f or m was not observed on affinity chromatography.
The ferredoxin/thioredoxin system of oxygenic photosynthesis, composed of ferredoxin, ferredoxin-thioredoxin reductase (FTR), and thioredoxin, links light to the regulation of photosynthetic enzymes. FTR plays a central role in this system by catalyzing the transfer of electrons from ferredoxin and protons to thioredoxin, which, in turn, regulates the activity of target proteins through the reduction of specific disulfide groups. In chloroplasts, the thioredoxins linked to FTR are of two types, f and m, that selectively regulate key enzymes (1–3).
Since the discovery of a role for thioredoxin in the activation of fructose-1,6-bisphosphatase (1), the number of known thioredoxin-linked enzymes has steadily increased (see refs. 2 and 3 for recent reviews). Most of the presently known thioredoxin-linked proteins were identified either by chance or by following up on prior observations (i.e., metabolite changes reported for in vivo light/dark transition experiments or increased enzyme activity found in vitro after adding DTT, a nonphysiological substitute for thioredoxin). By our count, studies during the past 25 years have led to the identification of a total of 16 soluble thioredoxin-linked proteins in chloroplasts.
To gain further insight on the extent of its influence, we have applied a combination of affinity chromatography and proteomics to isolate and identify thioredoxin-linked proteins in chloroplasts. The affinity procedure takes advantage of the mechanism by which thioredoxin reduces a specific regulatory disulfide (4–8). The mechanism requires the formation of a transient heterodisulfide bond between thioredoxin and enzyme before complete reduction of the targeted disulfide. Mutation of one of the two cysteines of the thioredoxin active site (the one buried in the molecule) stabilizes the normally transient heterodisulfide, thereby covalently linking the target protein to thioredoxin via a bond that can be cleaved by DTT. This property has been used earlier to generate covalent complexes between thioredoxin f and both phosphoribulokinase and fructose-1,6-bisphosphatase (4, 5). Further experiments in which the buried cysteine of the active site of extraplastidic thioredoxin h was mutated to serine led to the identification of a peroxidase as a target protein in yeast two-hybrid experiments (6). Recently, mutated thioredoxin m was immobilized on a resin and used in conjunction with proteomics to screen chloroplast stroma for potential target proteins (7). This approach led to the identification of eight thioredoxin targets: five known (rubisco activase, sedoheptulose-1,7-bisphosphatase, glyceraldehyde-3-phosphate dehydrogenase, glutamine synthetase, and 2-Cys peroxiredoxin) and three previously undescribed (peroxiredoxin-Q, cyclophilin, and rubisco small subunit). A similar experiment with mutated thioredoxin h led to the identification of a peroxiredoxin from Chlamydomonas reinhardtii (8).
In the present study, we have bound mutant thioredoxins f and m individually to affinity columns to isolate thioredoxin targets. Proteins trapped from spinach chloroplast stroma were identified by mass spectrometry. This approach enabled us to confirm the identity of more than half of the known soluble thioredoxin-regulated enzymes and to more than double the number of potential thioredoxin-interacting proteins. Many of the potential targets contain conserved cysteines and are members of processes not previously known to be redox regulated.
Materials and Methods
Materials.
CNBr-activated Sepharose 4B and activated thiol Sepharose 4B were purchased from Amersham Pharmacia Biosciences. Criterion Precast gels and IPG (immobilized pH gradient gel) strips were purchased from Bio-Rad. Other chemicals were obtained from commercial sources and were of the highest quality available.
Recombinant WT and Mutant Thioredoxin Proteins.
Recombinant WT thioredoxin f and mutant thioredoxins f C49S and m C40A were overexpressed in Escherichia coli BL21 DE3 pLysS and purified from cell extracts (2, 5, 9).
Isolation of Stroma from Spinach Chloroplasts.
Stroma was obtained from intact chloroplasts prepared from greenhouse-grown spinach (10).
Preparation of Thioredoxin-Sepharose 4B Resin.
Mutant or WT thioredoxin (10 mg) was coupled to the CNBr-activated Sepharose 4B according to the manufacturer's instructions.
Separation of Stromal Proteins on Thioredoxin-Sepharose Affinity Column.
Chloroplast stroma (5 mg of protein) was applied to the thioredoxin f or m column equilibrated in buffer (50 mM Tris⋅HCl, pH 7.5/50 mM NaCl). The column was then extensively washed with the same buffer and an increased salt concentration (500 mM NaCl) to remove noncovalently bound proteins. Subsequently, the potential target proteins linked by the newly formed heterodisulfide were eluted with above buffer containing 10 mM DTT and 0.5 M NaCl. Each elution step was continued until the absorbance at 280 nm reached almost zero. The DTT-eluted fractions with protein were concentrated and washed with buffer (50 mM Tris⋅HCl, pH 7.5) on Ultrafree centrifugal devices (Millipore, 5 kDa molecular mass cut-off). The amount of protein collected on addition of DTT to the WT thioredoxin f column was <50 μg, the mutated thioredoxin f column 150 μg, and the mutated thioredoxin m column 250 μg. The increased recovery with mutant thioredoxin m could reflect its greater stability relative to the f-type (5). Similar elution steps were performed with the activated thiol Sepharose column, and the addition of DTT yielded an extremely low amount of protein (≈10 μg).
Control Columns.
A control experiment with WT thioredoxin f was necessary because of the presence of a free surface-exposed cysteine capable of forming a heterodimer with stromal enzymes (11). Only three proteins were found in the fraction eluted with DTT (rubisco activase, the 28-kDa ribonucleoprotein, and elongation factor Tu), all of which were also present in the corresponding fraction obtained with the mutant. Moreover, the amount of each of the three proteins was lower by a factor of 3 with the WT thioredoxin f column than with the mutant counterpart, indicating that the additional free cysteine in thioredoxin f did not effectively form a stable complex with the target enzymes. For reasons not understood, two enzymes (choline monooxygenase and glucose-6-phosphate isomerase) were found mainly in the DTT-eluted fraction obtained with WT vs. the mutant thioredoxin f column.
An independent control experiment with activated thiol Sepharose was designed to identify proteins forming a nonspecific heterodisulfide. The quantity of protein eluted by DTT was extremely low, and only one protein was clearly identified that was also found with the mutant thioredoxin f and m Sepharose columns: phosphoglycerate kinase. This enzyme is not discussed further below.
2D Gel Separation.
Isoelectric focusing and SDS/PAGE were performed by using the Protean IEF Cell and Criterion Precast System (Bio-Rad) according to the instructions of the manufacturer. Isoelectric focusing was carried out by using a pH range from 3 to 10. The second dimension was developed with a 10–20% acrylamide gradient SDS/PAGE. Gels were stained with Coomassie brilliant blue R-250.
Protein Spot Excision and Digestion.
Protein spots were excised by using a Bio-Rad spot cutter according to the manufacturer's instructions, destained (12), and in-gel digested with trypsin (13) with a Massprep digestion robot (Micromass, Beverly, MA). Tryptic peptides were extracted from the gel pieces with 1% formic acid/2% acetonitrile.
Protein Identification.
Tryptic-digested peptides were analyzed by high performance liquid chromatography-tandem mass spectrometry, and the tandem mass spectra were interpreted by using the sequest algorithm and the nonredundant protein database from the National Center for Biotechnology Information, as described (14).
Results and Discussion
Overview.
The chloroplast stromal proteins found to interact with thioredoxins f and m owing to their capability to bind to mutant derivatives are discussed in relation to the extent of the data collected for each. The previously unknown thioredoxin-target proteins were detected with at least two different tryptic peptides that matched plant proteins not known to be linked to thioredoxin. Known thioredoxin target proteins include peptides with multiple matches to known thioredoxin-regulated enzymes.
A total of 35 thioredoxin-linked proteins participating in 18 different chloroplast processes were identified in this study (Table 1). Most of these, 26, represent potential thioredoxin targets that are members of either previously unrecognized, 15, or established, 11, thioredoxin-linked processes. Thus, in addition to the confirmation of 9 known thioredoxin-linked enzymes (functional in 5 chloroplast processes), this study led to the identification of 26 potential previously unrecognized target proteins that function in 15 different chloroplast processes: 5 established and 10 not previously known to be linked to thioredoxin. Because they often form a thioredoxin-reducible disulfide, we have designated the number of conserved cysteines identified in the 26 previously unknown targets with amino acid sequence alignment using clustalw. Thus, 17 of the 26 showed conserved cysteines (see Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). For some of the remaining nine proteins, the database was not sufficiently developed to permit the assignment of conserved amino acids. Each of the identified target proteins is discussed below in relation to its associated pathway or reaction. Because the analysis was conducted by using a protein database, we detected a minimal number of potential targets of unknown function. One particularly promising candidate stood out (SwissProt no. Q9SGT3). Likely, there are others.
Table 1.
Summary of chloroplast thioredoxin-linked processes and target proteins identified in this study
| Trx-linked processes | No. of processes | No. of target proteins
|
|
|---|---|---|---|
| Potential | Confirmed | ||
| Unrecognized | 10 | 15 | — |
| Established | 8 | 11 | 9 |
| Total | 18 | 26 | 9 |
The 26 proteins found to be thioredoxin targets are listed in Table 2. These proteins function in the 10 processes not previously known to be linked to thioredoxin (isoprenoid, tetrapyrrole and vitamin biosynthesis, protein assembly/folding, protein and starch degradation, glycolysis, HCO /CO2 equilibration, plastid division, and DNA replication/transcription) and in 5 established thioredoxin-regulated processes (Calvin cycle, nitrogen and sulfur metabolism, translation, and pentose phosphate cycle).
/CO2 equilibration, plastid division, and DNA replication/transcription) and in 5 established thioredoxin-regulated processes (Calvin cycle, nitrogen and sulfur metabolism, translation, and pentose phosphate cycle).
Table 2.
Potential chloroplast thioredoxin target proteins identified by proteomic analysis
| Unrecognized processes | Trx f* | Trx m* | Established processes | Trx f* | Trx m* |
|---|---|---|---|---|---|
| Isoprenoid biosynthesis | Calvin cycle | ||||
| GcpE protein | 8 (0) | 5 (0) | Transketolase† | 19 (15) | 3 (2) |
| DXP reductoisomerase† | 2 (0) | 0 | Triose phosphate isomerase† | 1 (1) | 6 (1) |
| Tetrapyrrole biosynthesis | Ribulose-P 3-epimerase† | 3 (3) | 0 | ||
| GSA aminomutase† | 4 (0) | 4 (0) | Nitrogen metabolism | ||
| Uroporphyrinogen decarboxylase† | 2 (0) | 3 (0) | P-glycerate dehydrogenase | 0 | 2 (0) |
| Magnesium chelatase† | 1 (0) | 2 (0) | Sulfur metabolism | ||
| Vitamin biosynthesis | Cysteine synthase† | 10 (5) | 6 (4) | ||
| Thiamin biosynthesis protein† | 2 (0) | 3 (0) | Translation | ||
| Thiazole biosynthetic enzyme† | 3 (0) | 2 (0) | 28 kDa ribonucleoprotein | 6 (5) | 1 (1) |
| Protein assembly/folding | Elongation factor Tu† | 6 (0) | 10 (0) | ||
| 70 kDa heat shock protein† | 22 (10) | 11 (6) | Elongation factor g | 3 (0) | 1 (0) |
| Rubisco binding protein (α and β)† | 16 (0) | 7 (0) | 30S ribosomal protein S1† | 7 (7) | 7 (7) |
| Protein degradation | Ribosomal protein S6 (PrpS6) | 2 (2) | 2 (1) | ||
| ATP dependent clp protease† | 21 (0) | 18 (0) | Pentose phosphate cycle | ||
HCO /CO2 equilibration /CO2 equilibration |
6-P-gluconate dehydrogenase | 0 | 2 (2) | ||
| Carbonic anhydrase† | 9 (6) | 14 (4) | |||
| Starch degradation | |||||
| β-Amylase | 2 (0) | 0 | |||
| Glycolysis | |||||
| Enolase | 0 | 3 (0) | |||
| DNA replication/transcription | |||||
| ATP-dependent DNA helicase | 3 (0) | 0 | |||
| Plastid division | |||||
| FtsZ protein | 2 (0) | 2 (0) |
The SwissProt ID number for each protein is from the plant with maximal matches. The ID numbers of the enzymes from top to bottom (left column), starting with GcpE protein, are Q9FF59, Q9SP64, P31593, Q42967, P16127, O82392, Q38814, O50036, P08926 (α) and P08927 (β), P31541, P16016, P16098, Q9LEJ0, Q9ZVG0, Q9LRC5. The corresponding numbers for the right column, starting with transketolase, are O20250, P48496, Q43157, Q9FSS6, P32260, P28644, Q43467, P34811, P29344, P82403, Q94KU2.
The first number represents the total number of different peptides isolated from the indicated affinity matrix that match the identified protein irrespective of the organism from which it was described. The number in parentheses corresponds to the number of different peptides detected that matches a protein from spinach.
Contains conserved cysteines. The numbers of conserved cysteines are given in Table 4. The thioredoxin disulfide target site for those proteins with a single conserved cysteine would be generated by the formation of an intermolecular disulfide bond, as is the case for 1-Cys perioxredoxin (6).
Potential Thioredoxin Target Proteins: Unrecognized Processes.
The potential proteins linked to chloroplast thioredoxin are listed in Table 2. They fall into two groups: the first, with matches to both thioredoxins, and the second, with matches to only one. It is possible that, in certain cases, the distinction between the two groups is due to sequence differences in spinach proteins and counterparts described in current databases.
Isoprenoid biosynthesis.
In plants, two pathways are known for the biosynthesis of isopentenyl diphosphate, the universal precursor of a spectrum of isoprenoid products (15). The cytosol houses the acetate/mevalonate pathway, whereas chloroplasts use the recently elucidated mevalonate-independent route known as the glyceraldehyde 3-phosphate/pyruvate or 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway. Chloroplast isopentenyl diphosphate biosynthesis starts with glyceraldehyde 3-phosphate and pyruvate, thus, ultimately yielding plastid isoprene products directly from the Calvin cycle and glycolysis.
The GcpE enzyme presently recognized to be thioredoxin-linked was recently described as a member of the MEP pathway in E. coli (16) and Arabidopsis (17). GcpE seems to catalyze one of the last steps of isopentenyl diphosphate formation, i.e., the reduction of 2-C-methyl-d-erythritol 2,4-cyclodiphosphate to 1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate. This reaction seems to require the repetitive reduction of a disulfide for catalysis (18). Thus, it is possible that, there, thioredoxin acts as a substrate, as is the case for methionine sulfoxide reductase and peroxiredoxin. More work is required to confirm this role and to determine whether thioredoxin also acts as a regulator of GcpE.
A second member of the chloroplast isoprenoid biosynthesis pathway was also identified as being possibly linked to thioredoxin, 1-deoxy-d-xylulose 5-phosphate (DXP) reductoisomerase. This enzyme catalyzes the reduction and isomerization of 1-deoxy-d-xylulose 5-phosphate to form 2-C-methyl-d-erythritol 4-phosphate (19), the first committed step of the pathway. The reaction takes place just after a branch point in which its 1-deoxy-d-xylulose 5-phosphate substrate also serves as a precursor for thiamin and pyridoxal biosynthesis. Such a central position in the isoprenoid pathway could explain its regulation by thioredoxin. Chloroplast 1-deoxy-d-xylulose-5-phosphate reductoisomerase has five conserved cysteines.
The finding that two of the enzymes of the chloroplast isoprenoid biosynthesis pathway are linked to thioredoxin raises the possibility that the pathway is light dependent. It becomes interesting to know whether the DTT routinely added to preparations of these enzymes has a specific activating effect (19, 20).
Tetrapyrrole biosynthesis.
The biosynthesis of tetrapyrrole (porphyrins and chlorins) via the C5-pathway, a process taking place entirely in chloroplasts, was found to have three potential thioredoxin-linked members: glutamate-1-semialdehyde 2,1-(GSA)-aminomutase, uroporphyrinogen decarboxylase, and magnesium chelatase. The first of these, glutamate-1-semialdehyde 2,1 aminomutase, catalyzes the formation of δ-aminolevulinic acid, the first committed precursor of the C5-pathway (21). Other evidence supports such a role for thioredoxin in this series of reactions. For example, the activity of glutamate-1-semialdehyde aminomutase in etiolated barley seedlings increases dramatically in the light whereas the level of the corresponding mRNA remains constant (21). Further, it has been observed that DTT alleviates the inhibition of the synthesis of an intermediate of the pathway, protoporphyrin IX under oxidizing conditions (22). Finally, the second potential target, uroporphyrinogen decarboxylase, catalyzes a reaction in common with heme biosynthesis, and the third, magnesium chelatase, catalyzes the insertion of magnesium, the first dedicated step of chlorophyll synthesis (21) that is considered pivotal to plastid-to-nucleus signaling (23, 24). The present results thus link chloroplast thioredoxins to the regulation of both tetrapyrrole biosynthesis and nuclear gene expression. Chloroplast glutamate-1-semialdehyde aminomutase, uroporphyrinogen decarboxylase, and magnesium chelatase (subunit i) have respectively four, one, and four conserved cysteines.
Vitamin biosynthesis.
Vitamin biosynthesis represents another chloroplast process that, according to Table 2, is linked to thioredoxin. Thus, two enzymes functional in thiamin biosynthesis (thiamin biosynthesis protein and thiazole biosynthetic enzyme) were identified as thioredoxin targets. As noted above, the thiamin biosynthesis pathway starts with the 1-deoxy-d-xylulose 5-phosphate, a precursor also shared with isoprenoid and pyridoxal biosynthesis (15). Chloroplast thiamin biosynthesis protein and thiazole biosynthetic enzyme have nine and three conserved cysteines, respectively.
Protein assembly/folding.
Heat shock proteins and chaperones act in the assembly and folding of proteins as well as in the transport of nuclear-encoded precursors into chloroplasts (25). Through the regulation of Hsp70 and rubisco-binding protein, identified in Table 2, thioredoxin could, therefore, modulate both the rate of protein maturation and its transport into chloroplasts. Higher plant chloroplast Hsp70 and rubisco-binding subunits α and β exhibit two, one, and four conserved cysteines, respectively.
Protein degradation.
The trapping of the ATP-binding or regulatory subunit of clp protease introduces a role for thioredoxin in chloroplast protein turnover. The ATP-dependent clp proteases have been recently recognized in chloroplasts (26). A link to thioredoxin is in accord with the known ability of light to stimulate chloroplast proteolysis (27). Plant isoforms of ATP-dependent clp proteases (ATP-binding subunit) contain two conserved cysteines.
Bicarbonate/CO2 equilibration.
Carbonic anhydrase has long been known to catalyze the reversible hydratation of CO2. The primary function of chloroplast carbonic anhydrase is to provide CO2 for rubisco in photosynthesis. At present, no evidence is available for the redox regulation of this well studied enzyme. Nonetheless, it has been shown that the activity of the oxidized form of the enzyme could be partially restored by reducing agents (28). Chloroplast carbonic anhydrases have five conserved cysteines, of which three are specific to chloroplast isoforms.
Starch degradation.
One enzyme active in starch degradation (β-amylase) was identified as a potential thioredoxin target. Based on antisense analysis, chloroplast β-amylase is required for the nocturnal degradation of transitory starch accumulated during the day (29). The thioredoxin-mediated regulation of β-amylase could thus possibly effect a decrease in the rate of starch degradation in the light, as occurs with the oxidative pentose cycle.
Glycolysis.
Chloroplasts contain enzymes of glycolysis that enable them to generate pyruvate (30). Enolase catalyzes a reaction in the latter phase of the pathway, the dehydration of 2-phosphoglycerate to form phosphoenolpyruvate, the immediate precursor of pyruvate. Experiments with extracts of ice plant leaves revealed an increase in the activity of this enzyme after diamide treatment vs. a decrease after DTT incubation (31).
DNA replication/transcription, plastid division.
Because they undergo division, chloroplasts must contain the enzymatic machinery to facilitate DNA replication and transcription. One such enzyme is ATP-dependent DNA helicase (32) which was identified as a thioredoxin target. The related finding that FtsZ protein is a target could link thioredoxin to plastid division (33).
Potential Thioredoxin Target Proteins: Established Processes.
The potential chloroplast targets functional in pathways known to be thioredoxin-linked are given in Table 2.
Calvin cycle.
Transketolase, which catalyses the transfer of a two-carbon unit from either sedoheptulose 7-phosphate or fructose 6-phosphate to glyceraldehyde 3-phosphate, participates in the regeneration of ribulose 1,5-bisphosphate. In contrast to the five enzymes of the Calvin cycle known to be linked to thioredoxin (2), there is no evidence for a role of redox regulation with transketolase (34). As discussed below, experiments on CO2 fixation with intact chloroplasts have shown, however, that the enzyme is sensitive to H2O2 (35). It is possible, therefore, that preliminary oxidation is necessary to observe a redox effect on activity. This condition may have been achieved in our experiment owing to isolation of the stroma in the absence of a reducing agent. Amino acid sequence alignment shows four conserved cysteines in different transketolase isoforms of chloroplasts.
According to Table 2, two other members of the Calvin cycle (triose phosphate isomerase and ribulose phosphate 3-epimerase) are also potentially thioredoxin regulated. Both catalyze reversible reactions, the isomerisation of glyceraldehyde 3-phosphate to dihydroxyacetone phosphate and conversion of xylulose 5-phosphate to ribulose 5-phosphate. Primary sequence alignments of triose phosphate isomerase and ribulose phosphate 3-epimerase showed three and two conserved cysteines in chloroplast isoforms, respectively.
Sulfur metabolism.
One enzyme functional in sulfate assimilation was identified in the DTT-eluted fraction of the mutant thioredoxin columns, namely, cysteine synthase (o-acetylserine [thiol]lyase). Derived from O-acetylserine and hydrogen sulfide, cysteine is the final product of sulfate assimilation. Regulation of cysteine synthase could, therefore, indirectly control the rate of sulfur assimilation (36). Interestingly, this enzyme requires DTT for activity (37). Chloroplast isoforms of cysteine synthase have three conserved cysteines.
Nitrogen metabolism.
An enzyme of nitrogen metabolism has been identified as a potential target for thioredoxin: phosphoglycerate dehydrogenase, which catalyzes the oxidation of 3-phosphoglycerate to phosphohydroxypyruvate in the first step of the serine biosynthesis. Because serine is also the precursor in the biosynthesis of glycine and cysteine, regulation of phosphoglycerate dehydrogenase would link thioredoxin to both nitrogen and sulfur assimilation.
Translation-related proteins.
Thioredoxin has been reported to regulate translation via an interaction with a protein disulfide isomerase, RB60 (38). In the present investigation, we have identified five additional translation-related proteins as potential thioredoxin targets: 28-kDa ribonucleoprotein, elongation factor Tu, elongation factor g, ribosomal protein S1, and ribosomal protein S6. A link to thioredoxin accounts for the known capability of light to activate translation (38) and to stabilize mRNA (39). The disulfide isomerase activity described for elongation factor Tu implies reversible oxido-reduction of an intermolecular disulfide bond formed between the two uniquely conserved cysteines (40). Chloroplast ribosomal protein S1 shows two conserved cysteines.
Pentose phosphate cycle.
The oxidative pentose phosphate cycle represents a principal mechanism whereby chloroplasts degrade sugars and generate NADPH in the dark. The first member of the cycle, glucose-6-phosphate dehydrogenase, has long been known to be reductively deactivated by thioredoxin. The present results indicate that 6-phosphogluconate dehydrogenase, which catalyzes the second step of the cycle (irreversible oxidative decarboxylation of 6-phosphogluconate to ribulose 5-phosphate), is also targeted by thioredoxin. It is noteworthy that, during purification, the addition of β-mercaptoethanol (a monothiol) stabilized the plastidic isoform of 6-phosphogluconate dehydrogenase whereas DTT was far less effective and could possibly have inactivated the enzyme (41). Similar to glucose-6-phosphate dehydrogenase, it is plausible, therefore, that the reduction of a regulatory disulfide by a dithiol inactivates 6-phosphogluconate dehydrogenase.
Confirmed Thioredoxin Target Proteins in Established Processes.
Table 3 shows that nine proteins were confirmed as thioredoxin targets. Thus we were able to trap more than half (9 of 16) of the soluble proteins reported to be linked to thioredoxin in chloroplasts. All are enzymes (or subunits) associated with five different processes: Calvin cycle, nitrogen metabolism, C4 cycle/malate valve, fatty acid biosynthesis, and oxidative stress.
Table 3.
Chloroplast thioredoxin target proteins confirmed by proteomic analysis
| Confirmed proteins | Trx f* | Trx m* |
|---|---|---|
| Calvin cycle | ||
| Sedoheptulose bisphosphatase | 7 (5) | 9 (8) |
| Phosphoribulokinase | 8 (3) | 2 (2) |
| GAP dehydrogenase | 26 (13) | 38 (19) |
| Rubisco activase | 21 (17) | 32 (20) |
| Rubisco small subunit | 5 (4) | 7 (5) |
| Nitrogen metabolism | ||
| Glutamine synthetase | 10 (0) | 9 (0) |
| C4 cycle/malate valve | ||
| NADP-malate dehydrogenase | 4 (2) | 3 (1) |
| Fatty acid biosynthesis | ||
| Acetyl-CoA carboxylase | 4 (0) | 3 (0) |
| Stress related protein | ||
| 2-Cys peroxiredoxin | 13 (0) | 10 (0) |
One prominent thioredoxin-linked enzyme of the Calvin cycle, fructose-1,6-bisphosphatase, was not detected in the DTT eluant from the thioredoxin columns. This failure was likely due to the ability of the third cysteine present in fructose-1,6-bisphosphatase to form a disulfide with the cysteine participating in the heterodisulfide bonding with thioredoxin, thus displacing the enzyme from the mutant column. This property has been observed in other studies (42, 43).
For unclear reasons, an additional six chloroplast proteins for which there is evidence for a link to thioredoxin were not observed in this study: glucose-6-phosphate dehydrogenase (44), RB60 (38), cyclophilin (7), 5′-adenylylsulfate reductase (45), and 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (46). There was also no evidence of methionine sulfoxide reductase, an enzyme using thioredoxin as a substrate (47). It is possible that these enzymes were not trapped on the column owing to the participation of an associated cysteine, as with fructose-1,6-bisphosphatase or, alternatively, were not identified owing to a lack of sufficient homology with proteins in the database.
Specificity of Thioredoxin f and m.
An interesting point emerging from this study is the apparent lack of specificity shown by the column-bound mutant f- and m-type thioredoxins. Whereas most previously known enzymes identified in Table 3 are preferentially regulated by thioredoxin f in vitro (2, 3), specificity is not reflected in the results obtained above with the mutated proteins. With few exceptions, no significant specificity for thioredoxin f or m was seen with either the previously unknown or known target proteins (Tables 2 and 3). These findings agree with a similar study based on a mutant form of thioredoxin m in which serine replaced the buried cysteine (7). In that case, the authors found that the mutant thioredoxin m protein trapped several chloroplast targets known to prefer thioredoxin f, which itself, when mutated, was ineffective in binding target enzymes under their conditions. Such a lack of thioredoxin specificity in the earlier (7) and present study can be explained by assuming that the replacement of one cysteine of the active site by either serine or alanine abolished specificity by inducing a slight change in the microenvironment. The absence of specificity could also be due to the high concentration of thioredoxins bound to the Sepharose, a factor that would favor the formation of a heterodisulfide regardless of enzyme preference. These conditions may possibly also promote the formation of heterodisulfide bonds with proteins that normally seem not to be targeted by thioredoxin (J. Beckwith, personal communication).
Oxidative Regulation: A Possible Role for Chloroplast Thioredoxins.
Owing to lack of mobility, plants must adapt to unanticipated changes in the environment. The most regular, if not dramatic, change is the alternation of day and night, a transition recognized by the chloroplast thioredoxin system. Environmentally induced stresses are deleterious and more sporadic (e.g., drought, heat, cold, and high light). Most, if not all, result in an increase in the production of reactive oxygen species that impede photosynthesis (48, 49).
Carbon dioxide assimilation is among the first chloroplast processes affected when plants experience stress. The Calvin cycle typically slows down with the onset of stress, well before photosynthetic electron flow and ATP synthesis (50, 51). This change seems to enhance survival by enabling the plant to redirect energy resources to antioxidant defense.
One means to achieve the observed effects lies in the sensitivity of the Calvin cycle to H2O2, a reactive oxygen product formed in most stresses [measurements show a 50% inhibition of photosynthetic CO2 assimilation by isolated chloroplasts at 10 μM H2O2 (35)]. It is likely that stress-induced increases in H2O2 initiate a survival response through the oxidation of sulfhydryl groups critical to the activity of certain enzymes. Accordingly, several thioredoxin-linked enzymes of the Calvin cycle have been found to be sensitive to H2O2 (52). Early results provided evidence that two of the previously undocumented thioredoxin-linked enzymes, transketolase and possibly ribulose-P 3-epimerase, are sensitive to H2O2 and are sites of H2O2-dependent inhibition (35). The presence of conserved cysteines in these chloroplast enzymes is consistent with such a role.
Transketolase is a participant in both the Calvin and oxidative pentose phosphate cycles. Furthermore, its products and substrates are the starting point of several other chloroplast pathways. A decrease in transketolase activity thus affects not only the Calvin cycle, where the enzyme seems to be rate limiting, but also carbohydrate, amino acid, and natural product metabolism (53). As a result of its multiple roles, transketolase is therefore an excellent candidate for an oxidative type of regulation that leads to its reversible deactivation, with an attendant slowing down of the associated metabolic processes. According to this interpretation, when the stromal environment returns to normal (i.e., becomes less oxidizing), thioredoxin would reduce the oxidized deactivated form of transketolase, thereby relieving inhibition and restoring full catalytic activity. It remains to be seen whether the enzyme is also regulated by light.
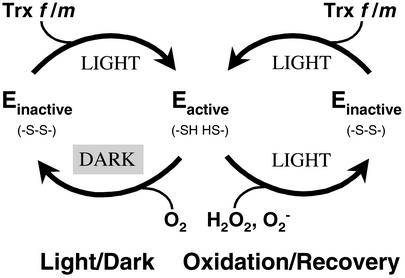
The present data suggest that, in addition to classical light/dark modulation, thioredoxin acts in a previously unrecognized oxidative type of regulation in chloroplasts. The latter function provides a reversible mechanism for the deactivation of photosynthetic CO2 assimilation in response to newly formed oxidants and for a subsequent reactivation with the return of agreeable cellular conditions, all in the light (Fig. 1). This oxidant-induced regulation allows the chloroplast to slow down light-dependent biosynthesis and accelerate carbohydrate degradation (e.g., oxidative pentose phosphate cycle) under constant illumination conditions, thereby minimizing energy loss and marshalling available biochemical resources for antioxidant defense. In the case of porphyrins and derived chlorins, impeding biosynthetic enzymes may also minimize the capability of products of the pathway to form free radicals under oxidative conditions.
Figure 1.
Role of chloroplast thioredoxin in the reversible regulation of enzymes as a result of light/dark transition (Left) or oxidation in the light (Right). In addition to light/dark regulation, thioredoxin provides a means to inactivate biosynthetic and accelerate degradative processes in the presence of deleterious oxidants while plants are maintained in the light. When oxidants are removed, metabolism returns to its normal state.
Other thioredoxin-linked enzymes are also candidates for oxidative regulation. One of the previously unknown targets, carbonic anhydrase, loses activity under oxidative conditions in a manner partially reversed by DTT (28). In addition, purified rubisco, which is inhibited by cold treatment, was restored to a fully active state by incubation with DTT (54). This cold-induced response could result from an oxidation of the small subunit of the enzyme that was also identified as a thioredoxin target in a previous study (7). The present findings suggest that rubisco and carbonic anhydrase [two extensively studied enzymes not known to be directly activated by light (the activation of the rubisco requires rubisco activase)] could be linked to thioredoxin to effect the slowing down of the Calvin cycle under stress. Thioredoxin-linked enzymes that have long been known to be regulated by light could also be subject to oxidative control.
Concluding Remarks
If confirmed in future studies, the present findings will have more than doubled the number of chloroplast proteins and processes known to be linked to thioredoxin. The proteomic approach will have amplified the influence of thioredoxin on light/dark modulation and extended its role to what seems to be a previously unrecognized oxidative type of regulation. The work thus opens the door to the application of emerging proteomic technologies to identify the large number of chloroplast disulfide proteins that are reduced in the light (55) and in so doing to help define the thioredoxin-linked component of the disulfide proteome (56).
Supplementary Material
Acknowledgments
We thank M. Lange, J. Yates, S. Reinbothe, and R. Wolosiuk for helpful comments on the manuscript and C. Deciu for assistance with data analysis. This work was supported by funds from the Torrey Mesa Research Institute. Y.B. and P.S., respectively, acknowledge a fellowship and a grant from the Swiss National Science Foundation.
References
- 1.Buchanan B B. Annu Rev Plant Physiol. 1980;31:341–374. [Google Scholar]
- 2.Schürmann P, Jacquot J-P. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:371–400. doi: 10.1146/annurev.arplant.51.1.371. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan B B, Schürmann P, Wolosiuk R A, Jacquot J-P. Photosynth Res. 2002;73:215–222. doi: 10.1023/A:1020407432008. [DOI] [PubMed] [Google Scholar]
- 4.Brandes H K, Larimer F W, Hartman F C. J Biol Chem. 1996;271:3333–3335. doi: 10.1074/jbc.271.7.3333. [DOI] [PubMed] [Google Scholar]
- 5.Balmer Y, Schürmann P. FEBS Lett. 2001;492:58–61. doi: 10.1016/s0014-5793(01)02229-3. [DOI] [PubMed] [Google Scholar]
- 6.Verdoucq L, Vignols F, Jacquot J-P, Chartier Y, Meyer Y. J Biol Chem. 1999;274:19714–19722. doi: 10.1074/jbc.274.28.19714. [DOI] [PubMed] [Google Scholar]
- 7.Motohashi K, Kondoh A, Stumpp M T, Hisabori T. Proc Natl Acad Sci USA. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyer A, Haslekas C, Miginiac-Maslow M, Klein U, Le Marechal P, Jacquot J-P, Decottignies P. Eur J Biochem. 2002;269:272–282. doi: 10.1046/j.0014-2956.2001.02648.x. [DOI] [PubMed] [Google Scholar]
- 9.Schürmann P. Methods Enzymol. 1995;252:274–283. doi: 10.1016/0076-6879(95)52030-9. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett S G, Grossman A R, Chua N-H. In: Methods in Chloroplast Molecular Biology. Edelman M, Hallick R B, Chua N-H, editors. Amsterdam: Elsevier Biomedical; 1982. pp. 1081–1092. [Google Scholar]
- 11.del Val G, Maurer F, Stutz E, Schürmann P. Plant Sci. 1999;149:183–190. [Google Scholar]
- 12.Gharahdaghi F, Weinberg C R, Meagher D A, Imai B S, Mische S M. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 13.Shevchenko A, Chernushevich I, Wilm M, Mann M. Methods Mol Biol. 2000;146:1–16. doi: 10.1385/1-59259-045-4:1. [DOI] [PubMed] [Google Scholar]
- 14.Koller A, Washburn M P, Lange M, Andon N L, Deciu C, Haynes P A, Hays L, Schieltz D, Ulaszek R, Wei J, et al. Proc Natl Acad Sci USA. 2002;99:135–146. doi: 10.1073/pnas.172183199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lichtenthaler H K. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:47–65. doi: 10.1146/annurev.arplant.50.1.47. [DOI] [PubMed] [Google Scholar]
- 16.Campos N, Rodríguez-Concepción M, Seemann M, Rohmer M, Boronat A. FEBS Lett. 2001;488:170–173. doi: 10.1016/s0014-5793(00)02420-0. [DOI] [PubMed] [Google Scholar]
- 17.Qerol J, Campos N, Imperial S, Boronat A, Rodríguez-Concepción M. FEBS Lett. 2002;514:343–346. doi: 10.1016/s0014-5793(02)02402-x. [DOI] [PubMed] [Google Scholar]
- 18.Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Proc Natl Acad Sci USA. 2001;98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwender J, Müller C, Zeidler J, Lichtenthaler H K. FEBS Lett. 1999;445:140–144. doi: 10.1016/s0014-5793(99)00849-2. [DOI] [PubMed] [Google Scholar]
- 20.Lange B M, Croteau R. Proc Natl Acad Sci USA. 1999;96:13714–13719. doi: 10.1073/pnas.96.24.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinbothe S, Reinbothe C. Eur J Biochem. 1996;237:323–343. doi: 10.1111/j.1432-1033.1996.00323.x. [DOI] [PubMed] [Google Scholar]
- 22.Manohara M S, Tripathy B C. Planta. 2000;212:52–59. doi: 10.1007/s004250000363. [DOI] [PubMed] [Google Scholar]
- 23.Beck C F. Protist. 2001;152:175–182. doi: 10.1078/1434-4610-00056. [DOI] [PubMed] [Google Scholar]
- 24.Surpin M, Larkin R M, Chory J. Plant Cell. 2002;14,Suppl.:S327–S338. doi: 10.1105/tpc.010446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson-Constan D, Akita M, Keegstra K. Biochim Biophys Acta. 2001;1541:102–113. doi: 10.1016/s0167-4889(01)00148-3. [DOI] [PubMed] [Google Scholar]
- 26.Clarke A K. Ann Bot (London) 1999;83:593–599. [Google Scholar]
- 27.Stieger P A, Feller U. J Exp Bot. 1997;48:1639–1645. [Google Scholar]
- 28.Johansson I-M, Forsman C. Eur J Biochem. 1993;218:439–446. doi: 10.1111/j.1432-1033.1993.tb18394.x. [DOI] [PubMed] [Google Scholar]
- 29.Scheidig A, Fröhlich A, Schulze S, Lloyd J R, Kossmann J. Plant J. 2002;30:581–591. doi: 10.1046/j.1365-313x.2002.01317.x. [DOI] [PubMed] [Google Scholar]
- 30.Hoppe P, Heintze A, Riedel A, Creuzer C, Schultz G. Planta. 1993;190:253–262. [Google Scholar]
- 31.Anderson L E, Li A D, Stevens F J. Phytochemistry. 1998;47:707–713. doi: 10.1016/s0031-9422(97)00659-6. [DOI] [PubMed] [Google Scholar]
- 32.Tuteja N, Phan T N, Tewari K K. Eur J Biochem. 1996;238:54–63. doi: 10.1111/j.1432-1033.1996.0054q.x. [DOI] [PubMed] [Google Scholar]
- 33.Reski R. Trends Plant Sci. 2002;7:103–105. doi: 10.1016/s1360-1385(02)02232-x. [DOI] [PubMed] [Google Scholar]
- 34.Teige M, Melzer M, Süss K-H. Eur J Biochem. 1998;252:237–244. doi: 10.1046/j.1432-1327.1998.2520237.x. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser W. Biochim Biophys Acta. 1976;440:476–482. doi: 10.1016/0005-2728(76)90035-9. [DOI] [PubMed] [Google Scholar]
- 36.Hell R, Jost R, Berkowitz O, Wirtz M. Amino Acids. 2002;22:245–257. doi: 10.1007/s007260200012. [DOI] [PubMed] [Google Scholar]
- 37.Rolland N, Ruffet M L, Job D, Douce R, Droux M. Eur J Biochem. 1996;236:272–282. doi: 10.1111/j.1432-1033.1996.00272.x. [DOI] [PubMed] [Google Scholar]
- 38.Trebitsh T, Danon A. Proc Natl Acad Sci USA. 2001;98:12289–12294. doi: 10.1073/pnas.211440698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baginsky S, Gruissem W. Nucleic Acids Res. 2002;30:4527–4533. doi: 10.1093/nar/gkf561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richarme G. Biochem Biophys Res Commun. 1998;252:156–161. doi: 10.1006/bbrc.1998.9591. [DOI] [PubMed] [Google Scholar]
- 41.Krepinsky K, Plaumann M, Martin W, Schnarrenberger C. Eur J Biochem. 2001;268:2678–2686. doi: 10.1046/j.1432-1327.2001.02154.x. [DOI] [PubMed] [Google Scholar]
- 42.Chiadmi M, Navaza A, Miginiac-Maslow M, Jacquot J-P, Cherfils J. EMBO J. 1999;18:6809–6815. doi: 10.1093/emboj/18.23.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balmer Y, Stritt-Etter A-L, Hirasawa M, Jacquot J-P, Keryer E, Knaff D B, Schürmann P. Biochemstry. 2001;40:15444–15450. doi: 10.1021/bi011646m. [DOI] [PubMed] [Google Scholar]
- 44.Wenderoth I, Scheibe R, Von Schaewen A. J Biol Chem. 1997;272:26985–26990. doi: 10.1074/jbc.272.43.26985. [DOI] [PubMed] [Google Scholar]
- 45.Bick J-A, Setterdahl A T, Knaff D B, Chen Y, Pitcher L H, Zilinskas B A, Leustek T. Biochemistry. 2001;40:9040–9048. doi: 10.1021/bi010518v. [DOI] [PubMed] [Google Scholar]
- 46.Entus R, Poling M, Herrmann K M. Plant Physiol. 2002;129:1866–1871. doi: 10.1104/pp.002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoshi T, Heinemann S H. J Physiol. 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray E A, Bailey-Serres J, Weretilnyk E. In: Biochemistry and Molecular Biology of Plants. Buchanan B B, Gruissem W, Jones R L, editors. Rockville, MD: Am. Soc. Plant Physiol.; 2000. pp. 1189–1197. [Google Scholar]
- 49.Bartosz G. Acta Physiol Plant. 1997;19:47–64. [Google Scholar]
- 50.Weis E. Planta. 1981;151:33–39. doi: 10.1007/BF00384234. [DOI] [PubMed] [Google Scholar]
- 51.Lawlor D W, Cornic G. Plant Cell Environ. 2002;25:275–294. doi: 10.1046/j.0016-8025.2001.00814.x. [DOI] [PubMed] [Google Scholar]
- 52.Takeda T, Yokota A, Shigeoka S. Plant Cell Physiol. 1995;36:1089–1095. [Google Scholar]
- 53.Henkes S, Sonnewald U, Badur R, Flachmann R, Stitt M. Plant Cell. 2001;13:535–551. doi: 10.1105/tpc.13.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tenaud M, Jacquot J-P. J Plant Physiol. 1987;130:315–326. [Google Scholar]
- 55.Crawford N A, Droux M, Kosower N S, Buchanan B B. Arch Biochem Biophys. 1989;271:223–239. doi: 10.1016/0003-9861(89)90273-7. [DOI] [PubMed] [Google Scholar]
- 56.Yano H, Kuroda S, Buchanan B B. Proteomics. 2002;2:1090–1096. doi: 10.1002/1615-9861(200209)2:9<1090::AID-PROT1090>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.