Abstract
Replication of hepatitis C virus (HCV) RNA in virus-infected cells is believed to be catalyzed by viral replicase complexes (RCs), which may consist of various virally encoded nonstructural proteins and host factors. In this study, we characterized the RC activity of a crude membrane fraction isolated from HCV subgenomic replicon cells. The RC preparation was able to use endogenous replicon RNA as a template to synthesize both single-stranded (ss) and double-stranded (ds) RNA products. Divalent cations (Mg2+ and Mn2+) showed different effects on RNA synthesis. Mg2+ ions stimulated the synthesis of ss RNA but had little effect on the synthesis of ds RNA. In contrast, Mn2+ ions enhanced primarily the synthesis of ds RNA. Interestingly, ss RNA could be synthesized under certain conditions in the absence of ds RNA, and vice versa, suggesting that the ss and ds RNA were derived either from different forms of replicative intermediates or from different RCs. Pulse-chase analysis showed that radioactivity incorporated into the ss RNA was chased into the ds RNA and other larger RNA species. This observation indicated that the newly synthesized ss RNA could serve as a template for a further round of RNA synthesis. Finally, 3′ deoxyribonucleoside triphosphates were able to inhibit RNA synthesis in this cell-free system, presumably through chain termination, with 3′ dGTP having the highest potency. Establishment of the replicase assay will facilitate the identification and evaluation of potential inhibitors that would act against the entire RC of HCV.
Hepatitis C virus (HCV) is the major cause of non-A, non-B transfusion-associated hepatitis and accounts for a significant proportion of viral hepatitis cases worldwide (9, 18). Although HCV infection is resolved in some subjects, up to 80% of infected individuals develop chronic HCV infection. HCV is a positive-sense RNA virus belonging to the Flaviviridae family (5). The life cycle of HCV consists of several important processes that occur primarily in the cytoplasm of the host cells. Among these important processes, RNA replication is a key step in the viral proliferation cycle and is thus an important target for antiviral development. The genome of HCV consists of approximately 9,600 bases, with a single open reading frame encoding a polyprotein of around 3,010 amino acids. The open reading frame is flanked by 5′ and 3′ untranslated regions, which are important for translation and RNA replication. The polyprotein is cleaved both co- and posttranslationally by cellular and virally encoded proteases into at least 10 different structural and nonstructural proteins (1, 9, 18).
The NS5B protein of HCV has been shown to possess RNA-dependent RNA polymerase activity in vitro. However, recombinant NS5B alone lacks the strict template specificity required for RNA synthesis (2, 6-8, 11, 13-15, 21, 27, 28). The lack of template specificity is presumably due to the absence of other viral or host factors that constitute the viral replicase complexes (RCs) in vivo. It is believed that RCs, rather than NS5B protein alone, are involved in catalyzing RNA synthesis during viral RNA replication. HCV RCs contain, in addition to NS5B, other viral nonstructural proteins (e.g., NS3/4A, NS4B, and NS5A) and possibly host factors. These additional components may play important roles in modulating polymerase activity, conferring template specificity and ensuring RNA synthesis with necessary fidelity during the replication process. So far, detailed characterization of HCV replicase activity in the context of a complex has not been accomplished, mainly due to the absence of a robust HCV cell culture system. The recent successful establishment of a HCV replicon cell culture provided a useful source for isolation of viral RCs (3, 12). In this study, we characterized the activities of the RCs in crude membrane fractions isolated from HCV subgenomic replicon cells. Isolation of the HCV replicase and determination of its activity are expected to provide a useful system for evaluation of potential inhibitors targeting HCV RNA replication.
Active RCs present in a membrane fraction isolated from HCV subgenomic replicon cells.
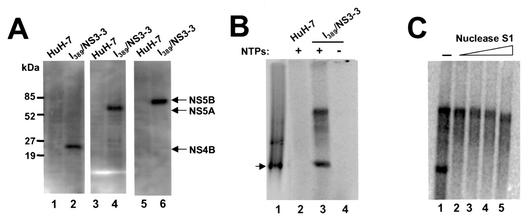
HCV subgenomic replicon cells (I389/NS3-3 cell line) (10, 12) were used to isolate the viral RC. Previous immunoelectron microscopic studies on the same replicon cells revealed that the majority of HCV viral proteins were associated with membranes of the endoplasmic reticulum (ER) (17). HCV NS5B protein was also reported to be located on the ER or an ER-like compartment in U-2 OS human osteosarcoma-derived cells expressing all HCV proteins (20). These findings provide strong evidence that HCV replication occurs through membrane-associated RCs, as in the case of several other positive-strand RNA viruses, including poliovirus, flock house virus, and flaviviruses (4, 23, 25, 26). We isolated a crude membrane fraction from subgenomic replicon cells by Dounce homogenization and high-speed centrifugation (22). Western blotting analysis confirmed that HCV nonstructural proteins (NS4B, NS5A, and NS5B) were present in this membrane preparation (Fig. 1A, lanes 2, 4, and 6). This analysis also revealed that approximately 20% of the HCV nonstructural proteins were retained in the membrane fraction (data not shown). No viral protein was detected in the membrane fraction isolated in parallel from the control HuH-7 cells (Fig. 1A, lanes 1, 3, and 5). An RNA replication assay was then used to determine whether the RCs in this membrane fraction were enzymatically active. Standard assay mixtures contained 50 mM HEPES (pH 7.3); 10 mM KCl; 10 mM MgCl2; 20 U of RNase inhibitor; 10 μg of actinomycin D per ml; 0.5 mM (each) ATP, GTP, and CTP; 10 μCi of [α-33P]-UTP; and 0.5 μl of the membrane fraction in a total volume of 20 μl. The reaction mixtures were incubated at 30°C for 2 h, followed by phenol-chloroform extraction and ethanol precipitation. As shown in Fig. 1, two major RNA products with different mobilities were detected (Fig. 1B, lane 3), with one of these corresponding to single-stranded (ss) subgenomic replicon RNA (Fig. 1B, lane 1). No product was detected with the membrane fraction isolated from the control HuH-7 cells (Fig. 1B, lane 2), indicating that the labeled products were specific to replicon cells. Formation of the products also required the presence of all nucleoside triphosphate substrates (ATP, CTP, and GTP, in addition to the labeled UTP) (Fig. 1B, lane 4), suggesting that it is not likely that they were produced by possible terminal nucleotidyl transferase activity associated with the membrane preparation.
FIG. 1.
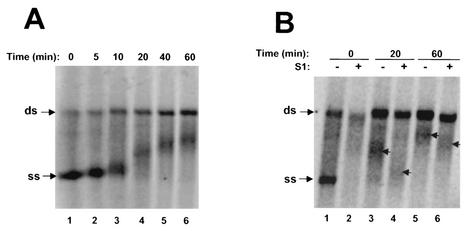
Characterization of HCV RCs in a crude membrane fraction isolated from the HCV subgenomic replicon cell line I389/NS3-3 (10, 12). (A) Western blot analysis to determine whether HCV nonstructural proteins NS4B (lanes 1 and 2), NS5A (lanes 3 and 4), and NS5B (lanes 5 and 6) were present in membrane fractions isolated from replicon cells and parental HuH-7 cells. The membrane extracts (6 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Molecular weight standards are shown on the left. (B) In vitro RNA replication directed by HCV RCs. The RNA replication assay was carried out for 2 h at 30°C. The RNA products were extracted with phenol-chloroform, ethanol precipitated, and separated on a 1% agarose gel. After electrophoresis, the gel was dried prior to autoradiography. Radiolabeled subgenomic replicon RNA transcripts synthesized from the I389/NS3-3 cDNA clone by T7 polymerases were used as the marker (lane 1), and membrane fractions either from HuH-7 cells (lane 2) or from the replicon cells (lanes 3 and 4) were used in the assay. Cold nucleoside triphosphate substrates (ATP, CTP, and GTP) were omitted from the reaction mixture to obtain the results shown in lane 4. The arrow indicates the position of ss subgenomic replicon RNA. (C) Nuclease S1 treatment of the labeled products synthesized by the RC. RNA products synthesized from the standard reaction were treated with increasing concentrations of nuclease S1 (1.2 to 32 U/μl). After digestion by S1, the RNA was recovered and separated on a 1% agarose gel. The upper and lower bands represent ds and ss RNA products, respectively.
Nuclease S1 treatment (specific for ss RNA regions) was used to determine the nuclease sensitivity of the labeled products. As shown in Fig. 1C, the monomer-length RNA product was highly sensitive to nuclease S1 treatment, whereas the larger RNA product was resistant to the treatment. This result indicated that the viral RC produced ss and double-stranded (ds) RNA products. Taken collectively, our data demonstrated that the membrane fraction isolated from the replicon cells contained active RC, which could utilize endogenous subgenomic replicon RNA as a template to synthesize both ss and ds RNA products.
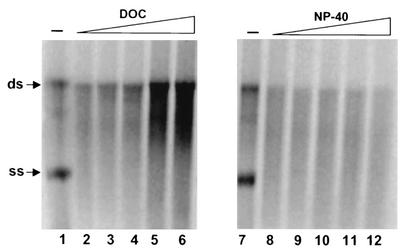
Pretreatment of the crude membrane fraction with mild detergents (deoxycholate or Nonident P-40) had different effects on the syntheses of ss and ds RNA products. As shown in Fig. 2, ss RNA formation was significantly inhibited by low concentrations of both detergents, suggesting that membrane structure was absolutely required for ss RNA synthesis by the viral RC. In contrast, synthesis of ds RNA was less sensitive to detergent treatment. In fact, higher concentrations of deoxycholate (range, 2 to 4%) could enhance the production of ds RNA product (Fig. 2, lanes 4 to 6).
FIG. 2.
Effect of detergent treatment on replicase activity. Membrane fractions were treated with various concentrations of deoxycholate (DOC) or Nonident P-40 (NP-40) for 30 min at 4°C. After the treatment period, a standard replication reaction mixture was added to a final volume of 20 μl, and the resulting mixture was incubated for 2 h at 30°C. The labeled RNA products were recovered by ethanol precipitation and separated on a 1% agarose gel. The positions of ss and ds RNA are indicated on the left. Lanes 1 and 7, no detergent; lanes 2 to 6 and 8 to 12, detergent at concentrations ranging from 0.25 to 4%.
Different effects of divalent cations on RNA synthesis.
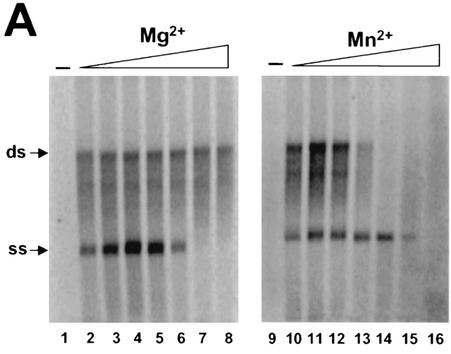
As in the case of recombinant NS5B protein, divalent cations were required for the replicase activity, as no product was detected in the absence of Mg2+ or Mn2+ (Fig. 3A, lanes 1 and 9). Interestingly, Mg2+ and Mn2+ showed different effects on the syntheses of ss and ds RNA products. Mg2+ specifically stimulated the synthesis of ss RNA, with optimal concentrations at around 5 mM, though at concentrations of up to 80 mM it had little effect on the synthesis of ds RNA (Fig. 3A). However, at higher concentrations (>20 mM), Mg2+ showed an inhibitory effect on ss RNA synthesis. In contrast, low concentrations of Mn2+ showed a stimulatory effect on the synthesis of ds RNA, with the optimal concentration at about 0.5 mM. Higher concentrations of Mn2+ (>3 mM for ss RNA and >0.5 mM for ds RNA) showed a strong inhibitory effect on RNA synthesis. This result is different from that observed with the recombinant NS5B protein, which preferred Mn2+ ions for maximal enzymatic activity (7, 13). Furthermore, the results shown in Fig. 3 also revealed that synthesis of ss RNA could occur under certain conditions (e.g., higher Mn2+ concentrations) in the absence of ds RNA production (Fig. 3A, lanes 13 to 15). Similarly, ds RNA could be synthesized (at higher Mg2+ concentrations) in the absence of ss RNA synthesis (Fig. 3A, lanes 7 to 8). This observation suggested that ss and ds RNA were likely generated either from different forms of replicative intermediates or from different types of RCs. This hypothesis was also supported by the earlier observation that the processes for synthesis of ss RNA and ds RNA showed different sensitivities to detergent treatment (Fig. 2).
FIG. 3.
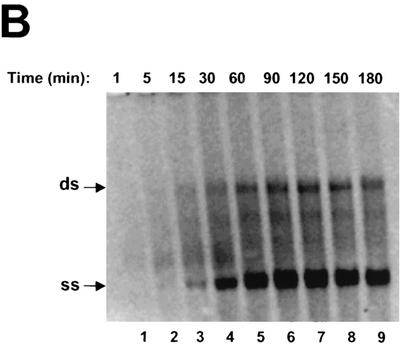
(A) Effects of divalent metal ions on RNA synthesis. Standard reaction mixtures were incubated at 30°C for 2 h in the presence of various concentrations of metal ions (see below). The labeled RNA products were recovered by ethanol precipitation, separated on a 1% agarose gel, and visualized by autoradiography. Lanes 2 to 8, Mg2+ in concentrations ranging from 1.25 to 80 mM; lanes 10 to 16, Mn2+ in concentrations ranging from 0.15 to 10 mM. (B) Time course of RNA synthesis directed by RC. Lanes 1 to 9, results of standard reactions carried out at 30°C and terminated at indicated time intervals by the addition of EDTA to a final concentration of 25 mM. The labeled products were recovered by ethanol precipitation and separated on a 1% agarose gel. The positions of ss and ds RNA are indicated with arrows at the left.
A time-course experiment was used to characterize the kinetics of RNA synthesis in this cell-free system. The reaction mixtures were incubated for various periods of times ranging from a few minutes to several hours. As shown in Fig. 3B, ss and ds RNA products started to appear at about 5 min, with production increasing over time and reaching a maximum at approximately 90 to 120 min. ss RNA was made in a large excess over ds RNA. Interestingly, the two RNA products were synthesized almost simultaneously. This is somewhat different from what has been observed for the RCs of flock house virus. In the case of that virus, ds RNA product was synthesized first and then served as the intermediate for synthesis of ss RNA (25). Our results further strengthened the hypothesis that synthesis of ss RNA by the HCV RC in this cell-free system was independent of that of ds RNA.
Newly synthesized ss RNA as the template for a further round of RNA synthesis.
To determine the template activity of the newly synthesized RNA product, a pulse-chase experiment was performed. Briefly, reaction mixtures were incubated at 30°C for 60 min in the presence of [α-33P]-UTP (in addition to cold ATP, CTP, and GTP). After the labeling reaction, a large excess of cold UTP (300-fold over labeled UTP) was added to the reaction and the reaction mixtures were further incubated for various time intervals. As shown in Fig. 4A, the newly synthesized ss RNA was chased into product larger than the template size, which increased in size over time. A small proportion of the radioactivity was also chased into ds RNA, as indicated by increasing intensity of the ds RNA. This result indicated that the newly synthesized ss RNA could serve as template for the synthesis of ds RNA.
FIG. 4.
Newly synthesized ss RNA can serve as template for further rounds of RNA synthesis. (A) Labeled ss RNA was chased into ds RNA and RNA species longer than monomeric replicon RNA. RNA products were pulse-labeled for 60 min in standard reactions (mixtures contained 10 μCi of [α-33P] UTP). After the labeling step, a 300-fold excess of unlabeled UTP was added to the reaction mixtures, and the labeled products were chased for 0, 5, 10, 20, 40, and 60 min (lanes 1, 2, 3, 4, 5, and 6, respectively). The positions of ss and ds RNA are indicated with arrows at the left. (B) Nuclease S1 treatment of the extended RNA products. The labeled products were chased for 0, 20 or 60 min in the presence of excess cold UTP. The reaction mixtures were then separated on a 1% agarose gel after no treatment (lanes 1, 3, and 5) or treatment with nuclease S1 (lanes 2, 4, and 6). The small arrows indicate the positions of the extended products with and without nuclease S1 treatment (as indicated, nuclease treatment resulted in smaller products).
The fact that most of the labeled ss RNA was chased into products less than the size of ds RNA prompted us to analyze further the nature of the extended RNA molecules by nuclease S1 treatment. As shown in Fig. 4B, nuclease treatment on the extended RNA products resulted in a reduction in size, suggesting that these molecules were partially ds, presumably representing various forms of replication intermediates. Due to the low product yield of this replication assay, we were unable to further characterize these partially ds molecules by conventional methods.
3′ dNTPs as inhibitors of HCV RC.
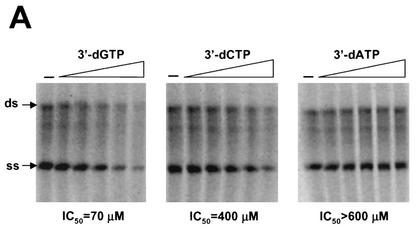
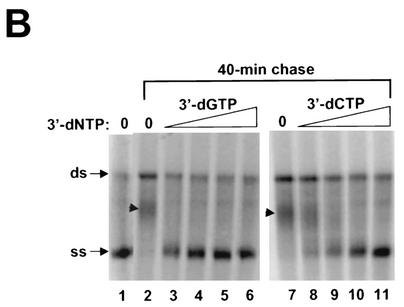
Establishment of the HCV replicase assay will help to identify and validate potential inhibitors targeting the RNA replication process. We first tested whether 3′ deoxyribonucleoside triphosphates (dNTPs) can inhibit replicase activity in a cell-free system. Since the hydroxyl group at the 2′ position (2′-OH) of rNTPs is specifically recognized by RNA polymerases and contributes to substrate specificity. It is conceivable that 3′ dNTPs, which retain the 2′-OH group, can serve as the substrate for HCV polymerase and subsequently inhibit RNA synthesis through chain termination. As shown in Fig. 5A, 3′ dGTP and 3′ dCTP inhibited both ss and ds RNA synthesis, with 50% inhibitory concentrations of 70 and 400 μM, respectively (the concentration of cold GTP or CTP in the reaction was 500 μM). In contrast, 3′ dATP showed no effect on the RNA synthesis at concentrations as high as 600 μM. To further demonstrate that 3′ dGTP and 3′ dCTP can prevent chain elongation during RNA synthesis, a pulse-chase experiment was performed in the presence of 3′ dGTP and 3′ dCTP. In this case, ss and ds RNA were labeled with [33P]-UTP for 60 min, after which a 300-fold excess of cold UTP and various concentrations (0 to 200 μM) of 3′ dGTP or 3′ dCTP were added. The labeled products were chased for an additional 40 min at 30°C. As shown in Fig. 5B, a majority of the labeled ss RNA was further extended into either ds RNA or RNA longer than the template length after the 40-min chase period (compare Fig. 5B, lanes 1 and 2). In the presence of the increasing concentrations of 3′ dGTP, no extension from ss RNA was detected and all the labeled ss RNA remained at the monomer size (Fig. 5B, lanes 3 to 6). Similarly, inhibition of chain elongation was also observed with 3′ dCTP (Fig. 5B, lanes 7 to 11), though it was less efficient than with 3′ dGTP. Taken collectively, these results demonstrated that 3′ dGTP and 3′ dCTP were able to inhibit RNA synthesis and prevent chain elongation directed by the HCV RC.
FIG. 5.
Inhibitory effect of 3′ dNTPs on RNA synthesis directed by HCV RCs. (A) Effects of 3′ dGTP, 3′ dCTP, and 3′ dATP on RNA synthesis. Standard reactions were performed at 30°C for 2 h in the presence of increasing concentrations (0 to 600 μM) of the indicated 3′dNTPs. The labeled RNA products were recovered by ethanol precipitation, separated on a 1% agarose gel, and visualized by autoradiography. Incorporation of [33P]-UMP was measured by phosphorimaging, and the concentrations resulting in a 50% reduction in labeled RNA were determined (IC50). (B) 3′ dGTP and 3′ dCTP prevent further elongation of the labeled ss RNA product. RNA products were pulse-labeled for 60 min in standard reactions containing 10 μCi of [α-33P]-UTP. The labeled products were chased for 40 min in the presence of excess cold UTP and increasing concentrations of the indicated 3′dNTP. Lanes 1, products from the standard pulse-labeling reaction prior to the chase step; lanes 2 to 6, products obtained with addition of 3′ dGTP at concentrations ranging from 0 to 200 μM; lanes 7 to 11, products obtained with addition of 3′ dCTP at concentrations ranging from 0 to 200 μM. The positions of ss and ds RNA are indicated with arrows at the left. The small arrowhead indicate the positions of the extended products.
HCV RC preparation could not use exogenous RNA as template.
So far, all our characterization of the HCV RC was based on the use of the endogenous replicon RNA as the template. We had also attempted to introduce exogenous RNA templates into the reaction mixture, after removal of the endogenous replicon RNA by micrococcal nuclease (MN) treatment (25), to assess whether the HCV RC was able to use exogenous RNA as a template. To this end, viral RC (0.5 μl) was treated with MN in a 10-μl reaction volume containing 50 mM HEPES (pH 7.3), 10 mM dithiothreitol, and 2 mM CaCl2 for 30 min at 30°C. MN was inactivated by addition of EGTA to a final concentration of 20 mM. The treated RC was then assayed for activity in the presence various exogenous RNA templates. Under these assay conditions, the HCV RC was not able to utilize exogenous RNA as template. One explanation for the failure was that certain viral or host proteins, which were essential for this activity, were removed during the preparation of the replicase extract. This is also consistent with the less robust nature of this replicase system in comparison with those of other positive-strand RNA viruses (10, 25). The other explanation is the inability of the added RNA templates to access the active site of the RC due to sequestration by membranes. Currently, we are focusing on improving the efficiency of the HCV replicase system by searching for additional factors necessary for its maximal activity.
We reported here that the membrane fraction isolated from HCV subgenomic replicon cells contained active viral RC. The RC could utilize the endogenous replicon RNA as template to synthesize both ss and ds RNA products. Several observations have pointed to the hypothesis that synthesis of ss RNA in this replicase assay was independent of that of ds RNA, as follows. (i) The processes for synthesis of ss and ds RNA possessed different sensitivities to detergent treatment, with that of ss RNA being highly sensitive to detergent treatment. (ii) Divalent metal ions showed different effects on synthesis of ss and ds RNA. Mg2+ ions primarily stimulated synthesis of ss RNA whereas Mn2+ ions mainly enhanced the synthesis of ds RNA. (iii) ss RNA could be synthesized under certain conditions (e.g., higher Mn2+ concentrations, as shown in Fig. 3A) in the absence of ds RNA production and vice versa. In addition, a pulse-chase experiment demonstrated that newly synthesized ss RNA was further chased into ds RNA and RNA species larger than the monomeric replicon RNA, indicating that the ss RNA could serve as a template for additional rounds of RNA synthesis. (iv) Finally, we showed that 3′ dNTPs, especially 3′ dGTP and 3′ dCTP, were able to inhibit synthesis of both ss and ds RNA and prevent elongation, probably by serving as chain terminators. This result suggested that HCV RCs were capable of using 3′ dNTPs as a substrate for incorporation. This finding might have an important implication in the design of antiviral drug against this important viral pathogen.
A significant difference between HCV RC and other viral RCs is that its activity was highly dependent on the presence of the endogenous replicon RNA. So far, no success has been accomplished in inducing the viral RC to utilize exogenous RNA molecules as template. A number of viral RCs isolated from infected cells with positive-stranded RNA viruses were able to initiate RNA synthesis from an exogenous RNA template. In the case of black beetle virus and tobacco mosaic virus, removal of the endogenous RNA template by treatment with MN was crucial for the viral RC to synthesize both positive- and negative-stranded RNAs from the exogenous RNA template (16, 19). However, the HCV RC failed to utilize exogenous RNA as a template after similar MN treatment, this even when the RNA template was introduced in the presence of Lipofectin (Invitrogen) (data not shown), which has been shown to induce the synthesis of positive-stranded RNA from the added RNA template for flock house virus RC (24). Relatively speaking, the activity of the HCV RC was not as robust as those of other viral RCs. This might reflect that the HCV RC was isolated from stable replicon cells rather than from cells that had actually been infected with the virus, as in the cases of other similar studies. Alternatively, the inability to use exogenous RNA as a template may be either due to the absence of certain cofactors which were removed during the preparation of the RC extract or simply because this cell-free replication assay needed to be further optimized. Nonetheless, establishment of this cell-free replication system using HCV RC rather than NS5B alone is expected to provide an essential tool for further investigating the mechanisms of HCV replication and for identifying compounds that specifically target the viral replication process of HCV.
Acknowledgments
We thank Wayne Cheney, Nigel Horscroft, Robert Hamatake, Jae Hoon Shim, and Jim Z. Wu for helpful discussions and Suhaila Naim for technical assistance.
This work was supported by Ribapharm, Inc.
REFERENCES
- 1.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 2.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. [DOI] [PubMed] [Google Scholar]
- 4.Bolten, R., D. Egger, R. Gosert, G. Schaub, L. Landmann, and K. Bienz. 1998. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent in situ hybridization. J. Virol. 72:8578-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-364. [DOI] [PubMed] [Google Scholar]
- 6.De Francesco, R., S. E. Behrens, L. Tomei, S. Altamura, and J. Jiricny. 1996. RNA-dependent RNA polymerase of hepatitis C virus. Methods Enzymol. 275:58-67. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yau, J. Y. N. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 9.Houghton, M. 1996. Hepatitis C viruses, p. 1035-1058. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Raven Press, New York, N.Y.
- 10.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 73:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 13.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh, J.-W., T. Ito, and M. M. P. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral. RNA J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh, J. W., G. T. Sheu, and M. M. Lai. 2000. Template requirement and initiation site selection by hepatitis C virus polymerase on a minimal viral RNA template. J. Biol. Chem. 275:17710-17717. [DOI] [PubMed] [Google Scholar]
- 16.Osman, T. A., and K. W. Buck. 1996. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J. Virol. 70:6227-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-960. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3 ed. Raven Press, New York, N.Y.
- 19.Saunders, K., and P. Kaesberg. 1985. Template-dependent RNA polymerase from black beetle virus-infected Drosophila melanogaster cells. Virology 147:373-381. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 21.Sun, X. L., R. B. Johnson, M. A. Hockman, and Q. M. Wang. 2000. De novo RNA synthesis catalyzed by HCV RNA-dependent RNA polymerase. Biochem. Biophys. Res. Commun. 268:798-803. [DOI] [PubMed] [Google Scholar]
- 22.Takeda, N., R. J. Kuhn, C. F. Yang, T. Takegami, and E. Wimmer. 1986. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J. Virol. 60:43-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, S. X., P. Ahlquist, and P. Kaesberg. 1992. Active complete in vitro replication of nodavirus RNA requires glycerophospholipid. Proc. Natl. Acad. Sci. USA 89:11136-11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, S. X., and P. Kaesberg. 1991. Synthesis of template-sense, single-strand Flockhouse virus RNA in a cell-free replication system. Virology 183:392-396. [DOI] [PubMed] [Google Scholar]
- 26.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]
- 27.Zhong, W., E. Ferrari, C. A. Lesburg, D. Maag, S. K. B. Ghosh, C. E. Cameron, J. Y. Lau, and Z. Hong. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:9134-9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong, W., A. S. Uss, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]