Abstract
Based on analyses of two-dimensional gel and cDNA microarrays, our laboratory and others have demonstrated that a number of genes show altered expression during development of cisplatin resistance (CP-r) in human cancer cells, including genes associated with DNA damage-repair, proto-oncogenes, apoptosis, stress-response, and transcription factors, etc. To verify these results and find genes that are directly responsible for CP-r, as opposed to reflecting a secondary response induced by cisplatin treatment or resulting from CP-r, we constructed a retroviral cDNA library in the vector pLNCX2 from KB-CP.5 (KCP.5), a cell line selected in one step after exposure to cisplatin at 0.5 μg/ml. Using a library of cDNAs (1.8 × 106 cDNA clones), and an intermittent cisplatin selection system to allow more effective functional cloning, eleven expressed cDNAs were identified in a primary pool of 93,000 transfected cell clones. Metallothionein 2A, a known CP-r gene, was among these 11 genes found in the transfectants after CP selection. Several other genes, including those encoding ribosomal proteins (e.g. RPL36) and heat shock protein (e.g. HSP10) were also found among the cisplatin-selected clones. Transfection of either the RPL36 cDNA or HSP10 cDNA conferred on KB-3-1 cells 2.5 to 3-fold resistance to cisplatin by clonogenic assays. A subsequent transfection also identified RPL36 as a CP-r gene. The finding that a ribosomal protein gene, RPL36 contributes to CP-r should stimulate study of the role of ribosomal proteins in multi-factorial mechanisms of cisplatin resistance.
ABBREVIATIONS: RPL36, ribosomal protein L36; HSP10, heat shock protein 10; CP-r, cisplatin-resistance; RT-PCR, reverse transcription-polymerase chain reaction
INTRODUCTION
Cisplatin (cis-Diamminedichloroplatinum II) has revolutionized chemotherapy by improving treatment of a wide spectrum of solid tumors. However, despite the high efficacy of the compound, the ability of cancer cells to become resistant to the drug remains a significant impediment to successful chemotherapy. Intensive efforts have been made through biochemical characterization, cellular, and genetic approaches to determine the basis of resistance and define genes that are involved in acquisition of cisplatin resistance. Recent studies using two-dimensional gel analysis (Shen et al., 1995), gene knockout (Niedner et al., 2001), differential display (Francia et al., 2004), subtractive hybridization (Yasui et al., 2004), and cDNA microarrays (Roberts et al., 2005) have documented that a large number of genes are either up-regulated or down-regulated in cisplatin-resistant cells, including genes that encode transcription factors, DNA damage-repair proteins, stress-response proteins, cell cycle checkpoints, apoptosis mediators, and transporters, (see reviews, Gottesman et al., 2002; Wang & Lippard, 2005).
Functional cloning and retroviral cDNA libraries have been applied to define genes responsible for drug resistance, apoptosis, etc. (Perez-Victoria et al., 2003; Mourtada-Maarabouni et al., 2004). In identifying genes related to cisplatin resistance, the approaches that have been used have a major drawback due to continuous stepwise challenge with increased cisplatin concentrations, and may reflect secondary changes in genotype and phenotype during multi-step selection of cisplatin-resistant cells. To explore genes primarily involved in cisplatin resistance, we inserted double stranded cDNA into a retroviral expression vector, pLNCX2, from cisplatin-resistant KB-CP.5 cells that were selected by a single step of cisplatin at 0.5 μg/ml. An intermittent cisplatin selection procedure was designed for functional cloning, and subsequent identification of genes related to cisplatin resistance was by PCR and sequencing. By this technique, eleven genes were found in the individual clones after functional cloning, including metallothionein, which has been reported to be associated with cisplatin resistance (Kelly et al., 1988), and a chaperone, heat shock protein 10 (HSP10), which had previously been found to be inducible by a metal salt, cadmium chloride (Lee et al., 2002). A ribosomal protein, RPL36, was identified in this work, and in a second, independent transfection, was shown to confer ~2.5-fold increased CP-r on transfected stable clones in comparison with the control, vector-only transfected cells. Therefore, this functional retroviral cloning system and modified intermittent selection provides a powerful tool for further cloning of genes able to confer CP-r.
MATERIALS AND METHODS
Cell lines and cell culture
The human epidermoid carcinoma cell lines KB-3-1, an early-stage cisplatin-resistant cell line KB-CP.5, which was isolated at 0.5 μg/ml of cisplatin from KB-3-1 (Liang et al., 2003) were studied in this work. The RetroPack PT67 cell line was purchased from Clontech (Palo Alto, CA). All cell lines were grown as monolayer cultures at 37°C in 5% CO2, using Dulbecco's modified Eagle medium with 4.5 g/l glucose (InVitrogen, Carlsbad, CA), supplemented with l-glutamine, penicillin, streptomycin and 10% fetal bovine serum (BioWhittaker, Walkersville, MD). Cisplatin (Sigma, St. Louis, MO) was added into the medium for KB-CP.5 (0.5 μg/ml).
Construction of a retroviral cDNA library
Double stranded cDNA was synthesized from a single step selection of the Cisplatin-resistant cell line KB-CP.5 which was maintained in medium containing cisplatin 0.5 μg/ml, using a BD SMART cDNA library construction kit (Clontech) as described by the manufacturer. The synthesized double stranded cDNA was digested with the restriction enzyme Sfi, and then column purified by a CHROMA SPIN-400. The lower cutoff cDNA size was 500 bp. Ligation of the Sfi-digested double stranded cDNA to a retroviral expression vector pLNCX2 (Clontech) with Sfi-cut was performed according to the manufacturer, and the library was named KB-CP.5/pLNCX2. The retroviral cDNA library was transformed into DH5j competent cells (Invitrogen), and resulted in 1.8 x10 6 6 clones (library complexity).
Retroviral transduction and programmed cisplatin selection
Retroviral supernatants were collected from KB-CP.5/pLNCX2-transfected retroviral packaging PT67 cells, and then used to infect the recipient KB-3-1 cells. After selection with G418, a pool of 96,000 clones containing the KB-CP.5 cDNA was established. A programmed selection with cisplatin is shown in detail in Fig. 1B.
Fig. 1.
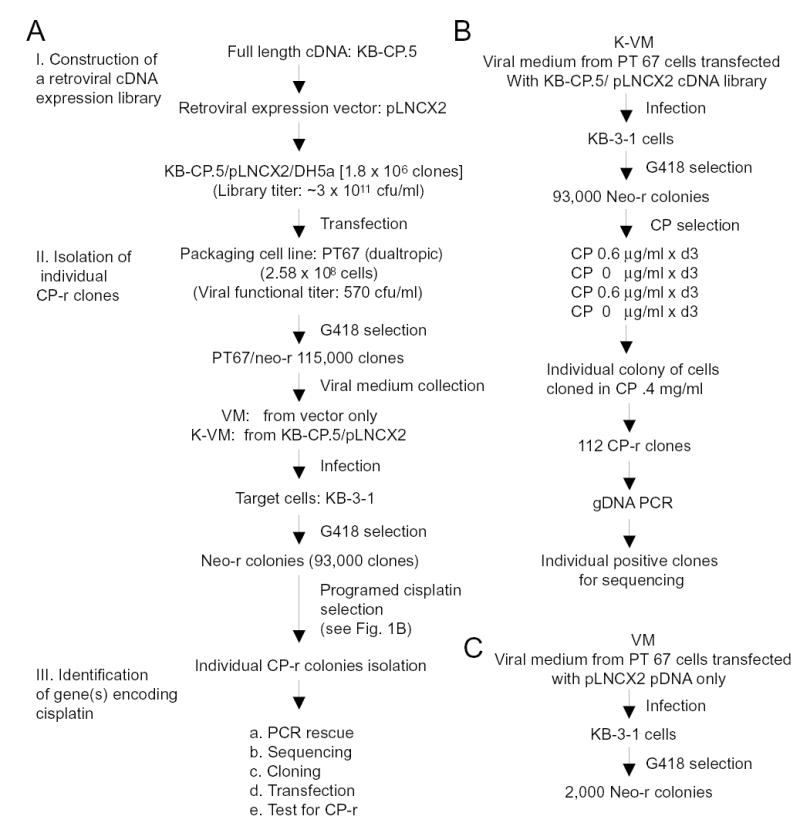
Flow charts. A, strategy to functionally clone CP-r genes using a retroviral cDNA library from an early stage cisplatin-resistant cell line, KB-CP.5 (human epidermoid carcinoma cells KB-3-1 cells selected with cisplatin 0.5 μg/ml). The flow chart shows the cloning system and is detailed in the text. B, flow diagram showing programmed/intermittent cisplatin selection for CP-r clones developed in this work, detailed in the text. C, flow chart showing isolation of 2,000 Neomycin-resistant clones from KB-3-1 cells, which served as a control.
Isolation of genomic DNA, PCR amplification, and sequencing
Genomic DNA was extracted by Miller’s method (Miller et al., 1988) from each clone from a total of 112 clones that were generated by programmed cisplatin selection. PCR amplification was performed in a total volume of 20 μl per reaction using BD Advantage 2 PCR kits (BD Biosciences, Palo Alto, CA). The retroviral vector pLNCX2 sequencing oligonucleotides were used as PCR primers for identification of inserts: 5’-AGCTCGTTTAGTGAACCGTCAGATC-3’(forward), and 5’-ACCTACAGGTGGGGTCTTTCATTCCC-3’(reverse). These two primers were also used for sequencing, which was performed at the Core facility of Center for Cancer Research, National Cancer Institute.
Preparation of RNA and RT-PCR
For determination of expression levels of genes of interest, RNAs were isolated from cells using an RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer. RT-PCR was performed using a GeneAmp kit (Applied Biosystems, Branchburg, NJ) as described by the manufacturer. Specific primers for tested genes are listed below: ribosomal protein RPL36, 5’-AAA TCC ATT GCC CGT GTT C-3’ (forward), and 5’-TCT TGG TCT TCA GGT TCT CC-3’(reverse); heat shock protein HSP10, 5’-AAGTTTCTTCCACTCTTTGACC-3’(forward) and 5’-TGAATCTCTCCACC-CTTTCC- 3’(reverse); γ-catenin, GAG AGT GTG CTG AAG ATT CTG (forward), and TGA TT CGT CCT TGT CAC C (reverse), which were used for verification of the quality and the size of genes in the library by PCR.
Gene transfection and assays of cell resistance levels to cisplatin, carboplatin and sodium arsenite
Full-length cDNA for the genes encoding RPL36 and HSP10 were purchased from ATCC (Manassas, VA). Both genes were inserted into a mammalian expression vector, pcDNA3.1 (Invitrogen) as described by the manufacturer. Gene transfection was done with Lipofectin (Invitrogen) following instruction of the manufacturer. Stable transfected clones were isolated after selection with G418. For testing cell sensitivities to cisplatin, carboplatin and sodium arsenite, cells were counted after 3 days using a Coulter Counter or Cell Counting Kit (CCK8) as described in the legend of Fig. 3. A clonogenic assay was used to further confirm ability to confer CP-r for the positive genes, such as RPL36 and HSP10. Cells were seeded into 60 mm dishes (Corning, NY). Cisplatin at a desired concentration was introduced into each dish prior to cell seeding. The medium was changed every three days with medium containing cisplatin at the desired concentration. Colonies were stained with methylene blue at the end of 12 days’ incubation, and then counted. Control cells were transfected with insert-free vector only. The values are means of triplicate determinations.
Fig. 3.
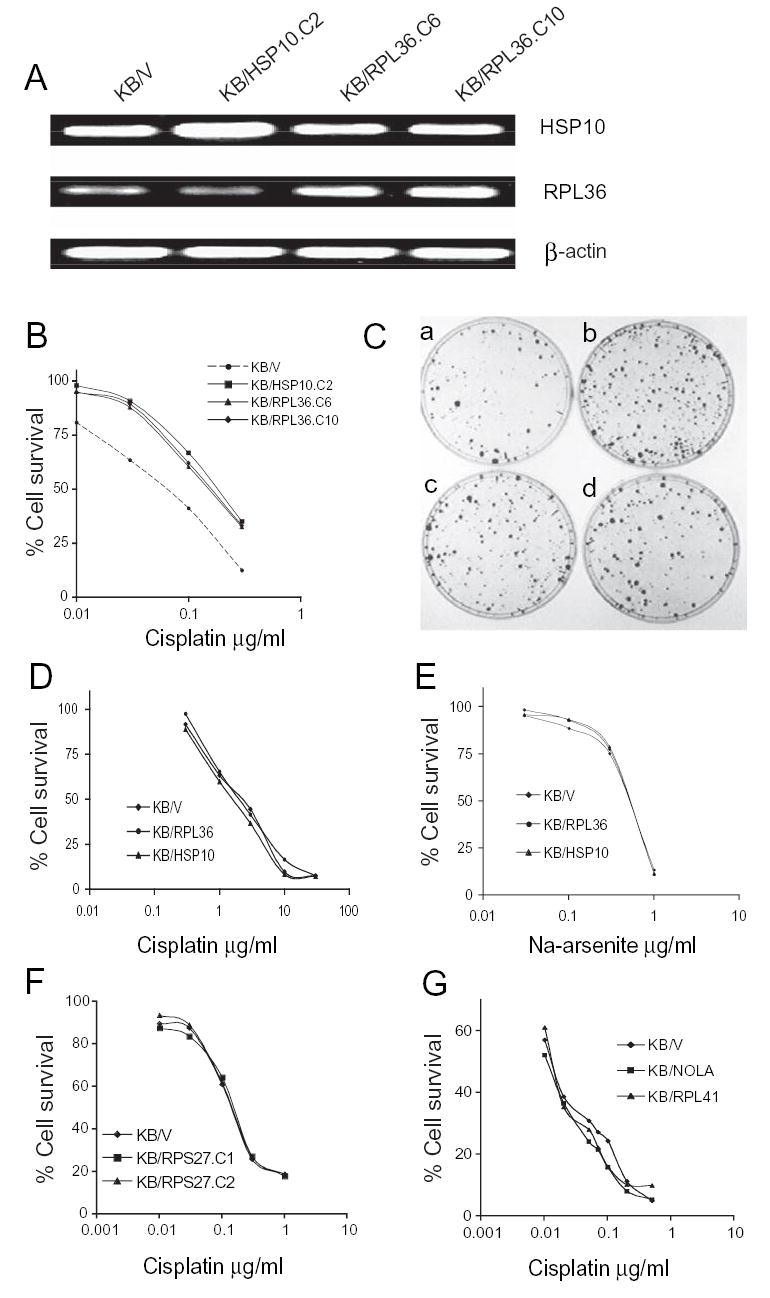
RT-PCR analysis. The specific primers for RPL36 and HSP10 are described in Materials and Methods. A, expression levels in transfectants: KB/V, KB-3-1 cells transfected with vector pcDNA3.1 only; KB/HSP10.C2, KB-3-1 cells were transfected with HSP10, clone 2; KB/RPL36.C6 and KB/RPL36.C10 cells were transfected with the RPL36 gene, two individual clones. ß-actin served as control. B and C, colony forming assay (CFA). B, killing curves measured at 12 days post cisplatin selection, methylene blue stained. KB/V, KB-3-1 cells transfected with vector pcDNA3; KB/HSP10.C2, KB cells transfected with HSP10/pcDNA3, clone 2; KB/RPL36.C6 and KB/RPL36.C10, KB cells transfected with RPL36/pcDNA3, 2 individual clones. C, the appearance of colonies in dishes of (a) KB/V; (b) KB/HSP10.C2, (c) KB/RPL36.C6; (d) KB/RPL36.C10 after exposure to cisplatin 0.3 μg/ml for 12 days as described in B. Fig. 3D, E, and G, cells were seeded at 5000 cells/well in a 96 well plate, and treated with the desired drug for 3 days, then cell numbers were determined by Cell Counting Kit-8 (CCK8, Dojindo Labs, Gaithersburg, MD) as described by the manufacturer. The concentrations of the drugs are indicated in the figures. Fig. 3F, cells were seeded at 5 x 104 cells/well in a 24 well/plate, and treated with cisplatin for 3 days, then counted using a Coulter Counter. The values are means of triplicate determinations.
RESULTS
Strategy for functional cloning
Fig. 1A shows a flow chart of construction of the library, isolation of CP-r clones and identification of genes related to CP-r. A retroviral cDNA expression library was established from an early stage (single step) CP-r cell line (KB-CP.5) into a retroviral expression vector, pLNX2. A total of 1.8 x 106 clones were pooled from KB-CP.5 cDNA/pLNCX2/DH5j. The ti ter of the unamplified library was ~ 3 × 1011 cfu/ml. The quality and insert size of the library is shown in Fig. 2A using the primers for pLNCX2 as described in Materials and Methods, indicating a distribution range of the inserts from 0.5 ~ 6 kb, and enriched between 0.7 to 1.8 kb. To determine if there were larger intact genes in this library, γ-catenin, a known cDNA of 2.3 kb, was generated by PCR amplification, and showed a full-length cDNA in the library (Fig. 2B).
Fig. 2.
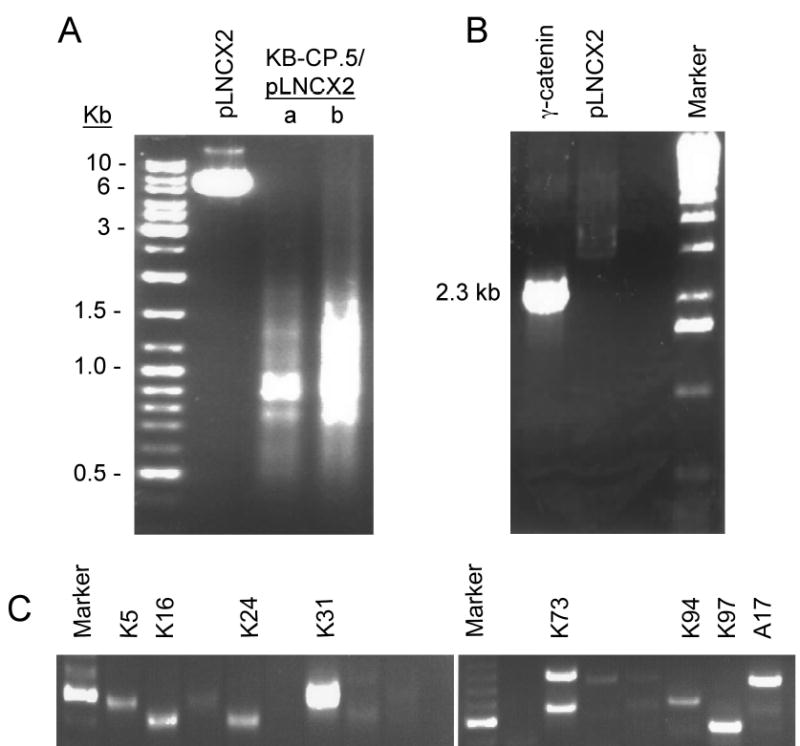
Images of PCR products. A, the KB-CP.5/pLNCX2 library was primed by specific oligonucleotides of the retroviral vector pLNCX2 as described in Materials and Methods, and then amplified by Advantage 2 PCR (Clontech). Two different loading amounts of KB-CP.5/pLNCX2 were applied as marked (a) 1/3 of loading and (b) full loading. B, γ-catenin was amplified by the specific primers described in Materials and Methods, showing the 2.3 kb of the full-length gene. C, images of genomic DNA PCR, showing inserts of different sizes in individual clones.
The RetroPack PT67 cell line was used to package the library. This cell line expresses a dualtropic/polytropic envelope, and the virus produced can infect a broad range of mammalian cells. The viral titer was determined by functional assay (G418 resistance) and resulted in 570 cfu/ml. Transfection of the KB-CP.5 cDNA retroviral library into PT67 cells yielded 115,000 neo-r colonies after G418 selection (Fig. 1A). The viral medium (K-VM) was collected from these cells, and then infected into the target KB-3-1 cells. A pool containing 93,000 neo-r clones was obtained after G418 selection (Fig. 1A). A control pool with 2000 neo-r clones was collected (Fig. 1C), using the vector only, without inserts.
An intermittent cisplatin selection was developed in this work as shown in Fig. 1B. Briefly, cells were intermittently selected with cisplatin at 0.6 μg/ml for 3 days, and then cultured in medium without the drug for another 3 days for recovery. A 2nd round of 3 days of cisplatin selection followed by a recovery period of three days was repeated. Cisplatin-resistant colonies appeared after a period of 12 days, and much larger colonies were seen in the group of KB-3-1 cells infected with the viral medium from the KB-CP.5/ pLNCX2 cDNA retroviral library (K-VM), while the control cells, which were infected with the viral medium from the vector without inserts, showed much smaller colonies (VM). 112 individual colonies that survived in the K-VM group were cloned and propagated in 0.4 μg/ml of cisplatin for genomic DNA preparation and further PCR determination. We tried to select cells continuously with cisplatin at different concentrations, but this approach was unsuccessful due to either cells being killed at high concentrations of cisplatin or spontaneously mutated at low concentrations, generating a high background of low-level cisplatin-resistant clones similar to the control. Therefore, a recovery period during the first two rounds of cisplatin selection seems to be important for functional gene cloning, particularly under the conditions described above.
Identification of genes in association with cisplatin resistance
All 112 cisplatin-resistant clones were analyzed by genomic PCR using the primers for the retroviral vector pLNCX2 as described in Materials and Methods, i.e., only the inserts containing the vector sequences at both ends could be detected in the clones. Fig. 2C shows fragment(s) of different sizes from 0.5 ~ 1 kb in these clones. After sequencing, eleven inserts were identified after BLAST match, and listed in Table 1. Interestingly, metallothionein 2A was one among the 11 genes observed, and this gene had been reported to be associated with resistance to cisplatin and other heavy metals (Kelly et al., 1988; Yang et al., 1994), demonstrating that the strategy applied in this work by functional cloning and using a retroviral cDNA library has the power to identify genes related to cisplatin resistance. It should be noted that there were several ribosomal protein genes in this pool, suggesting that overexpression of the ribosomal gene family might also play an important role in protecting cells from cisplatin-induced damage.
TABLE 1.
Positive clones identified by functional cloning and sequencing
| Clone | kb | Gene | mRNA | CDS |
|---|---|---|---|---|
| K5 | 0.55 | Metallothionein 2A | 451 bp | 76–261 |
| K16 | 0.5 | RPS27 (ribosomal protein S27) (Metallopanstimulin A) | 344 | 36–290 |
| K24 | 0.55 | ATP synthase, H+ transporting, Mitochondrial F0 complex, subunit d, FITO | 631 | 58–543 |
| K31 | 0.7 | RPL41 (ribosomal protein L41) | 478 | 84–161 |
| K73A | 1.0 | Nucleolar Protein family A, member 2 (H/ACA small nucleolar RNPs), | 879 | 87–548 |
| K73B | 0.7 | CGI-121, a novel PRPK | 660 | 64–591 |
| C104 | 0.65 | RPL36 (ribosomal protein L36) | 578 | 153–470 |
| A17 | 1.0 | Hypothetical protein FLJ21174 | 1062 | 200–847 |
| A22 | 0.9 | RPL13A (ribosomal protein L13A) | 1142 | 23–634 |
| A24 | 0.8 | HSP10 (heat shock protein 10) | 556 | 115–423 |
| A39 | 0.8 | Stratifin, SFN (14-3-3sq | 1309 | 49–795 |
| 0.7 | RPL36aL (Ribosomal protein L36a-like)* | 542 | 96–416 |
4 clones from B8/10-1A to D
We chose a ribosomal protein, L36 (RPL36), and a chaperone gene, HSP10, in our pool to evaluate if these two genes have roles in CP-r. Both RPL36 and HSP10 genes were inserted into a mammalian expression vector, pcDNA3.1, respectively, and then transfected into cisplatin-sensitive (CP-s) KB-3-1 cells separately. Individual clones were isolated from the transfectants after G418 selection. Fig. 3A shows that the RPL36 gene (middle panel) was overexpressed in two clones of RPL36- transfected cells, KB/RPL36.C6 and KB/RPL36.C10. Both the control cells (KB/V) which were transfected with vector only, and another transfected clone KB/HSP10.C2 showed much lower levels of the gene. In the upper panel, HSP10 was well-expressed in KB/HSP10.C2 cells, which were transfected with the HSP10 gene (about 2-fold higher than the control KB/V), and 2 other clones isolated from the RPL36 transfection. The lower panel shows similar expression levels of β-actin in these four clones, serving as loading controls.
The sensitivities of these clones to cisplatin were then determined by a clonogenic assay. The results are shown in Fig. 3B and C. Increased resistance to cisplatin (about 2.5 ~ 3-fold) was seen in the transfectants of RPL36, clone C6 and C10, and HSP10, clone C2, compared to the control vector-transfected only (Fig. 3C). In Fig. 3C, the amount and size of colonies in the clones expressing RPL36, clone C6 and C10 (c, d) or HSP10, clone C2 (b) were more numerous and larger than with the control vector only (a) after exposure to cisplatin 0.3 μg/ml for 12 days. These results demonstrate that, where overexpressed, the ribosomal protein RPL36 and the heat shock protein HSP10 may play roles in cisplatin resistance.
DISCUSSION
Recently, a variety of approaches have been applied in efforts to define genes that are related to cisplatin resistance. Our previous studies demonstrated that a pleiotropic defect occurred in the cisplatin-resistant cells, including defective influx of 14C-carboplatin, and other related heavy metals or unrelated compounds, mislocalization of membrane transporters, reduced expression of folate binding protein, and increased DNA hypermethylation (Liang et al., 2003; Shen et al., 2004) indicating a multifactorial mechanism involved in CP-r.
In this work, two genes, a ribosomal protein, RPL36, and a heat shock protein, HSP10, were identified to confer cisplatin resistance by 2.5 to 3-fold using a retroviral cDNA library and functional cloning. Although the function of these two genes in CP-r is unknown, their effect may plausibly be related to increased protein synthesis and protein stabilization to protect cells from the toxic effect of cisplatin, directly or indirectly, in association with regulation of proteins in DNA damage/repair, anti-apoptotic processes, or detoxification of cisplatin. We repeated the selection using a somewhat different protocol by which cells were selected with 1 jg/ml cisplatin for 24 hours, instead of intermittent exposure for a longer period of time, and then allowed to recover with drug for a period of 8–10 days till cloning. By this method, we further isolated more RPL36-related genes, such as RPL36aL, found in 4 individual clones from a pool of 30 (Table 1, last entry). RPL36aL has 90% identity to the coding region of the RPL36 gene. This result provides independent evidence that the selection of the RPL36 gene family members is not a random event, but reflects their association with CP-r. None of the transfected clones, however, were cross-resistant to carboplatin or sodium arsenite, as shown in Fig. D and E, respectively. This does not necessarily indicate a different mechanism of resistance, since the original parent cell line, KB-CP.5, was more resistant to cisplatin (up to 50-fold), while the RPL36- and HSP10-transfectants showed only 2.5 ~ 3-fold resistance.
None of the cDNAs isolated so far from the cDNA library made from these cells confers this phenotype on the full range of resistance seen in the KB-CP.5 cells. We assume, therefore, that this phenotype either results from expression of more than one gene, or that we have transfected genes whose overexpression confers CP-r by a mechanism different from that seen in KB-CP.5.
Heat shock proteins, HSP10, HSP 27, HSP60, and HSP70, etc., have been linked to stress response in protein folding and unfolding in drug resistance (Mandic et al., 2002; Shan et al., 2003; Zhao et al., 2005). Ribosomal proteins have recently been reported to be associated with regulation of apoptosis, multidrug resistance (Wendell et al., 2004), oncogenesis and chemotherapy (Zhang et al., 2004). Transfection of the ribosomal protein RPL23 into gastric cancer cells was reported to induce multidrug resistance, including CP-r, and to protect cells against vinblastine-induced DNA fragmentation, but had no effect on intracellular drug accumulation (Shi et al., 2004). Our finding in this work provides evidence that overexpression of RPL36 and HSP10 confers CP-r in KB-3-1 cells, but does not confer resistance to Na arsenite. The pattern of genes selected by cisplatin may reflect stress responses as well as cisplatin-specific resistance genes. Further functional cloning by selection with other agents, including carboplatin, would help to determine which genes are involved in general stress responses and which are unique to cisplatin.
Other genes were detected in the clones (Table 1), but RPS27, RPL41, and nucleolar protein family A, member 2 (NOLA), were tested individually and did not confer CP-r as shown in Fig. 3F, and G, respectively. Either these genes are not sufficient to confer CP-r, but may be necessary in the context of other patterns of gene expression, or they were simply false positive results from the screen applied here where the background (vector alone) was not zero. The other genes listed in Table 1 could not be tested initially because full-length cDNAs were not readily available. Nevertheless, these results indicate that the functions of RPL36 and HSP10 in CP-r cells and their potential role in human cancers need to be further elucidated.
Acknowledgments
We would like to thank George Leiman for editorial assistance.
Footnotes
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
References
- Francia G, Man S, Teicher B, Grasso L, Kerbel RS. Gene expression analysis of tumor spheroids reveals a role for suppressed DNA mismatch repair in multicellular resistance to alkylating agents. Mol Cell Biol. 2004;24:6837–6849. doi: 10.1128/MCB.24.15.6837-6849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- Kelly SL, Basu A, Teicher BA, Hacker MP, Hamer DH, Lazo JS. Overexpression of metallothionein confers resistance to anticancer drugs. Science. 1988;241:1813–1815. doi: 10.1126/science.3175622. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Nishio H, Ayaki H, Yamamoto M, Sumino K. Upregulation of stress response mRNAs in COS-7 cells exposed to cadmium. Toxicology. 2002;174:109–117. doi: 10.1016/s0300-483x(02)00045-8. [DOI] [PubMed] [Google Scholar]
- Liang XL, Shen DW, Garfield S, Gottesman MM. Mislocalization of membrane proteins associated with multidrug resistance in CP-r cancer cell lines. Cancer Res. 2003;63:5909–5916. [PubMed] [Google Scholar]
- Mandic A, Hansson J, Linder S, Shoshan MC. Cisplatin induces endoplasmic reticulum stress and nucleus-independent apoptotic signaling. J Biol Chem. 2002;278:9100–9106. doi: 10.1074/jbc.M210284200. [DOI] [PubMed] [Google Scholar]
- Miller MA, Korn D, Wang TS. The evolutionary conservation of DNA polymerase alpha. Nucleic Acids Res. 1988;16:7961–7973. doi: 10.1093/nar/16.16.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedner H, Christen R, Lin X, Kondo A, Howell SB. Identification of genes that mediate sensitivity to cisplatin. Mol Pharmacol. 2001;60:1153–1160. [PubMed] [Google Scholar]
- Mourtada-Maarabouni M, Kirkham L, Farzaneh F, Williams GT. Regulation of apoptosis by fau revealed by functional expression cloning and antisense expression. Oncogene. 2004;23:9419–9426. doi: 10.1038/sj.onc.1208048. [DOI] [PubMed] [Google Scholar]
- Perez-Victoria FJ, Gamarro F, Ouellette M, Castanys S. Functional cloning of the miltefosine transporter. A novel P-type phospholipid translocase from Leishmania involved in drug resistance. J Biol Chem. 2003;278:49965–49971. doi: 10.1074/jbc.M308352200. [DOI] [PubMed] [Google Scholar]
- Roberts D, Schick J, Conway S, Biade S, Laub PB, Stevenson JP, Hamilton TC, O'Dwyer PJ, Johnson SW. Identification of genes associated with platinum drug sensitivity and resistance in human ovarian cancer cells. Br J Cancer. 2005;92:1149–1158. doi: 10.1038/sj.bjc.6602447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan YX, Liu TJ, Su HF, Samsamshariat A, Mestril R, Wang PH. Hsp10 and Hsp60 modulate Bcl-2 family and mitochondria apoptosis signaling induced by doxorubicin in cardiac muscle cells. J Mol Cell Cardiol. 2003;35:1135–1143. doi: 10.1016/s0022-2828(03)00229-3. [DOI] [PubMed] [Google Scholar]
- Shen DW, Akiyama S-I, Schoenlein P, Pastan I, Gottesman MM. Characterization of high-level cisplatin-resistant cell lines established from a human hepatoma cell line and human KB adenocarcinoma cells: cross-resistance and protein changes. Br J Cancer. 1995;71:676–683. doi: 10.1038/bjc.1995.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DW, Su A, Liang XJ, Pai-Panandiker A, Gottesman MM. Reduced expression of small GTPases and hypermethylation of the folate binding protein gene in cisplatin-resistant cells. Br J Cancer. 2004;91:270–276. doi: 10.1038/sj.bjc.6601956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhai H, Wang X, Han Z, Liu C, Lan M, Du J, Guo C, Zhang Y, Wu K, Fan D. Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp Cell Res. 2004;296:337–346. doi: 10.1016/j.yexcr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- Yang YY, Woo ES, Reese CE, Bahnson RR, Saijo N, Lazo JS. Human metallothionein isoform gene expression in cisplatin-sensitive and resistant cells. Mol Pharmacol. 1994;45:453–460. [PubMed] [Google Scholar]
- Yasui K, Mihara S, Zhao C, Okamoto H, Saito-Ohara F, Tomida A, Funato T, Yokomizo A, Naito S, Imoto I, Tsuruo T, Inazawa J. Alteration in copy numbers of genes as a mechanism for acquired drug resistance. Cancer Res. 2004;64:1403–1410. doi: 10.1158/0008-5472.can-3263-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Berger SA. Increased calcium influx and ribosomal content correlate with resistance to endoplasmic reticulum stress-induced cell death in mutant leukemia cell lines. J Biol Chem. 2004;279:6507–6516. doi: 10.1074/jbc.M306117200. [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]


