Abstract
Two plasmids from which the sequences coding for the 36- and 95-kDa proteins of Carnation Italian ringspot virus (CIRV) could be transcribed in vivo in the yeast Saccharomyces cerevisiae under the control of the ADH1 promoter and terminator were constructed. The two proteins, which constitute the viral replicase, were correctly translated and integrated into membranes of the yeast cells. An additional plasmid was introduced in yeasts expressing the CIRV replicase, from which a defective interfering (DI) RNA (DI-7 RNA) could be transcribed under the control of the GAL1 promoter and terminated by the Tobacco ringspot virus satellite ribozyme, which cleaved 19 nucleotides downstream of the 3′ end of DI RNA. The DI-7 RNA transcripts were amplified by the viral replicase as demonstrated by the restoration of the authentic 3′ end, the requirement of a specific cis-acting signal at this terminus, the preferential accumulation of molecules with the authentic 5′ terminus (AGAAA), the synthesis of head-to-tail dimers, the presence of negative strands, and the incorporation of 5-bromo-UTP. Additionally, transformation with a dimeric construct of DI-7 RNA led to the synthesis of monomers, mimicking the activity of the viral replicase in plant cells.
Carnation Italian ringspot virus (CIRV) (genus Tombusvirus, family Tombusviridae) has a monopartite, linear, single-stranded RNA genome of 4,760 nucleotides (nt) in length (37). CIRV RNA contains five functional open reading frames (ORFs), of which ORF1 encodes a 36-kDa protein (p36) and ORF2 encodes a 95-kDa protein (p95) translated by readthrough of the amber stop codon. Whereas p95 contains the conserved motifs of RNA-dependent RNA polymerases (27), p36 contains the information to target the replicative virus machinery to the outer membranes of mitochondria, which are transformed into characteristic structures termed multivesicular bodies (8, 40, 42). The targeting function of p36 is maintained when the protein is transiently expressed in plant or yeast cells (41, 49). ORF3 codes for the 41-kDa coat protein, and the two small nested ORFs (ORF4 and -5) encode two proteins of 22 kDa (p22) and 19 kDa (p19), respectively, which have a role in virus movement and symptom expression in infected plants. Both p36 and p95 are translated directly from the viral genome, whereas translation of the coat protein, p22, and p19 requires the synthesis of two subgenomic RNAs (37).
The small size and the compact structure of the genomes of tombusviruses make them ideal for the study of the replication mechanisms of positive-strand RNA viruses. The roles of the different proteins encoded by the genomes of members of the genus Tombusvirus are known for several members of the genus. In particular, it was determined that the proteins encoded by the 5′-proximal ORFs (ORF1 and -2) are the only viral products required for replication in plant protoplasts (26, 39), and they complement each other when expressed from different RNAs (30, 32).
Since replicase proteins encoded by viral genomes may not be sufficient for replication, so that host proteins are likely to be involved in the construction of the replication machinery (48), in order to determine and analyze these host factors a yeast system which allowed the replication of a plant virus (Brome mosaic virus [BMV]) (24) and an animal virus (Flock house virus) (33) was developed. This system is based on Saccharomyces cerevisiae, whose molecular biology and genetics are well known (7).
To exploit the yeast system for studying the replication of tombusviruses, we thought it advantageous to use a subviral molecule as a replication template, rather than the genome itself. In fact, in tombusvirus infections, short forms of the viral genome, which are referred to as defective interfering (DI) RNAs, are produced (42). These molecules are smaller than genomic RNA (less than 800 nt in size), do not contain coding sequences, and are replicated more efficiently than the viral genome and only in the presence of the parent virus, which provides the trans-acting factors, encoded by ORF1 and ORF2, necessary for replication (26, 39). These properties make DI RNAs ideal replicons for studying tombusvirus replication in yeast. It is known that CIRV-infected tissues contain DI RNA molecules of two sizes (656 and 474 nt) (37), the larger of which, named DI-7, is composed of three sequence blocks (I, II, and III) derived entirely from genomic RNA. Block I is made up of the first 150 nt of the viral genome, block II comprises 136 nt derived from ORF2, and block III comprises 370 nt derived from the terminal portion of ORF4 and the complete 3′ noncoding region. This DI RNA was used to test the suitability of S. cerevisiae for the study of the tombusvirus replication.
MATERIALS AND METHODS
Yeast strain, transformation, and culture conditions.
S. cerevisiae strain YPH499 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) (46) was transformed with plasmid DNAs by using the lithium acetate-polyethylene glycol method (23). Following transformation, cells were grown and maintained on appropriate synthetic selective medium (SM) plates containing 2% dextrose (2). Relevant amino acids were omitted to maintain the selection for any plasmid used. For induction of CIRV DI-7, cells were first successively subcultured into SM containing 2% dextrose and 3% glycerol-0.1% dextrose and then induced in SM containing 3% glycerol-2% galactose and repressed in SM containing 3% glycerol-0.1% dextrose. Yeast cultures were grown at 26°C if not otherwise indicated.
Plasmid constructions.
All DNA manipulations were performed by standard procedures (2, 43). Site-directed mutagenesis on plasmids was performed with the QuikChange site-directed mutagenesis kit (Stratagene). All mutations were confirmed by DNA sequencing.
(i) Intermediate plasmids. (a) pA.
The 1.15-kb BamHI fragment containing the HIS3 selectable marker from plasmid YDp-H (5) (kindly provided by L. Palmieri) and the 2.2-kb EcoRI fragment containing the 2μm replication origin from plasmid Yep24 (6) (kindly provided by D. Botstein) were cloned into the BamHI and EcoRI sites, respectively, of pUC19. The SmaI and SacI restriction sites in the multicloning site of pUC19 were silenced by cutting the plasmid with both enzymes followed by treatment with T4 DNA polymerase and religation.
(b) pB3MI3S.
Plasmid pB3MI3 is a CEN4 yeast plasmid containing the TRP1 selectable marker and a derivative of BMV RNA3 terminated by the active sequence of the Tobacco ringspot virus satellite (TRSVs) ribozyme, flanked by the GAL1 promoter and ADH1 terminator signals (22). A SmaI restriction site was introduced into pB3MI3 by mutating the A residue of BMV RNA3 3′-terminal sequence (CCA) into a C residue, leaving unaltered the GAL1-dependent expression and the self-cleaving action of the TRSVs ribozyme. The mutation was performed with 5′-GAGACCCGGGAATTCGATACCCTGTCACCG-3′ and 5′-CGGTGACAGGGTATCGAATTCCCGGGTCTC-3′ oligonucleotides (the mutated bases are underlined).
(ii) CIRV p36 and p95 expression plasmids. (a) pA36K.
CIRV p36 was expressed in yeast from pA36K, a 2μm plasmid containing HIS3 as a selectable marker. To construct pA36K, first a 1-kb fragment encompassing the CIRV p36-coding sequence was generated by PCR from a CIRV cDNA full-length clone by using primers 5′-ATTAAGCTTCCCGGGATGGAGGGTTTGAAGGC-3′ and 5′-GTAAGCTTGAGCTCCCGGGCTATTTGACACCG-3′. The underlined sequences indicate the HindIII and SacI/HindIII restriction sites inserted upstream and downstream of the p36-coding sequence, respectively. The HindIII-digested fragment was then cloned into the unique HindIII site of pAHH5 (1) (kindly provided by G. Ammerer) between the ADH1 promoter and terminator. The 1.75-kb SphI fragment containing the expression cassette ADH1 promoter-ORF1-ADH1 terminator was subsequently excised and cloned into the unique SphI site of plasmid pA.
(b) YE95K.
The amber stop codon at the end of the p36-coding region cDNA was mutated into a TAT (tyrosine) codon with primers 5′-CGCACTAGGCCTCCATATTTGACACCGAGG-3′ and 5′-CCTCGGTGTCAAATATGGAGGCCTAGTGCG-3′ (mutated bases are underlined) by site-directed mutagenesis performed on a CIRV cDNA full-length clone. Subsequently, a 1.9-kb PCR fragment corresponding to the 3′-terminal CIRV p95-coding region was generated by using the primer 5′-GCTCGGGAGCTCAAGGGTAAGGATGG-3′ in conjunction with 5′-AATTGAGCTCAAGCTACGGCGG-3′ (the underlined sequence indicates a SacI restriction site inserted downstream of the p95-coding sequence). The SacI-digested fragment was then subcloned into SacI-restricted pA36K, thus replacing the 3′-end sequence of the p36-coding region. The 3.3-kb expression cassette ADH1 promoter-ORF2-ADH1 terminator was then cloned into the SphI site of YEplac181, a 2μm shuttle vector containing LEU2 as selectable marker (19).
(iii) Plasmids expressing CIRV DI-7. (a) pBDI-7.
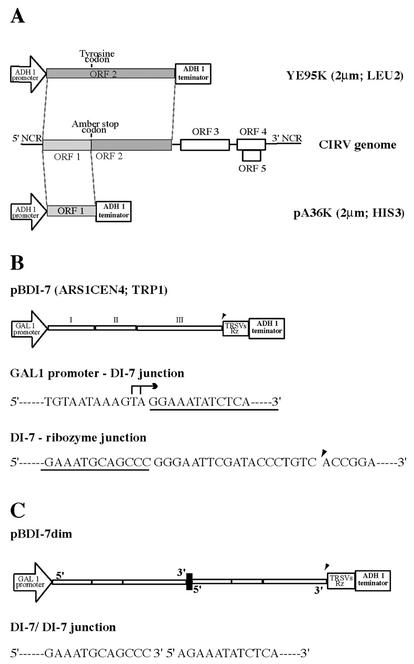
The CIRV DI-7 RNA was expressed in yeast from the plasmid pBDI-7, a CEN4 yeast plasmid containing TRP1 as a selectable marker. The plasmid was constructed by cloning the 0.65-kb BamHI (filled in)-SmaI fragment of pUC18 DI-7 (37), containing the DI-7 cDNA sequence, into the pB3MI3S vector restricted with SnaBI and SmaI (thus replacing the BMV RNA derivative with CIRV DI-7). Subsequently, the pUC18-derived extra bases situated between the GAL1 promoter and the 5′ end of CIRV DI-7 were deleted by site-directed mutagenesis with 5′-CAAATGTAATAAAAGTAGGAAATATCTCAGGATTTGACC-3′ and 5′-CGGTCAAATCCTGAGATATTTCCTACTTTTATTACATTTG-3′ oligonucleotides, thus creating the GAL1-CIRV DI-7 junction shown in Fig. 1B.
FIG. 1.
Schematic representation of CIRV genomic and DI RNAs and plasmids used in this work. (A) The ORF1 sequence of the CIRV genome (middle drawing) was cloned in plasmid pA36K, and the ORF2 sequence, containing a tyrosine codon in place of the ORF1 amber codon was cloned in plasmid YE95K. The plasmids contain the ADH1 promoter and terminator, the 2μm origin of replication, and the selectable markers HIS3 and LEU2, respectively. NCR, noncoding regions. (B) Diagrammatic representation of plasmid pBDI-7, containing the complete DI-7 sequence cloned under the control of GAL1 promoter and ADH1 terminator. I, II, and III indicate the three discontinuous sequence blocks derived from the viral genome. Plasmid pBDI-7 has the ARS1CEN4 origin of replication and TRP1 as a selectable marker. The 5′ terminus of DI RNA (underlined) was fused to the GAL1 promoter sequence, whose major transcription initiation sites are indicated by an arrow. The 3′ terminus (underlined) was fused to the TRSVs ribozyme, which leaves 19 nonviral nucleotides after self-cleavage at the position indicated by an arrowhead. (C) Dimeric head-to-tail DI-7 RNA was cloned in plasmid pBDI-7dim, which has the features of pBDI-7. The junction sequence is indicated.
(b) pBDI-7ΔG.
The G residue at position −4 of the 3′ terminus of the DI-7 cDNA in pBDI-7 was mutated into an A residue by using 5′-GGAACATTGCAGAAATGCAACCCGGGAATTCGATACCCTGTCACC-3′ and 5′-GGTGACAGGGTATCGAATTCCCGGGTTGCATTTCTGCAATGTTCC-3′ oligonucleotides.
(c) pBDI-7dim.
A PCR fragment of 0.65 kb corresponding to the DI-7 sequence was amplified on the DI-7 clone in pUC18 (37) by using oligonucleotides 5′-AGAAATATCTCAGGATTTGACCG-3′, homologous to the first 23 nt of the DI-7 sequence, and 5′-GGGCTGCATTTCTGCAAT-3′, complementary to the last 18 nt of the DI-7 sequence. The fragment was cloned into the unique SmaI site of pBDI-7, thus producing a clone containing a dimeric DI-7 cDNA with the natural 3′-5′ junction (Fig. 1C).
Protein and RNA analysis.
Mid-logarithmic-phase (optical density at 600 nm, 0.6 to 0.8) yeast cultures harboring selected plasmids as indicated, corresponding to approximately 2 units of optical density at 600 nm, were harvested by centrifugation and frozen on dry ice. Protein extraction and Western blot analysis were performed as previously described (38). Total RNA was prepared by the hot-phenol method (28) and resuspended in RNase-free water, and 2 μg of RNA was analyzed by Northern blotting (43). LiCl fractionation and two-cycle RNase protection assay were performed as described previously (13, 31). RNAs were detected by using 32P-labeled probes specific for positive-strand or negative-strand CIRV DI-7 RNA. Riboprobes were obtained from clones containing a 0.3-kb XbaI (position 360)-SmaI (in the polylinker) fragment or a 0.26-kb AccI (position 98)-XbaI (position 360) fragment from the full-length DI-7 clone cloned downstream of the T7 promoter in the transcription vector pTL7SN in the appropriate orientation (9). Both riboprobes were obtained by linearization with EcoRI and transcription of T7 RNA polymerase. RNA containing the TRSVs ribozyme sequence was detected by using the oligonucleotide 5′-GACAGGGTATCGAATTCCC-3′ labeled at the 5′ end with 32P (43). Hybridization signals were detected by autoradiography.
The DI-7 5′ and 3′ ends were determined as described by Hirzman et al. (21) and Shirako and Wilson (45), respectively.
Incorporation of BrUTP and detection of nascent RNA.
Intact yeast cells were permeabilized for 15 min on ice with 0.5% sodium N-lauroyl-sarcosine as described by Elion and Warner (16) and then incubated at 26°C for 15 min with 10 mM 5-bromo-UTP (BrUTP) in 10 mM Tris buffer (pH 7.4) containing 100 mM NaCl, 5 mM MgCl2, and 10 mM DTT. The cells were then processed for immunofluorescence as described below.
Immunofluorescence.
Fixation of cells with formaldehyde and immunofluorescence staining were performed as described previously (34, 36). Rabbit polyclonal antisera against the outer (TOM40) (3) and inner (YHM2) (10) mitochondrial membrane proteins were kindly provided by L. Palmieri. Mouse monoclonal antibody to BrUTP was from Sigma, and goat anti-rabbit Rhodamine Red-conjugated and goat anti-mouse Alexa Fluor 488-conjugated secondary antibodies were from Molecular Probes. Immunofluorescence images were obtained with a Nikon Eclipse E400 epifluorescence microscope.
RESULTS AND DISCUSSION
Expression of CIRV replicase proteins and DI-7 RNA transcription in yeast.
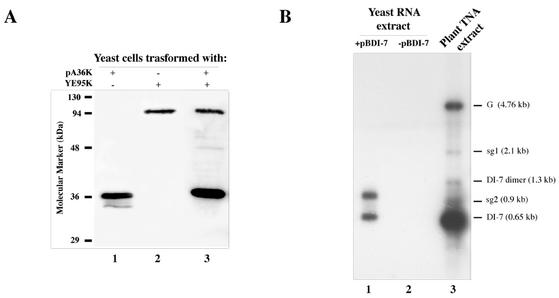
Yeast strain YPH499 cells transformed with the wild-type CIRV p95 gene (ORF2) expressed only p36 (ORF1) but not the fusion p95 (not shown), thus confirming the inability of this strain to read through the amber stop codon. Two plasmids were therefore prepared to express separately and individually p36 and p95; one (pA36K) contained the p36-coding sequence up to the amber stop codon, and the other (YE95K) contained the coding sequence of p95 in which the amber stop codon of ORF1 was replaced by a tyrosine codon. Both plasmids had the ADH1 promoter and terminator, the 2μm origin of replication, and HIS3 (pA36K) or LEU2 (YE95K) as a selectable marker (Fig. 1A). Yeast cells were transformed with pA36K and YE95K either separately, together, or with the same plasmids lacking the viral sequences; these cells are hereafter referred to as 36K+95K−, 36K−95K+, 36K+95K+, and 36K−95K−. Cells were grown in SM containing 2% dextrose for 16 h at 26°C, spheroplasted, lysed, and centrifuged at 30,000 × g to generate a supernatant and a pellet fraction. As shown in Fig. 2A, p36 and p95 were readily detected by Western blot analysis in the pellet fractions, regardless of whether they were expressed singly from plasmid pA36K (lane 1) or YE95K (lane 2) or in combination (lane 3). Comparable results were obtained in Western blot analysis of cells grown at 30°C (not shown). The occurrence of p36 and p95 in the pellet fractions and their absence in the supernatant (not shown) are in agreement with the notion that both proteins are integrated in the outer membrane of mitochondria (49).
FIG. 2.
Analysis of the expression of p36, p95, and DI-7 RNA in yeast. (A) Western blot analysis of protein extracts from yeasts transformed with plasmid pA36K (lane 1), plasmid YE95K (lane 2), or both (lane 3). The blot was probed with anti-36K antiserum, which recognizes both p36 and p95. (B) Northern blot analysis of RNA extracts from yeasts transformed with plasmid pBDI-7 (lane 1) or untransformed (lane 2) and from Nicotiana benthamiana-infected tissues (lane 3), showing the positions of genomic and subgenomic RNAs (G, sg1, and sg2, respectively) and monomeric and dimeric DI-7 RNA. The blot was probed with a 32P-labeled riboprobe recognizing the 3′ noncoding region of the CIRV genome.
CIRV DI-7 RNA was cloned in the low-copy ARS1CEN4-based plasmid pBDI-7 (Fig. 1B). The transcription cassette of this plasmid was composed of the GAL1 promoter, the DI-7 RNA sequence, the TRSVs ribozyme, and the ADH1 terminator. The 5′ end of CIRV DI-7 RNA cDNA was linked directly to the GAL1 promoter to reduce the number of extra nucleotides at this terminus, whereas the 3′ end was linked to TRSVs ribozyme, which cleaves 19 nt downstream of the viral 3′ end.
Yeasts were transformed with pBDI-7 and grown in SM with 2% galactose at 26 or 30°C. Two DI RNA-related bands were detected in Northern blot analysis of RNA extracts from cells grown at 26°C by using a positive-strand-specific riboprobe for DI-7 RNA (Fig. 2B). The fastest-migrating band, comigrating with DI-7 RNA synthesized in plants, was interpreted as containing unit-length DI-7 cleaved by the ribozyme, while the slowest-migrating band was likely made of uncleaved molecules with an approximate size of 1.2 kb containing the vector terminator sequence and the poly(A) tail. Northern blots of RNA extracts from cells grown at 30°C were comparable to those from cells grown at 26°C (not shown).
GAL1 promoter repression and maintenance of mitochondria.
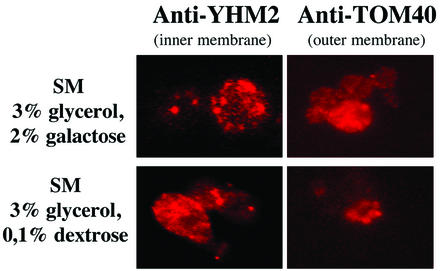
The replication of CIRV genomic and DI RNAs in plant cells takes place in vesicles formed by the outer membranes of mitochondria (14). In fact, both p36 and p95 contain and express targeting signals to mitochondria of plant and yeast cells (49). Attempts to set up a yeast system capable of replicating the CIRV genome or DI RNA had to be made with cells growing in medium containing little or no dextrose, since it is known that at dextrose concentrations of higher than 0.1%, mitochondria are reduced in number and have irregular shape and structure (47). On the other hand, transcription of pBDI-7, driven by the GAL1 promoter, must be repressed to avoid the possibility that the large mass of transcripts (see Fig. 2B) would obscure the potential presence of DI RNA molecules replicated by the constitutive viral replicase proteins. Since preliminary experiments indicated that as little as 0.1% dextrose was sufficient to repress transcription from the GAL1 promoter (not shown), 36K−95K−, 36K+95K−, 36K−95K+, and 36K+95K+ yeast transformants containing pBDI-7 were grown in parallel in SM containing 3% glycerol-2% galactose or 3% glycerol-0.1% dextrose. The morphology and distribution of mitochondria were then analyzed by immunofluorescence, using antibodies specific to the inner and outer membrane proteins (Fig. 3) or using the vital mitochondrion-specific stain MitoTracker (not shown). The general aspects of mitochondria and MitoTracker uptake were similar, regardless of the medium in which yeast cells were grown. In conclusion, since the medium containing 3% glycerol-0.1% dextrose was adequate for inhibiting induction by the GAL1 promoter and apparently did not alter the structure of mitochondria, it was chosen for the experiments described below.
FIG. 3.
Fluorescence light microscope visualization of mitochondria in yeast cells grown in SM containing 3% glycerol-2% galactose (upper panels) or 3% glycerol-0.1% dextrose (lower panels). Cells were immunolabeled by using antibodies to the inner (YHM2) or outer (TOM40) mitochondrial membrane proteins, which were detected with rhodamine-labeled secondary antibodies.
Accumulation of CIRV DI RNA in the presence of p36 and p95.
Yeast cells transformed with plasmid pBDI-7, expressing either p36 (36K+95K−), p95 (36K−95K+), both proteins (36K+95K+), or neither of them (36K−95K−), were precultured with two passages in SM containing 2% dextrose (first preculture) and SM containing 3% glycerol-0.1% dextrose (second preculture). The cells were then transferred in succession to GAL1-inductive medium (SM containing 3% glycerol-2% galactose) and then to GAL1-repressive medium (SM containing 3% glycerol-0.1% dextrose). These precultures proved to be necessary, since a low growth rate was consistently observed when yeast cells were moved directly from 2% dextrose- to 2% galactose-containing medium (not shown). Cells were grown at 26 and 30°C.
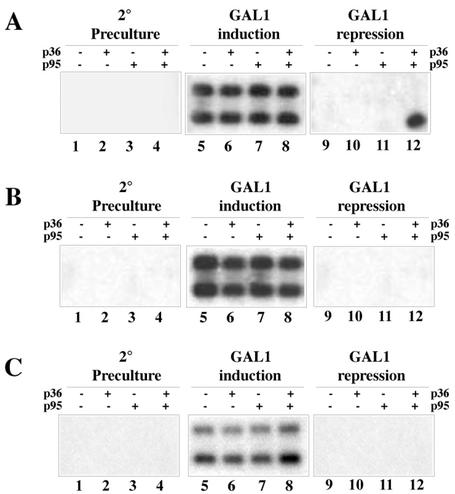
Total RNA was extracted and analyzed by Northern blotting. As expected, no GAL1-promoted DNA transcription was observed when yeasts were cultured in the absence of galactose (Fig. 4A and B, lanes 1 to 4), whereas the two bands shown in Fig. 2B were readily detected upon induction, regardless of the presence of the viral proteins and growth temperature conditions (Fig. 4A and B, lanes 5 to 8). By contrast, after repression of the GAL1 promoter at 26°C, only one band corresponding to DI-7 RNA was detected in extracts from 36K+95K+ cells (Fig. 4A, lane 12) but not in those from cells lacking both of these proteins (lane 9) or either one (lanes 10 and 11). No bands were detected in the extracts from cells grown at 30°C (Fig. 4B, lanes 9 to 12). The amount of DI-7 RNA in the 36K+95K+ sample was estimated to be 25 to 50 pg per μg of total RNA by comparison with blots in which known amounts of in vitro transcripts were hybridized with the same probe (not shown).
FIG. 4.
Northern blot analysis of extracts from yeasts transformed with plasmid pBDI-7 and expressing or not expressing p36 or p95 grown in SM containing 3% glycerol-0.1% dextrose (second preculture and GAL1 repression) or containing 3% glycerol-2% galactose (GAL1 induction) at 26°C (A and C) or 30°C (B). The blots in panels A and B were probed with a DI-7 RNA-specific probe; the blots in panel C were probed with a probe specific to TRSVs ribozyme sequence.
As mentioned above, RNA molecules giving the lower band present in the induction phase and the unique band found in the repression phase comigrated with authentic DI-7 RNA from infected plant cells. In order to discriminate between the DNA-derived transcripts cleaved by TRSVs ribozyme and the progeny from possible CIRV-based replication events, the same RNA preparations used for Fig. 4A were blotted and probed with a 32P-labeled oligonucleotide complementary to the residual 19 extra ribozyme bases after self-cleavage at the 3′ end of DI RNA (Fig. 1B). As shown in Fig. 4C, the labeled oligonucleotide hybridized both DI RNA species extracted from induced cells but not the unique species present in extracts from repressed 36K+95K+ cells (Fig. 4A, lane 12), thus indicating the absence of ribozyme-derived nucleotides at the 3′ end of this RNA.
To confirm this, a direct analysis of this terminus was carried out as described previously (43). Of the eight clones sequenced, four contained the terminal sequence CAGCCC, two contained CAGCCCC, and two contained CA. With the exception of the last two clones, which are probably artifactual, the clones clearly matched the authentic end of wild-type DI RNA (37).
Monomeric and dimeric DI RNAs.
The above-described Northern blot analysis revealed only the occurrence of DI RNA molecules corresponding in size to DI-7 RNA (Fig. 2B and 4A). However, it is known that replication of tombusvirus DI RNAs in plants results in the synthesis of head-to-tail dimers, in addition to the unit-length monomers (12, 17, 37) (see also Fig. 2B, lane 3).
To investigate whether DI-7 RNA dimers were present below the limits of detection by Northern blot analysis of repressed 36K+95K+ cells, cDNA to RNA extracted from these cells was amplified by using two oligonucleotides, one complementary to positions 106 to 128 and one homologous to positions 260 to 280 in the DI RNA sequence. A PCR product of the expected size (515 bp) was cloned, sequenced, and shown to contain the junction sequence of a dimeric DI RNA. Three out of six clones had the junction sequence GCCCGGAAA, two had GCCCCGAAA, and one had GCCCAGGAAA, indicating that while the sequence of the 3′ end of monomeric DI-7 RNA (underlined) was that of DI RNA replicating in plant cells, the 5′-end sequence was in no case identical to the authentic 5′ end (AGAAA) but rather was similar to the sequence in the plasmid pBDI-7 (GGAAA). The rest of the sequence reproduced exactly the DI RNA sequence.
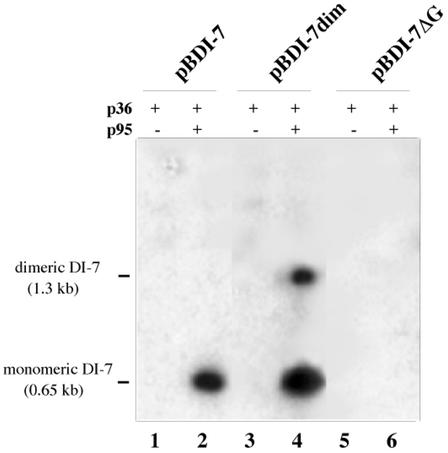
The occurrence of dimeric DI-7 RNA molecules synthesized from monomeric templates prompted us to investigate whether the reverse also occurred, i.e., whether monomeric DI RNA molecules were synthesized from a dimeric template. To do so, a cDNA to dimeric DI-7 RNA was synthesized and cloned to produce plasmid pBDI-7dim, which had essentially the same composition as pBDI-7 except that the DI sequence was doubled, with the dimer junction sequence CAGCCCAGAAA (the 3′ and 5′ termini of DI-7 RNAs are underlined and in boldface, respectively), and the 3′ end was fused to the TRSVs ribozyme (Fig. 1C). Yeasts 36K+95K+ and 36K+95K− containing plasmid pBDI-7dim were processed as before. Northern blot analysis showed accumulation of RNA molecules of unit-length size and double this size, with the latter being less represented (Fig. 5, lane 4).
FIG. 5.
Northern blot analysis of RNA extracts from 36K+95K+ (lanes 2, 4, and 6) and 36K+95K− (lanes 1, 3, and 5) cells transformed with pBDI-7 (lanes 1 and 2), pBDI-7dim (lanes 3 and 4), and pBI-7ΔG (lanes 5 and 6), showing synthesis of monomeric DI-7 RNA from the dimeric template (lane 4), which is indistinguishable from the progeny of the monomeric template (lane 2), and absence of progeny from the mutant pBI-7ΔG (lane 6).
Characterization of the 5′ end of DI RNA progeny.
The 5′ terminus of DI RNA in extracts from repressed 36K+95K+ cells cotransformed either with pBDI-7 or pBDI-7dim was investigated on clones obtained after tailing the 5′ ends of the first-strand cDNA products with dGTP. To this aim, terminal deoxynucleotidyl transferase was used, and the dG-tailed cDNA was amplified with an oligo(dC) primer and an oligonucleotide complementary to positions 100 to 118 in the DI-7 RNA sequence.
RT-PCR of pBDI-7 progeny gave two bands of ca. 120 and 800 nt, respectively. The smaller amplicon corresponded to the expected product from monomeric DI-7 RNA, whereas the 800-nt amplicon corresponded to a product amplified across the junction region of a newly synthesized dimer. The two bands were eluted separately, cloned, and sequenced. The 5′ termini of both DNA products were either GGAAA (three out of eight clones), AGGAAA (three clones), or, less frequently, GAAA (2 clones), and the 800-nt product contained the junction sequences GCCCGGAAA (two out of six clones), GCCCAGGAAA (two clones), or GCCCGAAA (two clones).
The 5′ ends of both large and small clones resulting from RT-PCR of pBDI-7dim progeny were AGAAA (six out of six clones), thus differing from the DNA-mediated transcripts (GGAAA). The 800-nt product contained only the junction sequence GCCCAGAAA (six out of six clones).
Requirement of a 3′ cis-acting signal for DI RNA accumulation.
The G residue at position −4 in the terminal sequence of tombusvirus genome is necessary for replication, as it cannot be deleted or replaced in infectious full-length clones or in DI RNA clones without loss of infectivity (11, 42). Even substitution with an A residue abolishes infectivity, notwithstanding the fact that this type of substitution stabilizes rather than weakens a stem in the secondary structure of the 3′ region necessary for replicase recognition (20). Accordingly, when G was mutated to A in plasmid pBDI-7, thus producing clone pBDI-7ΔG, which was transformed into 36K+95K+ and 36K+95K− yeasts, Northern blot analysis of RNA extracts from these cells revealed no accumulation of DI-7ΔG (Fig. 5, lanes 5 and 6).
Occurrence of negative-strand DI RNA.
RNA preparations from 36K+95K+ yeasts in the repression phase were analyzed by Northern blotting for the presence of negative-strand DI RNA with a negative-strand-specific probe. No bands were detected in any RNA preparations (not shown). Since a putative negative-strand RNA was likely to occur in the form of double-stranded RNA, a 4 M LiCl-insoluble fraction was prepared as described previously (13). Northern blot analysis failed to detect any band, regardless of whether a positive- or negative-strand-specific probe was used (not shown). The conclusion was that double-stranded and/or negative-strand RNA, if present, was below the detection limits of Northern blot analysis.
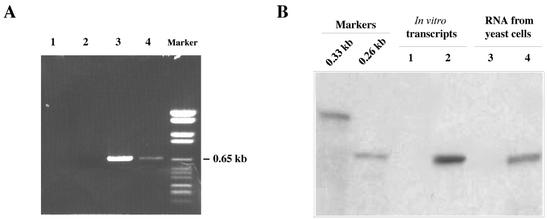
Alternative and more sensitive methods were therefore used to reveal the presence of negative-strand DI RNA. With one such method, RNA preparations were first treated with DNase, and a cDNA copy of DI RNA was then prepared by priming with an oligonucleotide complementary to the last 17 nt of the putative negative strand. Concurrently, cDNA was also prepared by priming with an oligonucleotide complementary to the last 17 nt of positive-strand RNA. The same two oligonucleotides were used for PCR amplification of both cDNAs. Figure 6A shows that DNA species of the expected size of the full-length DI RNA were obtained regardless of whether a cDNA copy of positive- or negative-strand RNA was used as the template. However, the PCR product derived from negative-strand cDNA (Fig. 6A, lane 4) was less abundant than that derived from positive-strand cDNA (Fig. 6A, lane 3). No amplification was obtained if uncopied RNA was used instead of cDNA (Fig. 6A, lane 1) or if primers were omitted (Fig. 6A, lane 2).
FIG. 6.
Detection of negative-strand DI-7 RNA in 36K+95K+ cells grown in GAL1 repression medium. (A) Ethidium bromide-stained gel of PCR products obtained by amplifying cDNA to positive-strand (lane 3) or negative-strand (lane 4) DI-7 RNA from 36K+95K+ cells. Lanes 1 and 2, control samples with no cDNA or no primers added, respectively. The marker lane contains DNA Molecular Weight Marker VI (Roche). (B) Two-cycle RNase protection assay of RNA extracts from 36K−95K− (lane 3) and 36K+95K+ (lane 4) cells. Lanes 1 and 2, results of two-cycle RNase protection assay of positive in vitro transcripts only (lane 1) or of a mixture of positive and negative in vitro transcripts (lane 2). Hybridization was with a 32P-labeled probe of 0.26 kb which was electrophoresed as a size marker.
A more sensitive assay for the presence of negative-strand DI RNA was based on two cycles of RNase protection (31), using RNA extracts (30 μg) from induced 36K−95K− and repressed 36K+95K+ yeasts cells. After initial hybridization and RNase treatment, double-stranded RNA was hybridized to a 32P-labeled riboprobe corresponding to nt 98 to 360 of the positive strand and treated again with RNases. Electrophoresis and autoradiography of the reaction products revealed the presence of a fragment of negative-strand RNA only in the extract from 36K+95K+ cells (Fig. 6B, lane 4). No signal was detected in the extract of 36K−95K− cells grown in the induction medium (Fig. 6B, lane 3), showing that no negative-strand RNA was present in these cells and that positive-strand DNA-derived transcripts did not protect the probe from the RNase digestion. Lanes 1 and 2 of Fig. 6B contained the reaction products of the assay using 30 μg of positive-strand DI RNA in vitro transcripts alone or in a mixture with 3 pg of negative-strand transcripts, respectively. Visual comparison of lanes 2 and 4 indicates that less than 0.1 pg of negative-strand RNA was present per μg of total RNA from 36K+95K+ cells.
In vivo labeling of nascent DI-7 RNA.
BrUTP incorporation has been used to detect sites of synthesis of plant virus RNA in both plant (15, 18, 35) and yeast (36) cells. This procedure was adopted to determine whether nascent DI-7 RNA in 36K+95K+ cells could be labeled with BrUTP after a short period of incorporation. To this effect, initial experiments were carried out with permeabilized spheroplasts, prepared as described by Schlenstedt et al. (44), which had been successfully used by Restrepo-Hartwig and Ahlquist (36) to localize the sites of BMV replication. Under our conditions, the method did not prove satisfactory, for the majority of the cells were too fragile to withstand manipulations.
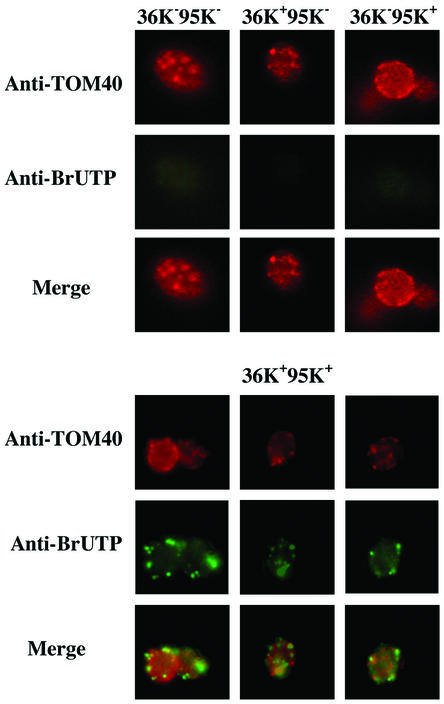
More suitable was the procedure described by Elion and Warner (16), in which yeast cells have their membranes permeabilized with sodium N-lauroyl-sarcosine while the cell wall is maintained. After the induction phase, yeast cells expressing p36 (36K+95K−), p95 (36K−95K+), neither p36 nor p95 (36K−95K−), or both p36 and p95 (36K+95K+) were grown for approximately 16 h in SM containing 3% glycerol-0.1% dextrose, collected by centrifugation, permeabilized, incubated with BrUTP for 10 min, fixed, and double labeled with a polyclonal antiserum to the mitochondrial marker TOM40 and a monoclonal antibody that recognizes BrUTP incorporated into RNA. As shown in Fig. 7, green fluorescence indicating the presence of BrUTP-containing RNA was found only in 36K+95K+ cells and not in cells lacking either one or both replicase proteins. The putative replication sites of DI-7 RNA appeared as punctate bodies, whose distribution, however, coincided only partially with the mitochondrial pattern.
FIG. 7.
Localization of bromo-RNA in 36K+95K+ cells after BrUTP incorporation. Cells were immunolabeled with mouse BrUTP-specific antibodies (middle panels, green) or rabbit antibodies against the mitochondrial outer membrane protein TOM40 (upper panels, red). Nascent RNA is present only in 36K+95K+ cells. The merged images (lower panels) show that the distribution of BrUTP-labeled RNA coincides only partially with the mitochondrial pattern.
In conclusion, the following lines of evidence suggest that the expression of CIRV p36 and p95 proteins in S. cerevisiae allowed the accumulation of a subviral RNA (DI-7 RNA), which contains the cis-acting signals recognized by the viral replicase.
(i) There was no synthesis of DI RNA in the absence of either one or both p36 and p95.
(ii) The requirement of a G residue at position −4 at the 3′ end of DI-7 RNA suggests that the accumulation of this RNA in 36K+95K+ yeast cells depends on the activity of the viral replicase. In fact, studies on the replication in planta of the related Cymbidium ringspot virus genomic and DI RNAs have shown that minus-strand synthesis initiates at this residue (11, 20). It was also suggested that C residues at the 3′ end are added posttranscriptionally, normally in the number of three but occasionally in excess (11, 42), which can explain why three or four Cs were found at the 3′ terminus of DI-7 RNA from 36K+95K+ cells.
(iii) The synthesis in yeast of dimeric forms of DI-7 RNA from a monomeric template and of monomeric forms from a dimeric template is a further indication of the possible activity of a viral RNA-dependent RNA polymerase. Dimers from monomers are synthesized when the viral replicase, before releasing a newly synthesized strand, begins a new round of replication on the same template, whereas monomers are synthesized from input dimeric molecules if the viral replicase recognizes signals to initiate and/or terminates transcription at the junction. On the other hand, that replication of DI-7 RNA did not begin at the 3′ end of the original template is confirmed by the observation that the DI-7 RNA progeny lack the 19 extra nucleotides of the TRSVs ribozyme. This ribozyme rather than that of Hepatitis delta virus (4, 22) was selected in order to have an indication of the activity of viral replicase as shown by the restoration of the DI RNA authentic 3′ end. The same type of reasoning led to the choice of having GG rather than the authentic AG at the 5′ end of the template. In fact, in vitro transcripts used for inoculation of plant cells normally begin with GG because they are obtained by using the T7 RNA polymerase, which has this preferential start. In these cells the 5′ end of the progeny is restored to AG (not shown). In yeast cells the progeny of pBDI-7 did not contain the authentic 5′ terminal sequence but contained either the input sequence GGAAA, or the partial modified sequences GAAA and AGGAAA, showing that these termini are compatible with replication. Conversely, the progeny of pBDI-7dim invariably contained the sequence AGAAA, suggesting that the replication of the downstream monomer (beginning AGAAA) overcomes other DI RNA molecules with the 5′-end sequence differing from the authentic one. In addition, new dimeric molecules are synthesized, as shown by the 5′-terminal sequence AGAAA, which outcompete the input dimers.
(iv) A remarkable similarity between the yeast and plant systems was found in the temperature requirement for the replication of DI RNA. Although a comparative investigation concerning CIRV DI RNA was not carried out, it is known from detailed studies with Tomato bushy stunt virus genomic and DI RNAs that temperatures above 27°C are detrimental to the replication of viral RNAs, which are barely detectable at 32°C (25). The dependence of the accumulation of DI-7 RNA in 36K+95K+ yeast cells points to similar mechanism of activity of the viral replicase in yeast and plant. In fact, the expression of the viral sequences per se was not influenced by the temperature. The choice of 26°C for all experiments with yeasts was dictated by a compromise between the optima for cell growth and DI RNA accumulation.
(v) BrUTP incorporation was detected only in the 36K+95K+ cells. However, the partial coincidence of BrUTP incorporation sites with mitochondria in 36K+95K+ yeast cells suggests that replication of DI-7 RNA may not involve only these organelles, as in plant cells, notwithstanding the reported mitochondrial localization of the p36 and p95 in yeast (49).
(vi) Negative-strand DI-7 RNA in 36K+95K+ yeast cells was not readily detected, since it occurred in very small amounts compared with positive-strand DI-7 RNA. In fact, the approximate ratio between negative and positive strands was estimated to be between 1:250 and 1:500. This ratio may be almost 1:1 in infected plant cells, where positive- and negative-strand RNAs accumulate in readily detectable double-stranded RNA (29). However, it should be noted that this holds true for infections where genomic RNA replicates with or without the synthesis of DI RNA. The only comparable situation in plant cells, where a DI RNA is replicated by nonreplicable virus replicase, is that of transgenic plant protoplasts expressing the replicase of Cymbidium ringspot virus, which is capable of replicating transfected DI RNA (26, 39). However, there are no data concerning the synthesis of negative-strand RNA in these cells. Therefore, the small amount of negative-strand RNA may be a characteristic of a replicative system in which the replicase is encoded by a sequence that is translatable but unable to replicate. Studies with the aim of using S. cerevisiae for the replication of the complete genome of CIRV are under way, which may also give an answer to this issue.
Acknowledgments
We thank G. P. Martelli for helpful suggestions and critical reading of the manuscript.
REFERENCES
- 1.Ammerer, G. 1983. Expression of genes in yeast using ADC1 promoter. Methods Enzymol. 101:192-201. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Baker, K. P., A. Schaniel, D. Vestweber, and G. Schatz. 1990. A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature 348:605-609. [DOI] [PubMed] [Google Scholar]
- 4.Been, M. D. 1994. cis- and trans-acting ribozymes from a human pathogen, hepatitis delta virus. Trends Biochem. Sci. 19:251-256. [DOI] [PubMed] [Google Scholar]
- 5.Berben, G., J. Dumont, V. Gilliquet, P. A. Bolle, and F. Hilger. 1991. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7:475-477. [DOI] [PubMed] [Google Scholar]
- 6.Botstein, D., S. C. Falco, S. E. Stewart, M. Brennan, S. Scherer, D. T. Stinchcomb, K. Struhl, and R. W. Davis. 1979. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene 8:17-23. [DOI] [PubMed] [Google Scholar]
- 7.Botstein, D., and G. R. Fink. 1988. Yeast: an experimental organism for modern biology. Science 240:1439-1443. [DOI] [PubMed] [Google Scholar]
- 8.Burgyan, J., L. Rubino, and M. Russo. 1996. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J. Gen. Virol. 77:1967-1974. [DOI] [PubMed] [Google Scholar]
- 9.Carrington, J. C., D. D. Freed, and C. S. Oh. 1990. Expression of potyviral polyproteins in transgenic plants reveals three proteolytic activities required for complete processing. EMBO J. 9:1347-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, J. H., S. J. Ha, L. R. Kao, T. L. Megraw, and C. B. Chae. 1998. A novel DNA-binding protein bound to the mitochondrial inner membrane restores the null mutation of mitochondrial histone Abf2p in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5712-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalmay, T., M. Russo, and J. Burgyan. 1993. Repair in vivo of altered 3′ terminus of cymbidium ringspot tombusvirus RNA. Virology 192:551-555. [DOI] [PubMed] [Google Scholar]
- 12.Dalmay, T., G. Szittya, and J. Burgyan. 1995. Generation of defective interfering RNA dimers of cymbidium ringspot tombusvirus. Virology 207:510-517. [DOI] [PubMed] [Google Scholar]
- 13.Diaz-Ruiz, J. R., and J. M. Kaper. 1978. Isolation of viral double-stranded RNAs using a LiCl fractionation procedure. Prep. Biochem. 8:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Di Franco, A., M. Russo, and G. P. Martelli. 1984. Ultrastructure and origin of cytoplasmic multivesicular bodies induced by carnation Italian ringspot virus. J. Gen. Virol. 65:1233-1237. [Google Scholar]
- 15.Dunoyer, P., C. Ritzenthaler, O. Hemmer, P. Michler, and C. Fritsch. 2002. Intracellular localization of the Peanut clump virus replication complex in tobacco BY-2 protoplasts containing green fluorescent protein-labeled endoplasmic reticulum or Golgi apparatus. J. Virol. 76:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elion, E. A., and J. R. Warner. 1986. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:2089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finnen, R. L., and D. M. Rochon. 1995. Characterization and biological activity of DI RNA dimers formed during cucumber necrosis virus coinfections. Virology 207:282-286. [DOI] [PubMed] [Google Scholar]
- 18.Gaire, F., C. Schmitt, C. Stussi-Garaud, L. Pinck, and C. Ritzenthaler. 1999. Protein 2A of grapevine fanleaf nepovirus is implicated in RNA2 replication and colocalizes to the replication site. Virology 264:25-36. [DOI] [PubMed] [Google Scholar]
- 19.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 20.Havelda, Z., and J. Burgyan. 1995. 3′ terminal putative stem-loop structure required for the accumulation of cymbidium ringspot viral RNA. Virology 214:269-272. [DOI] [PubMed] [Google Scholar]
- 21.Hirzman, J., D. Luo, J. Hahnen, and G. Hoborn. 1993. Determination of messenger RNA 5′-ends by reverse transcription of the cap structure. Nucleic Acids Res. 21:3597-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikawa, M., M. Janda, M. A. Krol, and P. Ahlquist. 1997. In Vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J. Virol. 71:7781-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, H., Y. Fulkuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 25.Jones, R. W., A. O. Jackson, and T. J. Morris. 1990. Defective-interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts. Virology 176:539-545. [DOI] [PubMed] [Google Scholar]
- 26.Kollar, A., and J. Burgyan. 1994. Evidence that ORF 1 and 2 are the only virus encoded replicase gene of cymbidium ringspot tombusvirus. Virology 201:169-172. [DOI] [PubMed] [Google Scholar]
- 27.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 28.Leeds, P., S. W. Peltz, A. Jacobson, and M. R. Culbertson. 1991. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translation termination codon. Genes Dev. 5:2303-2314. [DOI] [PubMed] [Google Scholar]
- 29.Martelli, G. P., D. Gallitelli, and M. Russo. 1988. Tombusviruses, p. 13-72. In R. Koenig (ed.), The plant viruses. Polyhedral virions with monopartite RNA genomes. Plenum Publishing Corporation, New York, N.Y.
- 30.Molinari, P., C. Marusic, A. Lucioli, R. Tavazza, and M. Tavazza. 1998. Identification of artichoke mottled crinkle virus (AMCV) proteins required for virus replication: complementation of AMCV p33 and p92 replication-defective mutants. J. Gen. Virol. 79:639-647. [DOI] [PubMed] [Google Scholar]
- 31.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 72:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, B. D., R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redding, K., C. Holcomb, and R. S. Fuller. 1991. Immunocolocalization of Kex2 protease identifies a putative late Golgi compartment in yeast Saccharomyces cerevisiae. J. Cell Biol. 113:527-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubino, L., J. Burgyan, and M. Russo. 1995. Molecular cloning and complete nucleotide sequence of carnation Italian ringspot tombusvirus genomic and defective interfering RNAs. Arch. Virol. 140:2027-2039. [DOI] [PubMed] [Google Scholar]
- 38.Rubino, L., A. Di Franco, and M. Russo. 2000. Expression of a plant virus nonstructural protein in Saccharomyces cerevisiae causes membrane proliferation and altered mitochondrial morphology. J. Gen. Virol. 81:279-286. [DOI] [PubMed] [Google Scholar]
- 39.Rubino, L., and M. Russo. 1995. Characterization of resistance to cymbidium ringspot virus in transgenic plants expressing a full-length viral replicase gene. Virology 212:240-243. [DOI] [PubMed] [Google Scholar]
- 40.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 41.Rubino, L., F. Weber-Lotfi, A. Dietrich, C. Stussi-Garaud, and M. Russo. 2001. The open reading frame-encoded (“36K”) protein of carnation Italian ringspot virus localizes to mitochondria. J. Gen. Virol. 82:29-36. [DOI] [PubMed] [Google Scholar]
- 42.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of Tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Schlenstedt, G., E. Hurt, V. Doye, and P. A. Silver. 1993. Reconstitution of nuclear protein transport with semi-intact cells. J. Cell Biol. 123:785-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirako, Y., and T. M. Wilson. 1993. Complete nucleotide sequence and organization of the bipartite RNA genome of soil-borne wheat mosaic virus. Virology 195:16-32. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevens, B. 1981. Mitochondrial structure, p. 471-504. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces. Life cycle and inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Strauss, J. H., and E. G. Strauss. 1999. With a little help from the host. Science 283:802-804. [DOI] [PubMed] [Google Scholar]
- 49.Weber-Lotfi, F., A. Dietrich, M. Russo, and L. Rubino. 2002. Mitochondrial targeting and membrane anchoring of a viral replicase in plant and yeast cells. J. Virol. 76:10485-10496. [DOI] [PMC free article] [PubMed] [Google Scholar]