Abstract
Infection with the Epstein-Barr virus (EBV) is often subclinical in the presence of a healthy immune response; thus, asymptomatic infection is largely uncharacterized. This study analyzed the nature of EBV infection in 20 asymptomatic immunocompetent hosts over time through the identification of EBV strain variants in the peripheral blood and oral cavity. A heteroduplex tracking assay specific for the EBV gene LMP1 precisely identified the presence of multiple EBV strains in each subject. The strains present in the peripheral blood and oral cavity were often completely discordant, indicating the existence of distinct infections, and the strains present and their relative abundance changed considerably between time points. The possible transmission of strains between the oral cavity and peripheral blood compartments could be tracked within subjects, suggesting that reactivation in the oral cavity and subsequent reinfection of B lymphocytes that reenter the periphery contribute to the maintenance of persistence. In addition, distinct virus strains persisted in the oral cavity over many time points, suggesting an important role for epithelial cells in the maintenance of persistence. Asymptomatic individuals without tonsillar tissue, which is believed to be an important source of virus for the oral cavity, also exhibited multiple strains and a cyclic pattern of transmission between compartments. This study revealed that the majority of patients with infectious mononucleosis were infected with multiple strains of EBV that were also compartmentalized, suggesting that primary infection involves the transmission of multiple strains. Both the primary and carrier states of infection with EBV are more complex than previously thought.
Epstein-Barr virus (EBV) is a human herpesvirus that establishes a life-long persistent infection in more than 90% of the world's population. Primary infection with the virus is usually asymptomatic but can result in the self-limiting disease infectious mononucleosis. EBV is associated with numerous pathologies that develop in B lymphocytes and epithelial cells, including Burkitt's lymphoma, Hodgkin's disease, AIDS-associated and posttransplant lymphomas, nasopharyngeal carcinoma, hairy leukoplakia, and several others (21).
EBV persists in memory B lymphocytes, which may expand in response to their cognate antigen and subsequently differentiate into plasma cells, in which viral reactivation and replication may occur (1, 2). The role of EBV replication in the maintenance of persistent infection is not well characterized. Viral proteins indicative of replication have been detected in lymphocytes in the tonsillar tissue, and replicating virus has been detected in sloughed oropharyngeal epithelial cells (1, 14, 17, 23, 29). The replication and subsequent release of virus into the oral cavity permit transmission of the virus to a new host and possibly reinfection of oropharyngeal epithelial cells and B lymphocytes. The virus detected in the oropharynx is thought to be released from epithelial cells or lymphocytes trafficking near mucosal surfaces; however, several studies have suggested that the prevalent viruses in the oral cavity and peripheral blood may be different (5, 22).
Several variants of EBV have been identified by polymorphisms in the viral genome. EBV types 1 and 2 are distinguished by sequence changes in EBV nuclear antigens 2 and 3 (EBNA2 and -3). Strain-specific changes have also been identified in EBNA1, BZLF1, and LMP1. LMP1 is quite variable, and seven phylogenetically distinct strains of LMP1 have been described that have signature changes in the coding sequence (6). Individual isolates differ by these specific base pair changes: the presence or absence of a 30-bp deletion, the number of 33-bp repeats, and a 15-bp insertion within one of the repeats (15). Based on these consistent sequence variations, a heteroduplex tracking assay (HTA) has been developed that analyzes a 264-bp region within the carboxy terminus of LMP1 and can distinguish each of the LMP1 variants (22).
In immunosuppressed individuals, infection with multiple strains and types of EBV is a frequent occurrence, and both types of EBV and multiple strains have been identified in the permissive lesion hairy leukoplakia (18, 22, 30). A single study that used markers such as the number of repeated sequence elements, restriction enzyme polymorphisms, or the presence or absence of the 30-bp deletion has shown that immunocompetent individuals may also harbor at least two strains (24). Infection with multiple strains may indicate that even a healthy individual's immune response is not sufficient to protect against secondary infections with additional EBV strains. Alternatively, individuals may be initially infected with multiple EBV strains. A recent study showed that in immunosuppressed individuals, the number and relative abundance of EBV strains changed over time (22).
In the current study, the EBV strains present in the oral cavity and peripheral blood were identified in healthy, asymptomatic carriers at multiple time points. The data revealed that most individuals harbor multiple EBV variants and that the relative abundance and presence of the strains differ over time. The prevalent EBV strain in the oral cavity was usually distinct from that in the peripheral blood; however, in most cases, all strains were eventually detected in the saliva and blood, suggesting transmission between the compartments.
MATERIALS AND METHODS
Patient samples.
Blood was collected from 20 asymptomatic EBV-positive carriers by a finger prick. Subjects ranged in age from 8 to 50 years old. Blood was collected by routine venipuncture from infectious mononucleosis patients, and lymphocytes were separated by a Ficoll step gradient. Throat wash samples were obtained by having subjects gargle with 5 ml of sterile phosphate-buffered saline.
Nucleic acid isolation.
DNA was purified from lymphocytes with the DNeasy tissue kit (Qiagen, Valencia, Calif.). DNA was harvested from 200 μl of whole blood or throat wash sample with the High Pure viral nucleic acid isolation kit (Roche Molecular Biochemicals, Indianapolis, Ind.).
Amplification of LMP1.
LMP1 was amplified from patient samples by nested PCR with 300 ng of DNA template in duplicate independent reactions. The reaction mixture contained 1 U of Taq DNA polymerase (Promega, Madison, Wis.); a 1:10 dilution of reaction buffer; a final concentration of 2.0 mM MgCl2; a final concentration of 10 mM each of a mix containing dATP, dGTP, dCTP, and dTTP (Promega); and 25 pmol of each primer in a 50-μl reaction volume. The first round of amplification with primers LMP-3UT, EBV genome coordinates 168017 to 168036 (ATCACGAGGAATTCAATGTGGCTTTTCAGCCTAG) (6) and FUC-HIND3, EBV genome coordinates 168427 to 168408 (ATCAGAGAGCTTTGACAATGGCCCACATGACC) (22) yielded a 411- or 381-bp product, depending on the presence or absence of a 30-bp deletion.
The second round of PCR with 6 μl of template from the first reaction was performed with primers FUE-ECO, EBV genome coordinates 168163 to 168183 (ATCACGAGGAATTCGTCATAGTAGCTTAGCTGAAC) (15) and FUC-HIND3, which yielded a 264- or 234-bp product.
The strain controls for the HTA were amplified from 200 ng of cloned DNA in one round of PCR with primers FUE-ECO and FUC-HIND3. Amplification was performed in a Robocycler Gradient 96 (Stratagene, La Jolla, Calif.) for 30 to 35 cycles for each round of PCR (1 min each of denaturation at 95°C, annealing at 55°C, and extension at 72°C). PCR products were analyzed by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining.
HTA.
The phylogenetically distinct LMP1 variants that have been identified include the prototypic undeleted strain B958, other undeleted strains (Ch2, AL, NC, and Med−), and strains with the 30-bp deletion (Med+ and Ch1) (6). Variants were previously named for the origin of the patient sample from which they were first isolated, but they are not restricted to that geographical region. The Ch1 and Ch2 strains were originally identified from Chinese nasopharyngeal carcinoma specimens, the AL strain was from an Alaskan case of nasopharyngeal carcinoma, and the Med strains were first identified in nasopharyngeal carcinoma specimens from the Mediterranean (6, 15, 26). The NC strain was originally identified in an infectious mononucleosis patient in North Carolina (6).
Clones of the carboxy terminus were prepared from each variant and used as positive controls for the HTA to determine the migration position of each variant. Each patient sample was analyzed with a probe of the undeleted Ch2 variant and the deleted Med+ variant to precisely identify the strains present. Ch2 and Med+ strain-specific probes were made as previously described (22). Briefly, clones of Ch2 and Med+ (EBV coordinates 168163 to 168427) were digested with EcoRI (New England Biolabs, Beverly, Mass.) to linearize the vector. The plasmid was end labeled for 30 min at room temperature in a reaction volume of 30 μl containing 25 μCi of [35S]dATP. The linearized vector was digested with HindIII (New England Biolabs) to release the radiolabeled fragment from the vector.
The heteroduplex formation reaction was performed with 8 μl of PCR product of a strain control or a patient sample, 1 μl of annealing buffer (1 M NaCl, 100 mM Tris-HCl [pH 7.5], 20 mM EDTA), and 1 μl of radiolabeled probe. The mixtures were denatured for 5 min at 100°C and allowed to reanneal for 4 min at 4°C. The reactions were separated in nondenaturing 10% polyacrylamide gels (Hoefer apparatus; Pharmacia Biotech, San Francisco, Calif.) and subsequently dried in a gel drier (Bio-Rad, Hercules, Calif.) and exposed to a Phosphorimager (Molecular Dynamics, Sunnyvale, Calif.).
Sequence analysis.
Sequence analysis was performed on a subset of samples to confirm the results of the HTA. From those samples that displayed a band with a consistent, unique migration, sequence analysis was performed on the sample that predominantly contained the strain of interest with the primer FUE-ECO. The samples were sequenced at the University of North Carolina-Chapel Hill Automated DNA Sequencing Facility on a model 3100 genetic analyzer (Applied Biosystems Division, Perkin-Elmer Cetus, Norwalk, Conn.) with the ABI Prism Dye Terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS (Applied Biosystems Division, Perkin-Elmer Cetus).
RESULTS
EBV strain profiles of asymptomatic carriers.
To identify and determine the relative abundance of the EBV strains present in blood and saliva, DNA was extracted from throat wash samples and total blood samples at multiple time points from 20 asymptomatic individuals and analyzed for the presence of LMP1 variants with the LMP1-HTA. HTA analysis is based on the principle that completely homologous DNA molecules (homoduplexes) migrate according to size in a polyacrylamide gel, whereas heteroduplexes with base pair differences will migrate more slowly due to malformations in the DNA helix. Seven phylogenetically distinct forms of LMP1 have been identified by their sequence differences and signature amino acid changes. These include the prototypic undeleted strain B958, other undeleted strains (Ch2, AL, NC, and Med−), and strains with the 30-bp deletion (Med+ and Ch1).
The LMP1-HTA could distinguish each of the LMP1 variants based on a consistent migration pattern when heteroduplexed to the Ch2 and Med+ probes (Fig. 1). In some of the patient samples, a few bands appeared as doublets, and some background bands were visible. These additional bands were presumed to be aberrantly migrating heteroduplexes, characteristic of the probe, as they were not present in analyses with both probes. Other samples contained bands with slight alterations in migration. These bands were shown to reflect variants that were detected with both probes and shown to have additional base pair changes from the known control strain sequences by sequence analyses (Fig. 2, throat wash sample day 112).
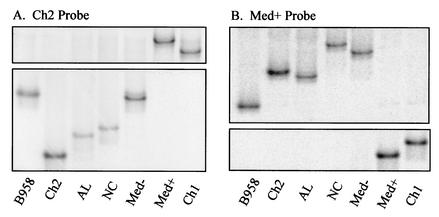
FIG. 1.
Migration position of LMP1 variants heteroduplexed to an undeleted and deleted probe. Only the regions of the gel that contained the heteroduplexes are shown. Undeleted strains bound to an undeleted probe migrate low in the gel, while deleted strains migrated high in the gel because of the large malformation of the DNA helix. In contrast, deleted strains bound to a deleted probe migrated more according to size due to relatively little mismatch, while undeleted strains migrated high in the gel. PCR-amplified control strains B958, Ch2, AL, NC, Med−, and Med+ were denatured with and reannealed to (A) the undeleted Ch2 probe and (B) the deleted Med+ probe.
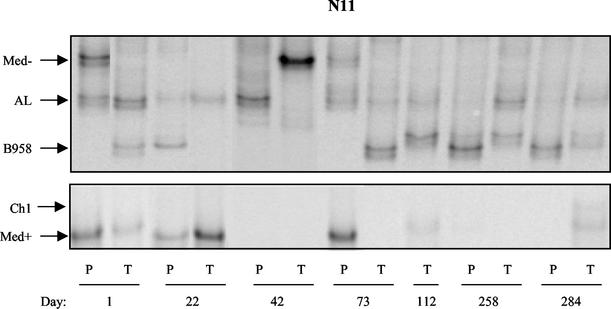
FIG. 2.
Strain profile of asymptomatic EBV carrier N11 determined by LMP1-HTA with the Med+ probe. Peripheral blood (P) and throat wash (T) samples were collected over 284 days. On day 112, a blood sample was not available. Strains were identified based on their migration in relation to that of known strain controls.
The LMP1-HTA has been previously shown to identify variants present in 1% of a mixed population (22). In order to determine the detection capabilities in a background of negative lymphocytes, EBV-negative DG75 cells were mixed with EBV-positive Raji cells, which contain 50 copies of the EBV episome per cell. The PCR conditions and the LMP1-HTA could easily detect one Raji cell in a background of 1.5 × 106 DG75 cells (data not shown).
A time course of samples that were collected during a 10-month period and analyzed by the LMP1-HTA (Med+ probe) is presented for subject N11 (Fig. 2). Each patient sample was also analyzed with the Ch2 probe to differentiate between the Med− and NC strains and the AL and Ch2 strains, which migrate to a similar position with the Med+ probe (data not shown). A blood sample was not available at day 112. At the first time point, strains detected in the peripheral blood included Med−, AL, and Med+, while strains in the oral cavity included AL, Med+, and B958. The Med− strain was not present in the throat wash sample on day 1 and was not detected in either compartment on day 22. However, on day 42, it appeared in the oral cavity but not in the blood. At the next collection, at day 73, it was present in the blood but not the throat wash sample and then was not detectable on days 112, 258, and 284.
The absence of Med− in the blood between time points may indicate that the infected cells carrying this strain had entered secondary lymphoid organs and were temporarily undetectable in the blood (13). The AL strain persisted in both the peripheral blood and oral cavity and was detected in each compartment at the majority of time points analyzed.
The B958 strain was detected only in the oral cavity at day 1, then only in the blood at day 22, and was not detectable in either compartment on day 42. B958 reappeared in the oral cavity at day 73 and was subsequently detected in the blood at the next time points tested, days 258 and 284. It is interesting that the appearance of B958 in the blood corresponded with an earlier detection of the strain in the oral cavity, similar to the pattern observed for Med−. A distinct variant of B958 marked by an additional base pair change (266A→T) was detected in the oral cavity at day 112, where it persisted for 5 months. The continued detection of this specific variant only in the oral cavity may indicate that EBV strains persist in oropharyngeal epithelial cells in a chronic lytic infection. The Med+ strain also exhibited a variable profile that increased and decreased in relative abundance in each compartment over time. The Ch1 strain was detected in the oral cavity at the final time point but was not detected in any of the previous samples.
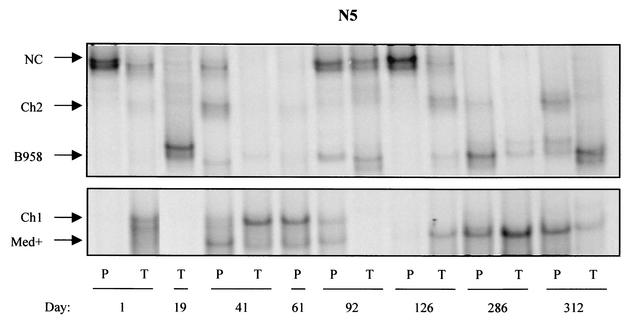
A striking dynamic strain profile was also detected in subject N5 (Fig. 3). Analyses of this subject revealed an actively changing strain profile where strains appeared, disappeared, and reappeared. Furthermore, strains appeared to be transmitted between compartments. Several strains, including NC, B958, and Med+, persisted over multiple time points. Samples were collected from this subject over 10 months; a blood sample was not available at day 19, and a throat wash sample was not available at day 61. At day 1, the NC strain was the only strain detected in the peripheral blood, while strains detected in the throat wash sample included NC, Ch1, Med+, and trace amounts of B958 and Ch2. At day 19, only a B958 variant (266A→T) was present in the throat wash sample. The strain profile in both compartments continued to change dramatically throughout the time course. At day 41, five strains were detected in the blood that were the same strains detected in the oral cavity at day 1, while the throat wash sample at day 41 predominantly contained Ch1 with trace levels of Med+, B958, and Ch2.
FIG. 3.
Strain profile of asymptomatic EBV carrier N5 determined by LMP1-HTA with the Med+ probe. Peripheral blood (P) and throat wash (T) samples were collected over 312 days. On day 19, a blood sample was not available, and on day 61, a throat wash sample was not collected. Strains were identified based on their migration in relation to that of known strain controls.
The blood sample at day 61 looked identical to the throat wash sample from day 41 with the notable absence of the NC strain. The NC strain reappeared and was abundant in both samples on days 92 and 126, but then disappeared and was not detectable on days 286 and 312. The blood sample from day 126 with abundant NC and trace levels of Med+ was nearly identical to the profile in the blood at day 1. The throat wash sample at day 126 contained NC, Ch2, B958, and Med+. These strains, Ch2, B958, and Med+, were subsequently detected in the blood, as indicated by the strain profile at day 286 and the final time point on 312. This may indicate that the peripheral blood lymphocytes have been infected with virus from the oral cavity. The cycling of strains between the oral cavity and peripheral blood reveals that virus may continuously traffic between compartments, which may be an important part of the virus life cycle.
Similar analyses of each of the 20 subjects in this study revealed that all subjects were infected with multiple strains. Infection with two to three EBV strains was detected in six subjects, four strains in eight subjects, and five to six strains in six subjects. The strain profiles of each subject frequently varied at individual time points, but some strains persisted at multiple time points. Clear compartmental differences between the oral cavity and peripheral blood were detected at individual time points in 16 of the 20 subjects. The predominance of a strain in one compartment and its absence in the other compartment likely reflect the limits of detection but also indicate clear differences in relative abundance in the two compartments.
The detection of unique strains in the oral cavity before their detection in the blood may represent the acquisition of a new strain from an exogenous source. Alternatively, unique strains in the oral cavity may represent virus produced by plasma cells in the oral lymphoid tissue that underwent selection and maturation upon stimulation with their cognate antigen. Viral DNA detected in the peripheral blood likely represents intracellular EBV from infected memory B lymphocytes and possibly cell-free viral DNA released from lymphocytes that have died due to immune surveillance.
At each collection, subjects completed a questionnaire regarding their perceived level of stress, any symptoms of ill health, including fatigue, sore throat, and runny nose, and if they had previously been diagnosed with infectious mononucleosis. Regardless of sickness or stress level, EBV was detected at each time point in both the oral cavity and peripheral blood. The strain profiles did not differ between subjects who had and had not experienced infectious mononucleosis. These analyses suggest that EBV infection involves the active exchange of virus between the peripheral blood and the oral cavity, where distinct virus strains increased and decreased in relative abundance and appeared to cycle between compartments.
EBV strain profiles of asymptomatic atonsillar carriers.
EBV is believed to persist in a latent infection in memory B cells, which may reactivate at mucosal surfaces. Viral replicative antigens have been detected in reactivated tonsillar lymphocytes. To determine if the absence of tonsillar tissue affected the presence of EBV or the strain profiles in the oral cavity, analyses of the asymptomatic EBV carriers included six subjects whose palatine tonsils and/or adenoids had been surgically removed at least 10 years prior to this study. The data indicate that subjects who had their palatine tonsils removed did not differ from other asymptomatic carriers with regard to the number of strains, compartmentalization, and transmission between compartments. However, these individuals retained other lymphoid tissues, including the tubal and lingual tonsils. In addition, regrowth of palatine tonsils may occur.
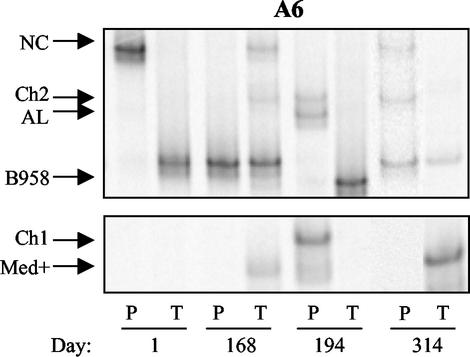
Subject A6 underwent a tonsillectomy that also included the removal of adenoidal tissue 20 years prior to enrollment in this study. Samples were collected over 11 months and analyzed by LMP1-HTA (Fig. 4). At day 1, NC was strikingly abundant in the blood, with trace levels of Ch2 and a B958 strain with a distinct migration pattern. Only this B958 strain, marked by a base pair change at position 266A→T, was detected in the throat wash sample at day 1, and it was subsequently the only strain detected in the blood on day 168. It was not detected in the blood or throat wash sample on day 194, but then reappeared in both compartments at day 314. The NC strain that was the prevalent strain in the blood at day 1 was not detected in the blood on day 168 but was present in the oral cavity. It was not detected in either sample on day 194 but reappeared in the blood on day 314. The throat wash sample on day 168 contained both NC and the distinct B958 and also contained Ch2 and Med+, which were detected for the first time. Ch2 and Med+ were both subsequently detected in the blood at day 194. The strains in the oral cavity and peripheral blood of this atonsillar subject were consistently different from each other at each time point. At day 314, Ch2 was present in the blood but not the throat wash sample, while Med+ was detected in the throat wash sample but not the blood.
FIG. 4.
Strain profile of asymptomatic atonsillar EBV carrier A6 determined by LMP1-HTA with the Med+ probe. Peripheral blood (P) and throat wash (T) samples were collected over 314 days. Strains were identified based on their migration in relation to that of known strain controls.
The AL and Ch1 strains were detected in the blood only at day 194, while a B958 strain, without the additional base pair change, was detected at high levels only in the oral cavity. These three strains were not subsequently detected at day 314.
Analyses of infectious mononucleosis subjects.
The complexity of EBV infection in immunocompetent asymptomatic carriers was unexpected. Whether individuals are subject to repeated infections throughout life or the immune response to EBV protects against secondary infections with similar and/or different strains is unknown. Because EBV is transmitted through saliva, the presence of multiple strains in throat wash samples suggested that multiple strains might be transmitted during primary infection. In order to investigate the number of strains acquired in the course of infectious mononucleosis, blood and throat wash samples were collected from 15 subjects on the day of diagnosis. These analyses revealed that 13 of 15 subjects were infected with multiple EBV strains, and each of the 13 subjects had compartmental differences in strain profiles (Table 1, Fig. 5). Four of 13 subjects were infected with two strains, a single different strain in each compartment, which may indicate that the virus unique to the oral cavity was more efficient at lytic replication and less efficient at establishing a latent infection in B lymphocytes. The peripheral blood of 5 of 13 subjects and the oral cavity of 6 of 13 subjects contained two or more strains, demonstrating that multiple strains can establish an infection in both compartments upon primary infection.
TABLE 1.
EBV strains in subjects with infectious mononucleosis
| Subject | Strain(s) detected
|
|
|---|---|---|
| Blood | Throat wash | |
| MO1 | Med+ | Med+ |
| MO2 | Ch1 | Ch1 |
| MO3 | Ch2 | B958 (266 A → T) |
| MO4 | Ch2 | Ch2, Med− |
| MO5 | Ch2 | Ch2, Med+, B958 (266 A → T) |
| MO6 | B958 | Ch1, NC, Med+, B958 |
| MO7 | Ch2 | Ch1, Med−, Med+, AL |
| MO8 | Ch2 | Ch1 |
| MO9 | Ch2 | Med− |
| MO10 | Med+ | Ch1 |
| MO11 | Ch2, Med− | Med− |
| MO12 | Ch1, B958 | Ch1, Med− |
| MO13 | Ch1, Ch2 | Ch1 |
| MO14 | Ch2, NC | NC |
| MO15 | Ch2, Med+, B958 | B958, Med− |
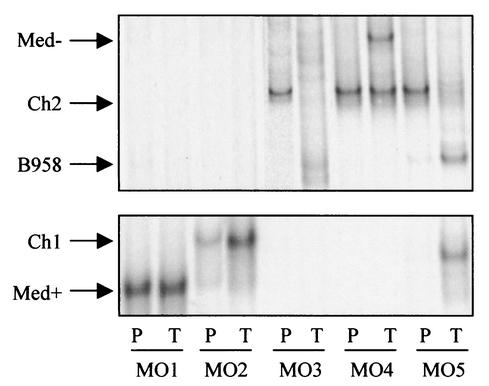
FIG. 5.
Strain profiles of five different infectious mononucleosis patients (MO1 to MO5) determined by LMP1-HTA with the Med+ probe. Peripheral blood (P) and throat wash (T) samples were collected at the day of diagnosis. Strains were identified based on their migration in relation to that of known strain controls.
Subjects MO1 and MO2 both contained a single deleted strain in both compartments (Fig. 5). In the blood and throat wash samples of MO1, Med+ was detected, and in subject MO2, Ch1 was detected. The blood of MO3 contained Ch2, while the throat wash sample had B958 and possibly several unique forms (Fig. 5). The blood of MO4 contained only Ch2, with both Ch2 and Med− detected in the throat wash sample (Fig. 5). Ch2 was the most abundant strain in the blood of MO5, with trace amounts of B958 and a distinct Ch1 strain. The throat wash sample contained the same strains at different relative levels compared to the blood; B958 and the distinct Ch1 were the most abundant, with a decreased level of Ch2 (Fig. 5).
DISCUSSION
The identification of strain variants is an effective approach to investigating virus transmission and persistence. EBV types can be identified by sequence divergence in the EBNA2 and EBNA3 genes. Another method of distinction called Ebnotyping distinguishes strains based on the size pattern of EBNA1, EBNA2, and EBNA3A, -3B, and -3C, which all contain repeat elements resulting in slightly different protein sizes. Ebnotyping has been used to study the transmission of virus within families (8). LMP1 is the most discriminating locus for EBV genotyping and has been widely used to identify strain variants by the presence or absence of a 30-bp deletion in the carboxy terminus.
In order to identify precisely the five undeleted and two deleted LMP1 strains, specific base pair changes in addition to the deletion must be considered. It is likely that numerous substrains exist that have additional base pair changes in addition to the sequence changes in the known strains, as demonstrated in this study by the identification of a B958 strain with an additional change at position 266. Previous studies have used cloning and sequencing to identify multiple LMP1 strains in tissue and blood samples (31). The LMP1 HTA enables the identification of all strain variants and reveals their relative abundance. This analysis allows accurate studies of transmission and persistence of the EBV strains represented by the LMP1 variants (22).
The presence of multiple strains in immunocompromised patients has been well described (3, 22, 34). The immune response generated during primary infection was thought to be effective in the prevention of subsequent superinfection with new viruses, while a weakened immune system was originally believed to be responsible for the susceptibility to superinfection with additional EBV strains. This is the first longitudinal study to characterize the changing nature of infection in the blood and oral cavity in human immunodeficiency virus-negative immunocompetent subjects. This study reveals that infection with EBV in the presence of a healthy immune response involves multiple strains with clear differences in relative abundance between the blood and oral cavity at a given time point. Although different strain profiles were detected at each time point, the strains demonstrated a corresponding rise and fall in relative abundance over time. For example, if a new strain was detected in the blood, it was often paralleled by a simultaneous decrease in abundance of another strain, which likely reflects a steady level of infected cells. The number of resting B lymphocytes infected with EBV has been reported to be relatively stable, ranging from 1 to 50 per 106 B cells; however, the life span of each infected cell is unknown (11). Memory B lymphocytes are the main cell type involved in persistent EBV infection; therefore, the maintenance of persistent EBV infection depends on the survival of an infected memory B cell or the continual reinfection of new cells that reseed the memory cell population.
The abundance of EBV-specific CD8+ T lymphocytes to viral proteins expressed in proliferating, latently infected lymphocytes suggests that expression of these proteins and B-cell proliferation may be common (12, 16). In addition, studies have revealed that in immunocompetent individuals, the T-cell response to lytic cycle antigens was more predominant than the responses to latent antigens, suggesting that reactivation to replication also occurs constantly (25, 28). In the tonsillar tissue in the oral cavity, reactivated lymphocytes undergoing viral replication have been detected. The subsequent release of virus into the oral cavity may allow infection of naïve B lymphocytes and epithelial cells at mucosal sites (2). Reactivated B lymphocytes that release virus into the oral cavity would die due to lytic viral replication; therefore, the persistent detection of specific strains in the oral cavity over time suggests that a chronic replicative phase may occur in epithelial cells. Thus, the oral cavity may be a temporary reservoir of virus for de novo infection of naive B lymphocytes. The two compartments are clearly related, since the strains were not restricted to one compartment, but the strains appeared to cycle in relative abundance between the oral cavity and peripheral blood. The variation in relative abundance of a given strain within the peripheral blood may indicate that the infected lymphocytes may enter secondary lymphoid tissue that was not sampled in this study.
The potential alternating infection of B lymphocytes and epithelial cells is conceivably an important component in the viral life cycle (4). A recent study has shown that the glycoproteins present in the virion envelope determine infection of the two cell types. Entry of EBV into B lymphocytes requires a tripartite glycoprotein complex containing gH, gL, and gp42, while entry into an epithelial cell requires a bipartite complex composed of gH and gL (32). The virus released from a B lymphocyte has less gp42 and therefore has a decreased efficiency for infection of B lymphocytes and an increased ability for infection of epithelial cells (4). Virus released from epithelial cells contains relatively more gp42 and can efficiently infect B lymphocytes (4). The regular cycling of virus between the oral cavity and peripheral blood in asymptomatic carriers suggests that this process occurs in vivo. In addition, the persistence of distinct strains in the oral cavity implicates the epithelial cell reservoir in the maintenance of persistent infection and in transmission to subsequent hosts.
The majority of EBV-infected B lymphocytes are detected in the peripheral circulation and in the oropharyngeal lymphoid tissues, specifically, the tonsils and adenoids that constitute Waldeyer's ring (13). Our investigation of asymptomatic carriers included analyses of subjects that had undergone tonsillectomies and/or adenoidectomies between childhood and adolescence. These subjects also exhibited multistrain profiles that appeared to cycle between compartments, similar to those of subjects who retained their tonsils and adenoids. The detection and persistence of distinct strains in the oral cavity in these subjects also suggests chronic infection at some oral site. Tonsillectomies involve removal of the lymphoid-rich palatine tonsils; however, the remaining tonsillar tissue may also harbor infected B lymphocytes that release virus into the oral cavity. It is also likely that replication in parotid epithelial cells is an important source of virus in the oral cavity and participates in the exchange of virus between compartments (33).
This study reveals for the first time that primary infection during infectious mononucleosis can involve multiple EBV strains. This is not surprising, based on the data presented in this study, which revealed multiple strains in the saliva of asymptomatic carriers, which presumably would be the inoculum to infect another person. The presence of multiple strains at primary infection has also been described for other viruses, including human immunodeficiency virus and human herpesvirus 6 (10, 20).
The majority of mononucleosis cases (13 of 15) also exhibited compartmentalization of strains between the blood and oral cavity. As the symptoms of infectious mononucleosis become apparent 4 to 8 weeks after exposure to EBV, viral dissemination and transmission between compartments has likely already occurred (19, 27). Therefore, it is quite striking that in 10 of 13 subjects, unique strains were detected in the oral cavity that were not detected in the blood, and the other three subjects (MO11, MO13, and MO14) contained unique strains in the blood that were not detected in the oral cavity. The compartmentalization of strains is similar to the infection of asymptomatic carriers described in this study. It will be important to follow patients after primary infection to determine if multiple strains persist and determine if any strains are unable to establish a persistent infection in the peripheral blood or oral cavity. In addition, longitudinal studies will assess the protective capacity of the immune response generated at primary infection by determining if new strains are subsequently acquired and transmitted to the blood.
The dramatically changing nature of infection in asymptomatic carriers has broad implications. Several studies have described compartmentalization of EBV variants in tumor tissues that are distinct from those in the blood at a particular time point (7, 9). The disappearance and reappearance of strains in asymptomatic carriers emphasize the importance of analyzing samples obtained from an individual at multiple time points before concluding that a particular variant is present or absent. This study also points to important considerations for future vaccine development. The continued release of virus into the oral cavity indicates that the high levels of cytotoxic lymphocytes directed against replicative proteins do not prevent virus replication. In addition, the cycling of strains from the oral cavity to the peripheral blood may indicate that B lymphocytes are consistently being reinfected in the oral cavity and then trafficking into the periphery. This would suggest that the natural mucosal immune response is not sufficient to protect an immunocompetent individual from repeated infections with the same or different strains.
Acknowledgments
We thank Joseph Pagano and Shannon Kenney for critical review of the manuscript.
This study was supported by U.S. Public Health Service grants DE11644 and CA32979 to N.R.-T. D.S.-G. is supported by NIH training grant DE07310.
REFERENCES
- 1.Anagnostopoulos, I., M. Hummel, C. Kreschel, and H. Stein. 1995. Morphology, immunophenotype, and distribution of latently and/or productively Epstein-Barr virus-infected cells in acute infectious mononucleosis: implications for the interindividual infection route of Epstein-Barr virus. Blood 85:744-750. [PubMed] [Google Scholar]
- 2.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 3.Berger, C., D. van Baarle, M. J. Kersten, M. R. Klein, A. S. Al-Homsi, B. Dunn, C. McQuain, R. van Oers, and H. Knecht. 1999. Carboxy terminal variants of Epstein-Barr virus-encoded latent membrane protein 1 during long-term human immunodeficiency virus infection: reliable markers for individual strain identification. J. Infect. Dis. 179:240-244. [DOI] [PubMed] [Google Scholar]
- 4.Borza, C., and L. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H. L., M. L. Lung, K. H. Chan, B. E. Griffin, and M. H. Ng. 1996. Tissue distribution of Epstein-Barr virus genotypes. J. Virol. 70:7301-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards, R. H., F. Seillier-Moiseiwitsch, and N. Raab-Traub. 1999. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 261:79-95. [DOI] [PubMed] [Google Scholar]
- 7.Faumont, N., T. Al Saati, P. Brousset, C. Offer, G. Delsol, and F. Meggetto. 2001. Demonstration by single-cell PCR that Reed-Sternberg cells and bystander B lymphocytes are infected by different Epstein-Barr virus strains in Hodgkin's disease. J. Gen. Virol. 82:1169-1174. [DOI] [PubMed] [Google Scholar]
- 8.Gratama, J. W., M. A. Oosterveer, G. Klein, and I. Ernberg. 1990. EBNA size polymorphism can be used to trace Epstein-Barr virus spread within families. J. Virol. 64:4703-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry, S., C. Sacaze, L. Berrajah, H. Karray, M. Drira, A. Hammami, J. Icart, and B. Mariame. 2001. In nasopharyngeal carcinoma-bearing patients, tumors and lymphocytes are infected by different Epstein-Barr virus strains. Int. J. Cancer 91:698-704. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson, A., S. Lindback, H. Gaines, and A. Sonnerborg. 1998. Characterization of the viral population during primary human immunodeficiency virus-1 infection. AIDS 12:839-847. [DOI] [PubMed]
- 11.Khan, G., E. M. Miyashita, B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1996. Is EBV persistence in vivo a model for B cell homeostasis? Immunity 5:173-179. [DOI] [PubMed] [Google Scholar]
- 12.Khanna, R., S. R. Burrows, M. G. Kurilla, C. A. Jacob, I. S. Misko, T. B. Sculley, E. Kieff, and D. J. Moss. 1992. Localization of Epstein-Barr virus cytotoxic T cell epitopes with recombinant vaccinia: implications for vaccine development. J. Exp. Med. 176:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laichalk, L., D. Hochberg, G. Babcock, R. Freeman, and D. Thorley-Lawson. 2002. The dispersal of mucosal memory B cells: evidence from persistent EBV infection. Immunity 16:745-754. [DOI] [PubMed] [Google Scholar]
- 14.Lemon, S., L. Hutt, J. Shaw, J. Li, and J. Pagano. 1977. Replication of EBV in epithelial cells during infectious mononucleosis. Nature 268:268-270. [DOI] [PubMed] [Google Scholar]
- 15.Miller, W. E., R. H. Edwards, D. M. Walling, and N. Raab-Traub. 1994. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J. Gen Virol. 75:2729-2740. (Erratum, 76:1305, 1995.) [DOI] [PubMed] [Google Scholar]
- 16.Murray, R. J., M. G. Kurilla, J. M. Brooks, W. A. Thomas, M. Rowe, E. Kieff, and A. B. Rickinson. 1992. Identification of target antigens for the human cytotoxic T cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J. Exp. Med. 176:157-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niedobitek, G., A. Agathanggelou, N. Steven, and L. Young. 2000. Epstein-Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. Mol. Pathol. 53:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palefsky, J. M., J. Berline, M. E. Penaranda, E. T. Lennette, D. Greenspan, and J. S. Greenspan. 1996. Sequence variation of latent membrane protein-1 of Epstein-Barr virus strains associated with hairy leukoplakia. J. Infect. Dis. 173:710-714. [DOI] [PubMed] [Google Scholar]
- 19.Papesch, M., and R. Watkins. 2001. Epstein-Barr virus infectious mononucleosis. Clin. Otolaryngol. 26:3-8. [DOI] [PubMed] [Google Scholar]
- 20.Pruksananonda, P., C. Hall, R. Insel, K. McIntyre, P. Pellett, C. Long, K. Schnabel, P. Pincus, F. Stamey, T. Dambaugh, et al. 1992. Primary human herpesvirus 6 infection in young children. N. Engl. J. Med. 326:1445-1450. [DOI] [PubMed] [Google Scholar]
- 21.Raab-Traub, N. 1996. Pathogenesis of Epstein-Barr virus and its associated malignancies. Semin. Virol. 7:315-323. [Google Scholar]
- 22.Sitki-Green, D., R. H. Edwards, J. Webster-Cyriaque, and N. Raab-Traub. 2002. Identification of Epstein-Barr virus strain variants in hairy leukoplakia and the peripheral blood by use of a heteroduplex tracking assay. J. Virol. 76:9645-9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sixbey, J. W., J. G. Nedrud, N. Raab-Traub, R. A. Hanes, and J. S. Pagano. 1984. Epstein-Barr virus replication in oropharyngeal epithelial cells. N. Engl. J. Med. 310:1225-1230. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava, G., K. Wong, A. Chiang, K. Lam, and Q. Tao. 2000. Coinfection of multiple strains of Epstein-Barr virus in immunocompetent normal individuals: reassessment of the viral carrier state. Blood 95:2443-2445. [PubMed] [Google Scholar]
- 25.Steven, N. M., N. E. Annels, A. Kumar, A. M. Leese, M. G. Kurilla, and A. B. Rickinson. 1997. Immediate early and early lytic cycle proteins are frequent targets of the Epstein-Barr virus-induced cytotoxic T cell response. J. Exp. Med. 185:1605-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sung, N. S., R. H. Edwards, F. Seillier-Moiseiwitsch, A. G. Perkins, Y. Zeng, and N. Raab-Traub. 1998. Epstein-Barr virus strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int. J. Cancer 76:207-215. [DOI] [PubMed] [Google Scholar]
- 27.Svedmyr, E., I. Ernberg, J. Seeley, O. Weiland, G. Masucci, K. Tsukuda, R. Szigeti, M. Masucci, H. Blomogren, and W. Berthold. 1984. Virologic, immunologic, and clinical observations on a patient during the incubation, acute, and convalescent phases of infectious mononucleosis. Clin. Immunol. Immunopathol. 30:437-450. [DOI] [PubMed] [Google Scholar]
- 28.Tan, L. C., N. Gudgeon, N. E. Annels, P. Hansasuta, C. A. O'Callaghan, S. Rowland-Jones, A. J. McMichael, A. B. Rickinson, and M. F. Callan. 1999. A re-evaluation of the frequency of CD8+ T cells specific for EBV in healthy virus carriers. J. Immunol. 162:1827-1835. [PubMed] [Google Scholar]
- 29.Tao, Q., G. Srivastava, A. C. Chan, L. P. Chung, S. L. Loke, and F. C. Ho. 1995. Evidence for lytic infection by Epstein-Barr virus in mucosal lymphocytes instead of nasopharyngeal epithelial cells in normal individuals. J. Med. Virol. 45:71-77. [DOI] [PubMed] [Google Scholar]
- 30.Walling, D. M., S. N. Edmiston, J. W. Sixbey, M. Abdel-Hamid, L. Resnick, and N. Raab-Traub. 1992. Coinfection with multiple strains of the Epstein-Barr virus in human immunodeficiency virus-associated hairy leukoplakia. Proc. Natl. Acad. Sci. USA 89:6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walling, D. M., N. Shebib, S. C. Weaver, C. M. Nichols, C. M. Flaitz, and J. Webster-Cyriaque. 1999. The molecular epidemiology and evolution of Epstein-Barr virus: sequence variation and genetic recombination in the latent membrane protein-1 gene. J. Infect. Dis. 179:763-774. [DOI] [PubMed] [Google Scholar]
- 32.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf, H., M. Haus, and E. Wilmes. 1984. Persistence of Epstein-Barr virus in the parotid gland. J. Virol. 51:795-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao, Q. Y., R. J. Tierney, D. Croom-Carter, D. Dukers, G. M. Cooper, C. J. Ellis, M. Rowe, and A. B. Rickinson. 1996. Frequency of multiple Epstein-Barr virus infections in T-cell-immunocompromised individuals. J. Virol. 70:4884-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]