Abstract
Rubella virus is an enveloped positive-strand RNA virus of the family Togaviridae. Virions are composed of three structural proteins: a capsid and two membrane-spanning glycoproteins, E2 and E1. During virus assembly, the capsid interacts with genomic RNA to form nucleocapsids. In the present study, we have investigated the role of capsid phosphorylation in virus replication. We have identified a single serine residue within the RNA binding region that is required for normal phosphorylation of this protein. The importance of capsid phosphorylation in virus replication was demonstrated by the fact that recombinant viruses encoding hypophosphorylated capsids replicated at much lower titers and were less cytopathic than wild-type virus. Nonphosphorylated mutant capsid proteins exhibited higher affinities for viral RNA than wild-type phosphorylated capsids. Capsid protein isolated from wild-type strain virions bound viral RNA more efficiently than cell-associated capsid. However, the RNA-binding activity of cell-associated capsids increased dramatically after treatment with phosphatase, suggesting that the capsid is dephosphorylated during virus assembly. In vitro assays indicate that the capsid may be a substrate for protein phosphatase 1A. As capsid is heavily phosphorylated under conditions where virus assembly does not occur, we propose that phosphorylation serves to negatively regulate binding of viral genomic RNA. This may delay the initiation of nucleocapsid assembly until sufficient amounts of virus glycoproteins accumulate at the budding site and/or prevent nonspecific binding to cellular RNA when levels of genomic RNA are low. It follows that at a late stage in replication, the capsid may undergo dephosphorylation before nucleocapsid assembly occurs.
Rubella virus (RV) is a member of the family Togaviridae, a group of positive-strand RNA viruses with relatively simple virion structures and replication schemes (10, 15, 44). The virus is highly teratogenic and causes devastating malformations in human fetuses when in utero infection occurs during the first trimester of pregnancy. Although congenital rubella syndrome is rare in western countries, it remains endemic in the developing world and parts of Eastern Europe (14, 40). Despite its medical importance and the widespread use of live RV vaccines, most aspects of virus replication and pathogenesis remain poorly understood.
Virions are comprised of three structural proteins: two membrane-spanning glycoproteins, E2 and E1, and a capsid protein (15, 34). The membrane glycoproteins are required for binding to host cells and membrane fusion during infection (10). During virus assembly, they are thought to play a major role in facilitating intracellular budding at the Golgi complex (17). Recently, we have focused our efforts on studying the roles of the capsid protein in virus replication. Our data suggest that the capsid is a multifunctional protein that has roles in virus-host interactions as well as replication. For example, the capsid interacts with multiple host cell proteins and appears to be proapoptotic in certain cultured cell lines (5, 8). One of the functions of the capsid in the assembly pathway is to package RV genomic RNA into nucleocapsids. Formation of RV nucleocapsids appears to be regulated and membrane dependent (10, 42). This mode of nucleocapsid assembly is unique among togaviruses and may be a consequence of the unusual posttranslational processing of the capsid protein. RV structural proteins are synthesized as a polyprotein precursor in the order NH2-capsid-E2-E1-COOH (33). During translocation of virus glycoproteins into the endoplasmic reticulum (ER), signal peptidase cleavage separates E2 from the capsid while leaving the E2 signal peptide attached to the carboxy terminus of the capsid (42). In addition to mediating membrane association of the capsid, the E2 signal peptide is required for the targeting of the capsid to the perinuclear region where virus budding occurs (25).
It has been reported that the capsid is modified by the addition of phosphate prior to virus assembly (11, 29), but the role of this posttranslational modification in virus replication has not previously been investigated for RV or other togaviruses. In the present study we report that serine 46 regulates capsid phosphorylation. Interestingly, serine 46 lies within the RNA binding site (amino acid residues 28 to 56) of the capsid (27). This region of the capsid is rich in arginine residues and binds to a packaging signal located between nucleotides 347 and 375 of the genomic RNA. Nonphosphorylated capsid has a higher affinity for genomic RNA, indicating that phosphorylation negatively regulates the RNA binding activity of this protein. We propose a model where phosphorylation of the capsid during the early stages of assembly prevents nonspecific binding of cellular RNA and/or premature assembly of nucleocapsids. In addition, dephosphorylation of capsid during the latter stages of virus assembly may be required for efficient nucleocapsid assembly and budding.
MATERIALS AND METHODS
Reagents, antibodies, and cDNA clones.
Reagents and supplies were from the following sources. Protein A- and G-Sepharose and glutathione-Sepharose were purchased from Pharmacia (Alameda, Calif.). Phenylmethylsulfonyl fluoride, fibronectin, sodium dodecyl sulfate (SDS), bovine serum albumin, and general lab chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.). [14C]-labeled protein standards were purchased from Amersham Corp. (Arlington Heights, Ill.). Radiolabeled inorganic phosphate H333PO4 (4,000 Ci/mmol) was purchased from ICN (Costa Mesa, Calif.). Media and sera for cell culture were purchased from Life Technologies-Invitrogen, Inc. or Sigma. Pwo polymerase and PerFectin transfection reagent were purchased from Roche Molecular Biochemicals (Laval, Canada) and Gene Therapy Systems, Inc. (San Diego, Calif.), respectively. Vero, COS, RK13, and BHK cells were obtained from the American Type Culture Collection (Manassas, Va.). The M33 strain of RV and an infectious cDNA clone (46) were kindly provided by Shirley Gillam (University of British Columbia). The C1 anti-capsid monoclonal antibody was a gift from Jerry Wolinsky (University of Texas, Houston). Purified phosphatases were purchased from Sigma or purified as described previously (30).
Cell culture.
BHK, COS, and Vero cells were cultured in Dulbecco's minimal essential medium (high glucose) containing 10% fetal bovine serum, 2 mM glutamine, 1 mM HEPES, and antibiotics. RK13 cells were cultured in minimal essential medium containing 10% fetal bovine serum, 2 mM glutamine, 1 mM HEPES, and antibiotics.
Plasmid construction.
Capsid constructs were generated by PCR by using Taq polymerase (Invitrogen-Gibco) and primers listed in Table 1. All new cDNA constructs that were used in this study were sequenced to verify their authenticity and ensure the absence of second-site mutations.
TABLE 1.
Primers used in this study
| Name | Sequence (5′-3′) | Underlined sequence | Bold sequence |
|---|---|---|---|
| Av11(F) | TACGGTGGGAGGTCTATATAGCA | NAa | NA |
| AV10cla | TTGATCATCGATGGGCACTGGAGTGGCAAC | ClaI | NA |
| Cap312(R) | GGGGATCCTATTGCATACGCGGGGGTTG | BamHI | Stop codon |
| Cap331(F) | CGCAATCCGCCACCATGGAGCAAAAGCTCATTTCTGAAGAGGACTTGCCGCGTATGCAAACCGG | Ribosome binding sequence | Annealing to capsid |
| Capsid(R) | GGTCAGATCTCTAGGCGCGCGCGGTGC | BglII | Stop codon |
| S45/46A(F) | CCGCGGCCGCCGCGACAGCGCGACGCCGCAACCTCCGGAGATGACTCCGGCCGTGACTCC | NotI | Serine-to-alanine change |
| S52/56A(F) | CCGCGGCCGCCGCGACAGCGCGACTCCAGCACCTCCGGAGATGACGCCGGCCGTGACGCCGGAGGGCCCCGC | NotI | Serine-to-alanine change |
| S45A(F) | CCGCGGCCGCCGCGACAGCGCGACGCCAGCACCTCCGGAGATGACTCCGGCCGTGACTCC | NotI | Serine-to-alanine change |
| S46A(F) | CCGCGGCCGCCGCGACAGCGCGACTCCGCAACCTCCGGAGATGACTCCGGCCGTGACTCC | NotI | Serine-to-alanine change |
| CapSal(R) | GTAAAATGCAGGTCGACG | NA | NA |
| CGST(F) | AGATCTGGATCCATGGCTTCCACTACCCCCATC | BamHI | Annealing to capsid |
NA, not applicable.
Capsid truncation mutants.
CapN was generated with the vector-specific forward primer AV11(F) and the reverse primer Cap312(R) by using pCMV5-CapE2SP (25) as a template. The resulting cDNA, which encodes the first 312 nucleotides of the capsid followed by a stop codon and a BamHI site, was digested with EcoRI and BamHI and then ligated into pCMV5 (2) that had been treated with EcoRI and BglII. CapC was generated with the forward primer Cap331(F) and the reverse primer Capsid(R) by using pCMV5-CapE2SP as the template. The resulting PCR product encodes a ribosome binding site followed by a translation start site, a c-myc tag, amino acids 107 to 300 of the capsid, the E2 signal peptide, and a stop codon followed by a BglII site. The PCR product was digested with EcoRI and BglII and ligated into pCMV5.
Capsid phosphorylation mutants.
All mutants were generated by PCR with pCMV5-CapE2SP (25) as the template. Four forward mutagenic primers [S45/46A(F), S52/56A(F), S45A(F), and S46A(F)], each specifying serine-to-alanine changes, were utilized in combination with the reverse primer CapSal(R). The resulting PCR products were digested with NotI and SalI and used to replace a 401-bp fragment in pCMV5-CapE2SP.
Introduction of capsid mutations into the RV infectious clone.
cDNA fragments encoding the desired mutants were subcloned into a plasmid encoding the entire rubella strain M33 genome (pBRM33) (46) by replacing the NotI and SphI fragments (nucleotides 6622 and 7242) within the capsid-encoding region.
GST-capsid fusion protein.
The capsid cDNA was amplified by PCR with primers CGSTF and AV10Cla by using pCMV5-CapE2SP as a template, digested with BamHI, and then ligated in frame to the 3′ end of the glutathione S-transferase (GST) cassette in the mammalian expression vector pEBG (31). Transcription of the GST fusion protein is driven by the EF-1α promoter.
Transfection of COS cells and 33P labeling of capsid.
COS cells (1.5 × 105) in 35-mm-diameter culture dishes were transfected with 2 μg of each capsid plasmid combined with 7 μl of PerFectin transfection reagent. The next day, cells were washed with phosphate-buffered saline (PBS) and then incubated with phosphate-free media (Life Technologies-Invitrogen, Inc.) for 30 min prior to the addition of 100 μCi of H333PO4 (4,000 Ci/mmol) (ICN). Cells were radiolabeled with phosphate for 12 to 14 h at 37°C, washed with cold PBS, and then lysed in RIPA buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing phosphatase inhibitors (1 mM sodium orthovanadate, 50 mM sodium fluoride, 5 mM tetrasodium pyrophosphate). Lysates were centrifuged at 17,000 × g for 10 min at 4°C to remove insoluble material, and the capsid was immunoprecipitated from the resulting supernatants with rabbit anti-capsid sera as described previously (5). All solutions contained phosphatase inhibitors. Immunocomplexes were denatured by heating at 95°C in 2× SDS gel loading buffer, separated by SDS-polyacrylamide gel electrophoresis (PAGE) on 10% acrylamide gels (23), and processed for fluorography.
In vitro RNA binding assay.
Capsid proteins were isolated from rubella virions, transfected COS cells, or infected cells by immunoprecipitation as described above. Virions were isolated from the precleared culture media of infected Vero cells by centrifugation at 100,000 × g for 60 min at 4°C. Where indicated, some capsid preparations were dephosphorylated prior to use in RNA binding experiments. Immunoprecipitated capsids were subjected to a final wash in dephosphorylation buffer (0.05 M Tris-HCl [pH 8.5], 1 mM EDTA) and then incubated for 12 to 15 h at 37°C in dephosphorylation buffer containing 100 U of calf intestine alkaline phosphatase (Roche Molecular Biochemicals). Samples were then separated by SDS-PAGE and transferred to 0.45-μm-pore-size nitrocellulose membranes (Bio-Rad Laboratories) by using a wet-transfer apparatus (Bio-Rad Laboratories) for 1 h at 280 mA.
Membranes were washed in probe buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 1× Denhardt's solution) for 10 min at room temperature followed by blocking in probe buffer containing 250 μg of baker's yeast tRNA (Roche Molecular Biochemicals)/ml for 1 h. The RNA probe used for Northwestern blots corresponded to nucleotides 1 to 4211 of the M33 genome sequence, a region that contains the RNA packaging signal (27). An EcoRI and EcoRV cDNA fragment from pBRM33 encompassing this region was ligated into the EcoRI and SmaI sites of pBluescriptSK+ (Stratagene). The resulting plasmid was linearized with BamHI and used as a template to generate noncapped RNA with the MEGAscript kit (Ambion, Inc.). Transcription reaction mixtures (20 μl) contained 1 μl of α-35S-labeled CTP (10 μCi, 600 Ci/mmol) (ICN). Membranes were incubated with 35S-labeled RV RNA for 1 h at room temperature in probe buffer and then washed in the same buffer three times at room temperature before exposure to a PhosphorImager.
In vitro dephosphorylation assays.
GST-capsid was transiently expressed in COS cells and labeled with [33P]orthophosphate as described above. Cells were lysed on ice with lysis buffer (1% NP-40, 350 mM NaCl, 2 mM EDTA, 50 mM Tris-Cl [pH 7.5]) plus phosphatase inhibitors. Radiolabeled GST-capsid was isolated on glutathione-Sepharose beads at 4°C. The beads were washed three times with cold lysis buffer and resuspended in a solution containing 50 mM Tris-Cl and 2 mM CaCl2 (pH 7.5). Aliquots of the beads were incubated with 1 μg of purified phosphatases (protein phosphatase 1A [PP1A], PP1B, PP2A, PP2B, or calf intestinal alkaline phosphatase) for 16 h at 37°C. Samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes before analysis on a PhosphorImager. Total levels of GST-capsid were determined by probing the membranes with rabbit anti-GST antibodies followed by enhanced chemiluminescence (ECL) detection.
Synthesis of infectious viral RNA and production of mutant viruses.
Full-length RV cDNA clones were linearized with HindIII and used as templates for transcription of capped RNAs with a mMessage mMachine kit (Ambion, Inc.). RNAs were quantitated by gel electrophoresis and spectroscopy at 260 nm before introduction into BHK cells by electroporation. Briefly, subconfluent BHK cells were trypsinized and then resuspended (107 cells/ml) in PBS. Wild-type and mutant viral RNAs (10 μg each) were added to 0.5-ml aliquots of BHK cell suspensions. The cell-RNA mixtures were transferred to a 0.2-mm-gap electroporation cuvette and incubated on ice for 10 min. Cells were electroporated at a 500-V 100-μF pulse in an Electro Cell Manipulator ECM600 (BTX Electronic Genetics). Immediately after the pulse, 1.0 ml of culture media was added to the cuvette. Cells were further diluted in 11.0 ml of culture media and distributed into six 35-mm-diameter culture dishes. Virus-containing culture media collected on consecutive days were clarified by centrifugation at 7,000 × g for 10 min before use or storage at −80°C.
Plaque assays and determination of cytopathic effect.
RK13 cells (2 × 105 cells) were infected with virus stocks in 35-mm-diameter dishes for 2 h, washed, and allowed to recover in culture media for 1 h. Cells were then overlaid with 0.5% agarose in culture medium and incubated at 35°C in a 5% CO2 atmosphere for 6 days. Plaques were visualized after staining with 4% neutral red solution (Sigma) for 3 h. The cytopathic effect was scored by examination of infected cultures by light microscopy after crystal violet staining. For staining, cells were washed with PBS and then fixed and stained with 0.05% crystal violet in 17% methanol for 2 h at room temperature. Excess stain was removed by washing the cells with distilled water. The stained cells were examined with a Zeiss Axioskop 2 microscope, and images were captured by using a Spot camera (Diagnostics Instruments, Inc.).
RESULTS
Identification of phosphorylated amino acid residues in capsid.
Previous work revealed that the RV capsid is phosphorylated prior to virus assembly (11). However, the significance of this posttranslational modification has been overlooked for most RNA viruses, including togaviruses. In order to determine if capsid phosphorylation is important for virus replication, it was necessary to map the phosphorylated amino acid residue(s) within this protein. Analysis of the capsid sequence with the NetPhos algorithm (http://www.cbs.dtu.dk/services/NetPhos/) revealed the presence of 10 potentially phosphorylated serine/threonine residues in the protein. Interestingly, six of these residues are clustered within the amino-terminally located RNA binding region (27). Two capsid constructs encoding the amino-terminal and carboxy-terminal regions of the capsid, CapN and CapC, respectively, were used to localize the phosphorylated amino acid residues within a specific region of the capsid (Fig. 1A). The proteins were transiently expressed in COS cells and immunoprecipitated from cells that had been labeled with H333PO4 for 12 h.
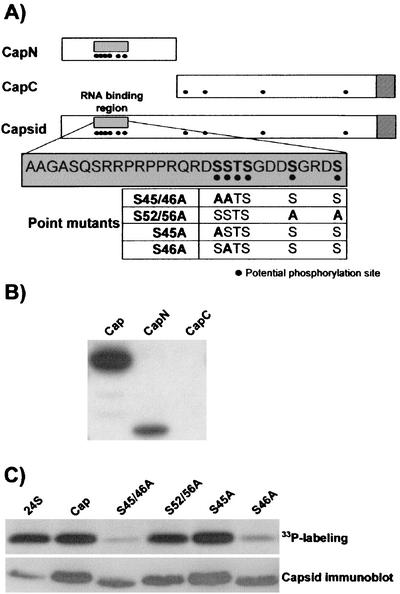
FIG. 1.
(A) Schematics of capsid constructs used in this study. CapN encompasses the amino-terminal 110 amino acids of the RV capsid protein. The genomic RNA binding region is contained in this construct (grey box). CapC contains an initiator methionine and a c-myc tag followed by amino acids 107 to 300 of the capsid, including the E2 signal peptide. The relative positions of potential phosphorylation sites are shown as black dots. Striped boxes at the ends of Capsid and CapC represent the signal peptide of the E2 glycoprotein. The sequence of the RNA binding region is shown in the expanded grey box, and potentially phosphorylated residues are shown in bold. A subset of the serine-to-alanine point mutants is shown at the bottom of the section. The mutants are named according to the amino acid number of the altered amino acids. For example, S46A is a mutant capsid protein in which serine 46 was changed to alanine. (B) Radioimmunoprecipitates prepared with rabbit anti-capsid or mouse anti-myc antibodies were subjected to SDS-PAGE and fluorography. Capsid and CapN, but not CapC, efficiently incorporate radiolabeled phosphate. The expression of each capsid construct was confirmed by immunoblotting (data not shown). (C) Radioimmunoprecipitates prepared from H333PO4-labeled cells expressing all RV structural proteins (24S), the capsid (Cap), or capsid proteins with point mutations within the RNA binding region were subjected to SDS-PAGE and fluorography (upper panel). Relative expression levels of the capsid proteins were determined by immunoblot analysis of the cell lysates (lower panel).
Both the capsid and CapN, which is the first 110 amino acids of the capsid, including the RNA binding domain, efficiently incorporated radiolabeled phosphate (Fig. 1B). In contrast, the CapC protein (amino acids 107 to 300) was not labeled with phosphate under these conditions. Immunoblot analysis confirmed that the CapC construct was stable and adequately expressed in the transfected cells (data not shown). Accordingly, the simplest interpretation of these results is that all of the phosphorylated residues in the capsid, or residues that are necessary for phosphorylation, are located within the first 110 amino acids of the protein. Conversely, the kinase that normally phosphorylates residues in the carboxy-terminal two-thirds of the protein may not recognize the truncated capsid (CapC) as a substrate. However, the results discussed below argue against this latter possibility.
Since there are a limited number of potential phosphorylation sites within the RNA binding region, we elected to use site-directed mutagenesis to identify which of the serine/threonine residues were essential for capsid phosphorylation. All six potential phosphorylated serine and threonine residues were mutated to alanine. The resulting mutant Cap-6S/T>A was expressed in transfected COS cells that were subsequently radiolabeled with phosphate as described above. Quantitation of the radioimmunoprecipitates with a PhosphorImager revealed that the amount of labeled phosphate incorporated by Cap-6S/T>A was approximately 50 times lower than that for the wild-type capsid (data not shown). This suggested that the majority of phosphorylation occurs within the RNA binding region of the capsid. Next, a series of single- and double-alanine mutants were constructed and analyzed in the same manner to determine which of the six serine/threonine residues in the RNA binding domain become phosphorylated (Fig. 1A). Mutation of serine 46 to alanine resulted in the most dramatic reduction in capsid phosphorylation (Fig. 1C). The loss of phosphorylation in S46A was comparable to that in the Cap-6S/T>A mutant (data not shown). In contrast, the mutation of serines 45, 52, and 56 (Fig. 1C) or threonine 47 or serine 48 (data not shown) to alanine had little or no effect on capsid phosphorylation. Low levels of 33P labeling were observed in all cases, which could mean that other amino acid residues in the capsid are phosphorylated, albeit at much lower levels. These results indicate that serine 46 of the capsid is critical for normal capsid phosphorylation.
Capsid phosphorylation is important for virus replication.
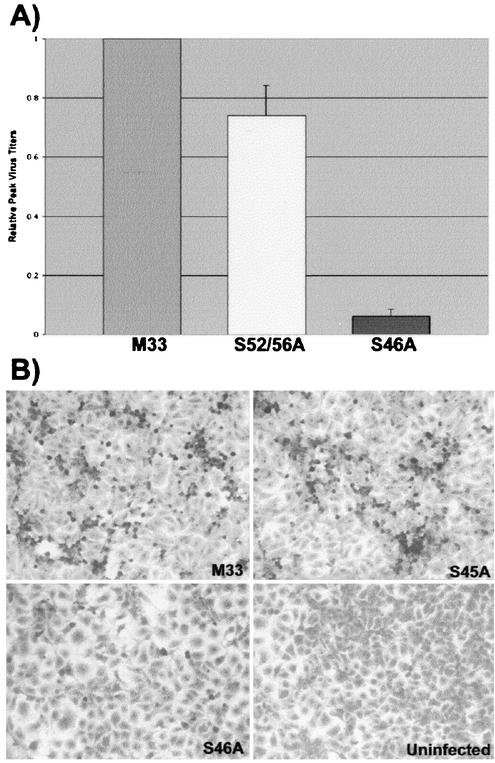
To examine the importance of capsid phosphorylation for virus replication, the serine-alanine mutants were expressed in the context of an infectious RV clone (46). Capped RV genomic RNAs from M33 and capsid mutant strains were synthesized in vitro, and equal amounts of RNA were electroporated into BHK cells. The media from transfected cells were collected at daily intervals, and virus titers were determined by plaque assay on RK13 cells. The titers for all viruses generally peaked between 2 and 4 days postelectroporation (data not shown). However, depending upon which day virus titers were determined, we found that titers of hypophosphorylated capsid-containing viruses were as much as 60-fold lower than those of virus strains with normally phosphorylated capsids. Figure 2A shows the relative virus titers on day 3 (postelectroporation), and it can be seen that titers of the S46A virus are more than 10-fold lower than those of wild-type M33 virus. In addition to the defect in virus replication, the cytopathic effect of mutant viruses was markedly reduced. RK13, a cell line that is exquisitely sensitive to the RV cytopathic effect, was infected with M33 and capsid mutant strains (at the same multiplicity of infection) and examined for the cytopathic effect after 2 to 4 days. Figure 2B clearly shows that the S46A strain is less cytopathic than the M33 and S45A strains. Similar results were obtained with other hypophosphorylated strains (data not shown). These results demonstrate that although capsid phosphorylation is not absolutely essential for virus replication, abolition of this process significantly impairs virus growth and reduces the cytopathic effects.
FIG. 2.
Reduction of capsid phosphorylation affects virus growth and cytopathogenicity. (A) Media from BHK cells expressing wild-type and mutant RV RNAs were harvested 3 days postelectroporation, and virus titers were quantitated by plaque assay with RK13 cells. Wild-type M33 titers were set to a value of 1, and mutant virus titers were normalized to this value. (B) RK13 cells were infected with M33, S45A, and S46A virus strains at a multiplicity of infection of 0.25. Four days postinfection, the cells were stained with crystal violet and photographed. Cells exhibiting the cytopathic effect are round and darkly stained.
Capsid phosphorylation negatively regulates RNA binding.
One of the functions of the capsid during virus assembly is to package genomic RNA into nucleocapsids. Since the bulk of phosphorylation occurs in the RNA binding region of the capsid, we reasoned that this posttranslational modification may regulate binding of genomic RNA and subsequent nucleocapsid assembly. The abilities of capsid phosphorylation mutants to bind viral RNA were tested by an in vitro RNA binding assay (27). Capsid proteins were immunoaffinity purified, separated by SDS-PAGE, and transferred to nitrocellulose membranes. The membranes were incubated with a radiolabeled RV-specific RNA probe that contains the capsid-binding packaging signal (27). These experiments revealed unexpectedly that hypophosphorylated capsids (S45/46A and S46A) bound RNA more efficiently than wild-type capsid or mutants (S52/56A and S45A) that retain normal levels of phosphorylation (Fig. 3A).
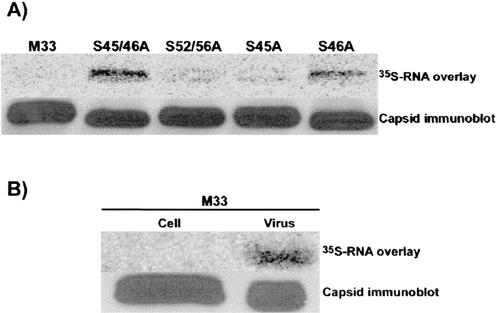
FIG. 3.
Phosphorylation of capsid negatively regulates RNA binding. (A) Capsid proteins were isolated from transfected COS cells by immunoaffinity purification, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were incubated with 35S-labeled RV-specific RNA and washed, and RNA binding to capsid proteins was detected with a PhosphorImager (upper panel). Relative capsid expression levels were determined by stripping the membranes and then reprobing them with a monoclonal antibody to capsid followed by ECL detection (lower panel). (B) Cell- and virion-associated capsids were isolated from cells infected with the M33 strain of RV. RNA binding to virion- and cell-associated capsids was determined as described for panel A.
Initially, these results were puzzling, since at some point, the RV capsid must interact with viral RNA to form nucleocapsids. In addition, a similar assay employed by Liu et al. was used to map the sites of interaction between capsid and genomic RNA (27). A critical difference between their experiments and ours was that the capsid protein used for the binding studies in the previous study was isolated from virions, whereas our assay employed capsid isolated from transfected cells. Similar to Liu et al., we observed that virion-derived wild-type capsid efficiently bound viral RNA (Fig. 3B). In contrast, capsid isolated from lysates of infected cells did not bind detectable amounts of RNA in the in vitro RNA binding assay.
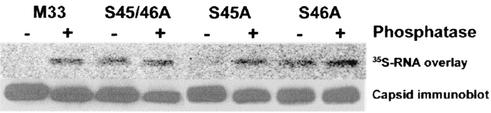
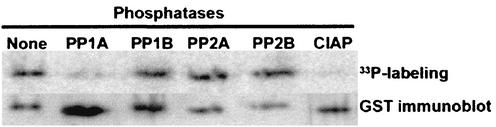
The results described above indicate that virion- and cell-associated capsids differ in some manner which influences their abilities to bind RNA. To ascertain whether differences in phosphorylation could account for these observations, we tested the ability of cell-associated capsids to interact with RNA after treatment with phosphatase. As expected, phosphatase treatment of the wild-type, S52/56A, and S45A capsids resulted in an increased binding of virus RNA (Fig. 4). However, the RNA-binding activities of the hypophosphorylated capsid mutants (S45/S46A and S46A) were not affected by phosphatase treatment. These results are consistent with a scenario where the capsid undergoes a dephosphorylation step prior to interacting with viral RNA and subsequent nucleocapsid formation. Based on sequence analysis, we hypothesized that serine 46 is a substrate for PP1A. Indeed, PP1A and the control phosphatase (calf intestinal alkaline phosphatase), but not other phosphatases (PP1B, PP2A, or PP2B), catalyzed removal of radiolabeled phosphate from the RV capsid in an in vitro phosphorylation assay (Fig. 5).
FIG. 4.
Dephosphorylated capsids exhibit higher affinities for virus genomic RNA. Purified capsids were treated with or without calf intestinal phosphatase, separated by SDS-PAGE, and then transferred to nitrocellulose membranes. The membranes were incubated with 35S-labeled RV-specific RNA and washed, and RNA binding to capsids was detected with a PhosphorImager (upper panel). Relative capsid expression levels were determined by stripping the membranes and then reprobing them with a monoclonal antibody to capsid followed by ECL detection (lower panel).
FIG. 5.
The capsid is dephosphorylated by PP1A in vitro. 33P-labeled GST-capsid was incubated with or without PP1A, PP1B, PP2A, PP2B, or calf intestinal phosphatase (CIAP) for 16 h at 37°C. Samples were then subjected to SDS-PAGE and transferred to a polyvinylidene difluoride membrane. The top panel shows 33P-labeled GST-capsid as detected by a PhosphorImager. The lower panel is an immunoblot with anti-GST antibody to show the total levels of GST-capsid protein in each lane.
DISCUSSION
Togaviruses have served as classical models for the study of virus assembly, replication, and entry into host cells (13, 41). However, other than ER- and Golgi-specific modifications of spike glycoproteins (16, 38, 39, 43), very little is known about posttranslational modification of virus antigens. Indeed, phosphorylation of virus structural proteins seems to have different effects depending upon the virus in question. In some cases, phosphorylation decreases the RNA binding activity of a given protein, whereas in other cases it has no effect (12, 20, 21, 24, 28, 45). The nonstructural protein NSP3 is modified by phosphorylation in a number of alphaviruses (26, 36), but to our knowledge, it is not known if alphavirus capsids are phosphorylated. Moreover, surprisingly little is known about how phosphorylation of virus proteins is regulated or if and how this modification is required for replication. This is particularly true of togaviruses. In the present study, we have identified serine 46 in the RNA binding region as the most important amino acid residue for phosphorylation of the RV capsid.
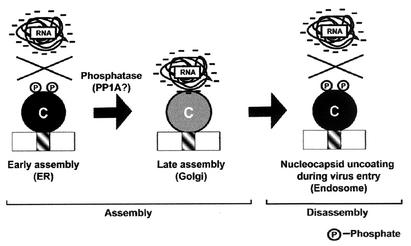
Our working model proposes that capsid phosphorylation negatively regulates its interaction with viral RNA early in the assembly pathway (Fig. 6). At a later step, dephosphorylation of the capsid is required before RNA binding and efficient nucleocapsid assembly can occur. Why is it important to regulate the RNA binding activity of the capsid? First, it is tempting to speculate that capsids which are free of RNA may be transported from the site of synthesis (ER) to the budding site (Golgi complex) with greater efficiency than fully formed nucleocapsids. In keeping with this hypothesis, previous studies showed that the capsid is still phosphorylated normally under conditions where its transport to the virus assembly site is blocked (11). Unlike alphavirus capsids, most RV capsid remains membrane bound (25, 42) and thus cannot simply move by diffusion from the site of synthesis to the budding site. Although it is not known how RV capsids are transported from the ER to the Golgi complex, vesicular transport is the most obvious mechanism. If this is the case, the assembly of bulky nucleocapsid structures on the ER surface would likely inhibit the recruitment of coat proteins needed for formation of transport vesicles that ferry the capsid to the Golgi complex. Second, in the early phase of replication, the ratio of virus genomic RNA to cellular RNA is presumed to be very low. Since nonphosphorylated capsid is a very basic protein, it may have a propensity to bind nonspecifically to nucleic acids with low affinity, a situation that could interfere with the binding of virus genomic RNA. The negative charge of the phosphate on serine 46 in the RNA binding region may prevent nonspecific electrostatic interaction with RNA. Alternatively, capsid phosphorylation may alter its conformation in a manner that inhibits RNA binding. For example, phosphorylation alters the conformation of the duck hepatitis virus capsid protein (47). However, we favor the first possibility because, based on analogy to alphavirus capsids, the RNA binding region of the RV capsid is located in a region of the protein which is structurally flexible (9).
FIG. 6.
Model for the roles of dynamic capsid phosphorylation in virus replication. During early assembly, the nascent capsid is phosphorylated on the surface of the ER. Phosphorylation serves to prevent nonspecific binding of cellular RNA and premature formation of nucleocapsids. Later in the replication cycle, structural proteins are transported to the Golgi complex where nucleocapsid assembly and virus budding takes place. The capsid is dephosphorylated by a Golgi-localized phosphatase (possibly PP1A) and is then able to bind viral RNA to form nucleocapsids. It is also possible that the capsid is dephosphorylated during transport to the Golgi complex. Upon virus entry, rephosphorylation of the capsid may promote the release of genomic RNA from the nucleocapsid.
The kinase(s) responsible for modification of capsid is currently unknown. Sequence analyses with algorithms such as Scansite (http://scansite.mit.edu/) or NetPhos (http://www.cbs.dtu.dk/services/NetPhos/) indicate that serine 46 is a potential substrate for cyclic AMP-dependent protein kinase (protein kinase A), Akt/protein kinase B, or calmodulin-dependent kinase. These kinases all have a broad range of substrates and are involved in aspects of cellular signaling. Their involvement in signaling processes necessitates that their activities or access to substrates is regulated. Restricting the localization of protein kinases is one way that eukaryotic cells modulate the activity of these enzymes (19, 32, 35). Differential localization of a given kinase can be mediated by binding to a variety of anchoring proteins, each with different targeting signals (18). Presumably, phosphorylation of nascent capsid is a process that occurs on the surface of the ER. With respect to protein kinase A and calmodulin-dependent kinase, both enzymes have been localized to the ER (22). However, we are unaware of any reports specifically documenting the association of active Akt/protein kinase B with the ER. Rather, activated forms of this kinase translocate to the plasma membrane after which they detach from this site and move to the nucleus (3).
The formation of RV nucleocapsids is generally coordinated with virus budding, and preformed nucleocapsids are rarely observed in RV-infected cells. In the context of our model (Fig. 6), this would require that capsid dephosphorylation occurs at the virus budding site and that nucleocapsid assembly is regulated by a Golgi-localized phosphatase. Thus, identification of the phosphatase that functions at the cellular level to dephosphorylate the capsid would provide insight into the control of virus assembly. In eukaryotes, the major groups of serine/threonine protein phosphatases display slightly different but sometimes overlapping preferences for substrates (reviewed in reference 6). For example, phosphorylated serines or threonines located downstream of multiple basic residues are preferred sites for PP1 and PP2A (1, 7). As serine 46 of the capsid is downstream of a series of arginines, it is a potential substrate for both of these enzymes. However, our results suggest that PP1A is the most likely candidate since this enzyme, not PP2A, efficiently dephosphorylates the capsid in vitro. In addition, PP1 isoforms are required for membrane trafficking steps near the RV budding site (37) and are thus in a position to dephosphorylate the RV capsid at the appropriate time and place. However, further studies are clearly required to verify this model.
We favor the theory that capsid phosphorylation is dynamic, a situation that could facilitate efficient binding of viral RNA and, just as importantly, release of RNA at the appropriate points in the replication cycle. During virus entry, phosphorylation of the capsid by a cellular kinase could destabilize the nucleocapsid, thereby mediating release of the genomic RNA (Fig. 6). This process would presumably permit better access of the genomic RNA to ribosomes, allowing translation of the nonstructural proteins to occur with greater efficiency. For example, encapsidated RNA of potato virus X is nontranslatable in vitro but can be rendered translatable after phosphorylation of the coat protein (4). In the rabies virus, viral transcription and replication are regulated by phosphorylation of the nucleoprotein (45). It will be important to determine whether capsid phosphorylation is a general mechanism that regulates nucleocapsid assembly for other positive-strand RNA viruses. We are currently studying this process by using a structurally related alphavirus. Finally, capsid phosphorylation might be important not only for virus replication but for virus-host interactions as well. RV strains with hypophosphorylated capsids are less cytopathic in certain cell types. In this respect, it will be of interest to determine whether the phosphorylation state of the capsid affects its interactions with host proteins.
Acknowledgments
We thank Shirley Gillam and Jerry Wolinsky for gifts of reagents and Marita Hobman for input on the manuscript. Margaret Hughes and Eileen Reklow are acknowledged for excellent technical assistance.
The work was funded by a grant to T.C.H. from the Canadian Institutes of Health Research. T.C.H. is the recipient of a Senior Medical Scholarship from the Alberta Heritage Foundation for Medical Research. L.M.J.L. and M.D.B. are supported by predoctoral studentship awards from the Alberta Heritage Foundation for Medical Research. M.D.B. is also the recipient of a predoctoral studentship award from the Canadian Institutes of Health Research.
REFERENCES
- 1.Agostinis, P., J. Goris, L. A. Pinna, F. Marchiori, J. W. Perich, H. E. Meyer, and W. Merlevede. 1990. Synthetic peptides as model substrates for the study of the specificity of the polycation-stimulated protein phosphatases. Eur. J. Biochem. 189:235-241. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, S., D. L. Davis, H. Dahlback, H. Jornvall, and D. W. Russell. 1989. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 264:8222-8229. [PubMed] [Google Scholar]
- 3.Andjelkovic, M., D. R. Alessi, R. Meier, A. Fernandez, N. J. Lamb, M. Frech, P. Cron, P. Cohen, J. M. Lucocq, and B. A. Hemmings. 1997. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 272:31515-31524. [DOI] [PubMed] [Google Scholar]
- 4.Atabekov, J. G., N. P. Rodionova, O. V. Karpova, S. V. Kozlovsky, V. K. Novikov, and M. V. Arkhipenko. 2001. Translational activation of encapsidated potato virus X RNA by coat protein phosphorylation. Virology 286:466-474. [DOI] [PubMed] [Google Scholar]
- 5.Beatch, M. D., and T. C. Hobman. 2000. Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J. Virol. 74:5569-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, P. T. 1997. Novel protein serine/threonine phosphatases: variety is the spice of life. Trends Biochem. Sci. 22:245-251. [DOI] [PubMed] [Google Scholar]
- 7.Donella-Deana, A., M. H. Krinks, M. Ruzzene, C. Klee, and L. A. Pinna. 1994. Dephosphorylation of phosphopeptides by calcineurin (protein phosphatase 2B). Eur. J. Biochem. 219:109-117. [DOI] [PubMed] [Google Scholar]
- 8.Duncan, R., A. Esmaili, L. J. Law, S. Bertholet, C. Hough, T. C. Hobman, and H. L. Nakhasi. 2000. Rubella virus capsid protein induces apoptosis in transfected RK13 cells. Virology 275:20-29. [DOI] [PubMed] [Google Scholar]
- 9.Forsell, K., M. Suomalainen, and H. Garoff. 1995. Structure-function relation of the NH2-terminal domain of the Semliki Forest virus capsid protein. J. Virol. 69:1556-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey, T. K. 1994. Molecular biology of rubella virus. Adv. Virus Res. 44:69-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garbutt, M., L. J. Law, H. Chan, and T. C. Hobman. 1999. Role of rubella virus glycoprotein domains in assembly of virus-like particles. J. Virol. 73:3524-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazina, E. V., J. E. Fielding, B. Lin, and D. A. Anderson. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helenius, A., and M. Marsh. 1982. Endocytosis of enveloped animal viruses. Ciba Found. Symp. 92:59-76. [DOI] [PubMed] [Google Scholar]
- 14.Hinman, A. R., B. S. Hersh, and C. A. de Quadros. 1998. Rational use of rubella vaccine for prevention of congenital rubella syndrome in the Americas. Rev. Panam. Salud Publica 4:156-160. [DOI] [PubMed] [Google Scholar]
- 15.Hobman, T. C. 2002. Rubella, p. 2849-2852. In T. E. Creighton (ed.), Wiley encyclopedia of molecular medicine. John Wiley & Sons, Inc., New York, N.Y.
- 16.Hobman, T. C., M. L. Lundstrom, and S. Gillam. 1990. Processing and transport of rubella virus structural proteins in COS cells. Virology 178:122-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobman, T. C., L. Woodward, and M. G. Farquhar. 1993. The rubella virus E2 and E1 spike glycoproteins are targeted to the Golgi complex. J. Cell Biol. 121:269-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, L. J., L. Wang, Y. Ma, K. Durick, G. Perkins, T. J. Deerinck, M. H. Ellisman, and S. S. Taylor. 1999. NH2-terminal targeting motifs direct dual specificity A-kinase-anchoring protein 1 (D-AKAP1) to either mitochondria or endoplasmic reticulum. J. Cell Biol. 145:951-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubbard, M. J., and P. Cohen. 1993. On target with a new mechanism for the regulation of protein phosphorylation. Trends Biochem. Sci. 18:172-177. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov, K. I., P. Puustinen, A. Merits, M. Saarma, and K. Makinen. 2001. Phosphorylation down-regulates the RNA binding function of the coat protein of potato virus A. J. Biol. Chem. 276:13530-13540. [DOI] [PubMed] [Google Scholar]
- 21.Kann, M., and W. H. Gerlich. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol. 68:7993-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosmopoulou, I., G. Koliakos, C. Haitoglou, D. Christodoulou, A. Dimitriadou, and A. Trakatellis. 1994. Rat liver endoplasmic reticulum protein kinases. Int. J. Biochem. 26:403-414. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lan, Y. T., J. Li, W. Liao, and J. Ou. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342-348. [DOI] [PubMed] [Google Scholar]
- 25.Law, L. M., R. Duncan, A. Esmaili, H. L. Nakhasi, and T. C. Hobman. 2001. Rubella virus E2 signal peptide is required for perinuclear localization of capsid protein and virus assembly. J. Virol. 75:1978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, G. P., M. W. La Starza, W. R. Hardy, J. H. Strauss, and C. M. Rice. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179:416-427. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Z., D. Yang, Z. Qiu, K.-T. Lim, P. Chong, and S. Gillam. 1996. Identification of domains in rubella virus genomic RNA and capsid protein necessary for specific interaction. J. Virol. 70:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maroto, B., J. C. Ramirez, and J. M. Almendral. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J. Virol. 74:10892-10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marr, L. D., A. Sanchez, and T. K. Frey. 1991. Efficient in vitro translation and processing of the rubella virus structural proteins in the presence of microsomes. Virology 180:400-405. [DOI] [PubMed] [Google Scholar]
- 30.McCready, T. L., B. F. Islam, F. J. Schmitz, H. A. Luu, J. F. Dawson, and C. F. Holmes. 2000. Inhibition of protein phosphatase-1 by clavosines A and B. Novel members of the calyculin family of toxins. J. Biol. Chem. 275:4192-4198. [DOI] [PubMed] [Google Scholar]
- 31.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mochly-Rosen, D. 1995. Localization of protein kinases by anchoring proteins: a theme in signal transduction. Science 268:247-251. [DOI] [PubMed] [Google Scholar]
- 33.Oker-Blom, C. 1984. The gene order for rubella virus structural proteins is NH2-C-E2-E1-COOH. J. Virol. 51:354-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oker-Blom, C., N. Kalkkinen, L. Kaariainen, and R. F. Pettersson. 1983. Rubella virus contains one capsid protein and three envelope glycoproteins, E1, E2a, and E2b. J. Virol. 46:964-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawson, T., and J. D. Scott. 1997. Signaling through scaffold, anchoring, and adaptor proteins. Science 278:2075-2080. [DOI] [PubMed] [Google Scholar]
- 36.Peranen, J. 1991. Localization and phosphorylation of Semliki Forest virus non-structural protein nsP3 expressed in COS cells from a cloned cDNA. J. Gen. Virol. 72:195-199. [DOI] [PubMed] [Google Scholar]
- 37.Peters, C., P. D. Andrews, M. J. Stark, S. Cesaro-Tadic, A. Glatz, A. Podtelejnikov, M. Mann, and A. Mayer. 1999. Control of the terminal step of intracellular membrane fusion by protein phosphatase 1. Science 285:1084-1087. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt, M. F. 1982. Acylation of viral spike glycoproteins: a feature of enveloped RNA viruses. Virology 116:327-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sefton, B. M. 1975. Virus-dependent glycosylation. J. Virol. 17:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semerikov, V. V., I. N. Lavrentyeva, V. F. Popov, M. A. Fletcher, and M. E. Kolotov. 2000. Rubella in the Russian federation: epidemiological features and control measures to prevent the congenital rubella syndrome. Epidemiol. Infect. 125:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons, K., H. Garoff, and A. Helenius. 1982. How an animal virus gets into and out of its host cell. Sci. Am. 246:58-66. [DOI] [PubMed] [Google Scholar]
- 42.Suomalainen, M., H. Garoff, and M. D. Baron. 1990. The E2 signal sequence of Rubella virus remains part of the capsid protein and confers membrane association in vitro. J. Virol. 64:5500-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waxham, M. N., and J. S. Wolinsky. 1985. A model of the structural organization of rubella virus virions. J. Infect. Dis. 7:S133-S139. [DOI] [PubMed] [Google Scholar]
- 44.Wolinsky, J. 1996. Rubella, p. 899-929. In B. N. Fields et al. (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 45.Wu, X., X. Gong, H. Foley, M. Schnell, and Z. Fu. 2002. Both viral transcription and replication are reduced when the rabies virus nucleoprotein is not phosphorylated. J. Virol. 76:4153-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao, J., and S. Gillam. 1999. Mutational analysis, using a full-length rubella virus cDNA clone, of rubella virus E1 transmembrane and cytoplasmic domains required for virus release. J. Virol. 73:4622-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, M., and J. Summers. 1994. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J. Virol. 68:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]