Abstract
The Bo17 gene of bovine herpesvirus 4 (BoHV-4) is the only viral gene known to date that encodes a homologue of the cellular core 2 β-1,6-N-acetylglucosaminyltransferase-mucin type (C2GnT-M). To investigate the origin and evolution of the Bo17 gene, we analyzed its distribution among BoHV-4 strains and determined the sequences of Bo17 from nine representative strains and of the C2GnT-M gene from six species of ruminants expected to encompass the group within which the gene acquisition occurred. Of 34 strains of BoHV-4, isolated from four different continents, all were found to contain the Bo17 gene. Phylogenetic analyses indicated that Bo17 was acquired from a recent ancestor of the African buffalo, implying that cattle subsequently acquired BoHV-4 by cross-species transmission. The rate of synonymous nucleotide substitution in Bo17 was estimated at 5 × 10−8 to 6 × 10−8 substitutions/site/year, consistent with previous estimates made under the assumption that herpesviruses have cospeciated with their hosts. The Bo17 gene acquisition was dated to around 1.5 million years ago. Bo17 sequences from BoHV-4 strains from African buffalo and from cattle formed two separate clades, estimated to have split about 700,000 years ago. Analysis of the ratio of nonsynonymous to synonymous nucleotide substitutions revealed a burst of amino acid replacements subsequent to the transfer of the cellular gene to the viral genome, followed by a return to a strong constraint on nonsynonymous changes during the divergence of contemporary BoHV-4 strains. The Bo17 gene represents the most recent of the known herpesvirus gene acquisitions and provides the best opportunity for learning more about this important process of viral evolution.
Members of the Herpesviridae form an extensive family of large DNA viruses whose natural hosts are humans, other mammals and vertebrates, and in one described case, an invertebrate (25). The Herpesviridae family has been divided into three subfamilies, the Alpha-, Beta-, and Gammaherpesvirinae. This subdivision was based initially on biological properties, but has subsequently been confirmed by molecular phylogenetic analyses, which have also suggested that members of the Herpesviridae family have generally evolved by cospeciation with their hosts (5, 19-21). A conserved core set of genes can be identified in all herpesvirus genomes (21). In addition, different herpesviruses contain a diverse collection of genes with homology to genes found in vertebrate genomes, which appear to have been acquired by herpesviruses from their hosts at different times in the past (24). These genes appear to have played an important role in the diversification and adaptation of herpesviruses. In principle, phylogenetic analysis of a herpesvirus gene and its cellular homologue from potential host species could provide information on the likely origin of the viral gene, the approximate date of viral gene acquisition, and the nature of any changes in the evolution of the gene subsequent to entering the virus genome. In practice, this type of analysis has never been completed for any herpesvirus cellular gene homologue because (i) homologous gene sequence data are not available for a sufficient number of host species closely related to the potential source species and (ii) in many cases, the gene acquisition appears to have been quite ancient and the host and viral sequences are now highly divergent.
Bovine herpesvirus 4 (BoHV-4) is a gammaherpesvirus which has been isolated throughout the world from healthy cattle as well as from those exhibiting a variety of diseases (29). Isolates of BoHV-4 have also been recovered from other ruminant species such as American bison (Bison bison) (32), African buffalo (Syncerus caffer) (26), and sheep (36). Sporadic isolations were reported in lions, cats (9), and owl monkeys (Aotus trivirgatus) (1). BoHV-4 has a B-type genome structure consisting of a long unique region (LUR) flanked by polyrepetitive DNA (prDNA) elements. Recently, we showed that the Bo17 gene located at the right end of the BoHV-4 LUR encodes a functional homologue of the cellular core 2 β-1,6-N-acetylglucosaminyltransferase-mucin type (C2GnT-M) (35, 40). C2GnT-M is a member of a family of homologous cellular enzymes playing crucial roles in glycan biosynthesis. This family of enzymes is involved in important physiological and pathological processes, such as development, immunodeficiency, and oncogenesis (11). The Bo17 gene of BoHV-4 is the only viral gene known to date that encodes a homologue of the β-1,6-N-acetylglucosaminyltransferase family (35).
In the present study, we investigated the origin and evolution of the Bo17 gene by comparing its sequence among various BoHV-4 strains and the sequences of its cellular homologue for a group of ruminant species encompassing the host lineage from which the viral gene may have been acquired. This enables us to identify the source of this herpesvirus cellular gene homologue, estimate the time when the gene acquisition occurred, and examine how the pattern of evolution of the gene has changed subsequent to its arrival in the viral genome. In addition, we were able to estimate the rate of nucleotide substitution in herpesviruses without making the assumption of host-virus cospeciation.
MATERIALS AND METHODS
Virus strains.
Thirty-four BoHV-4 strains isolated throughout the world and from different animal species were used in this study (Table 1).
TABLE 1.
BoHV-4 strains used in this study
| Strain(s)a | Host species | Reference or source | Country of isolation | Restriction profileb |
|---|---|---|---|---|
| V. test | Cattle (B. taurus) | 30 | Belgium | Movar-like |
| LVR140 | Cattle (B. taurus) | 37 | Belgium | Movar-like |
| LVR 260, LVR 500, MVR 706, MVR 732, OVR 610, QVR 2348, QVR 3138, QVR 3140, PVR 2475, AVR 1478, BVR 318, 31788, 99/2823, 99/3114, 00/2776, 00/3847, 00/6568 | Cattle (B. taurus) | C. Letellier (Cerva) | Belgium | Movar-like |
| MOVAR | Cattle (B. taurus) | 2 | Hungary | Movar-like |
| HB 420 | African lion (Panthera leo) | 7 | Hungary | Movar-like |
| Skin lesion | Cattle (B. taurus) | L. Egyed (Veterinary Medical Research Institute) | Hungary | Movar-like |
| Taiwan | Cattle (B. taurus) | 16 | Taiwan | Movar-like |
| FHV-2 | Cat (Felis sylvestris domesticus) | 9 | United States | Movar-like |
| DN599 | Cattle (B. taurus) | 22 | United States | DN599-like |
| 66-p-347 | American bison (B. bison) | 32 | United States | DN599-like |
| HVA-2 | Owl monkey (A. trivirgatus) | 1 | United States | DN599-like |
| M40 | Zebu (Bos indicus) | 23 | India | Unclassifiable |
| 108, 128, 130, BK6, B34, Buf. | African buffalo (S. caffer) | 26 | Kenya | Unclassifiable |
The BoHV-4 strains used in this study were isolated in different countries and from various host species. The names listed are those given to the BoHV-4 isolates in the original publications. The strains for which the Bo17 sequence was determined in the present study are presented in boldface type.
Three groups of BoHV-4 strains can be defined based on restriction profiles: the Movar-like strains, the DN599-like strains, and the unclassifiable strains.
DNA of ruminants.
Bovine (Bos taurus) DNA extracted from calf thymus was purchased from Sigma. DNAs from Yak (Bos grunniens) and African buffalo (Syncerus caffer caffer) were kindly provided by O. Hanotte (International Livestock Research Institute, Nairobi, Kenya). Sheep (Ovis aries) DNA was a gift from C. Charlier (University of Liège, Liège, Belgium). DNAs from red deer (Cervus elaphus) and giraffe (Giraffa camelopardalis) were extracted from subcutaneous tissues with the Easy-DNA kit (Invitrogen).
Amplification and sequencing of Bo17 and C2GnT-M genes.
The BoHV-4 Bo17 open reading frame (ORF) was amplified by PCR with the forward primer 5′-CAAGAGTGCTTGTACCAATGC-3′ and one of the following reverse primers, 5′-CTGGGCATGTTGGCACAGTCC-3′ or 5′-GTGGAGCTCCAAGAGACATG-3′, according to the viral strain. The entire C2GnT-M ORF from the bovine, yak, and African buffalo was amplified by PCR with the forward primer 5′-CTCTACTGATCTCCCATCCC-3′ and the reverse primer 5′-CCAGTACGCCCTGGGCATATT-3′, which hybridize upstream and downstream of the ORF, respectively. Two partially overlapping PCR products were generated to cover the entire sheep C2GnT-M ORF. The first was generated with the forward primer 5′-TCCTCCTCCACCTTGCCTGTGC-3′ and the reverse primer 5′-TCAAAGTTCAGTCCCATAGAT-3′, which hybridize upstream of and inside the ORF, respectively. The second amplicon was produced with the forward primer 5′-ATAGCCTGAAGCTGCCA-3′ and the reverse primer 5′-CTGGGCATGTTGGCACAGTCC-3′, which hybridize inside and downstream of the ORF, respectively. Two partially overlapping PCR products were also generated to cover the red deer and giraffe C2GnT-M ORFs. A first product was generated with the forward primer 5′-ATGAAGATGGCTGGGT-3′ and the reverse primer 5′-TCAAAGTTCAGTCCCATAGAT-3′, which correspond to the first 16 and the last 21 bases of the ORF, respectively. A second PCR product was generated with the forward primer 5′-ATAGCCTGAAGCTGCCA-3′ for the red deer or the forward primer 5′-GGCCCTCAAGATGTTGA-3′ for the giraffe and the partially degenerate reverse primer 5′-CTGGGCATGTTGGYACMWGNIN-3′. These forward primers and this reverse primer hybridize inside and downstream of the ORF, respectively. PCR products were sequenced on an ABI 377 automated sequencer (PE Biosystems).
Sequence analysis.
Sequences were aligned by eye. Ka and Ks, the numbers of nonsynonymous and synonymous substitutions per site, were estimated by using the method of Li (15). Phylogenetic trees were estimated by the neighbor-joining (NJ) (27), maximum parsimony (MP), and maximum likelihood (ML) methods. The NJ method was used with DNA distances corrected by Kimura's two-parameter method and with protein distances corrected by Kimura's empirical approach, as implemented in CLUSTAL W (31). For Ka and Ks distances, the NJ method was implemented by using NEIGHBOR in the Phylip package (J. Felsenstein [http://evolution.genetics.washington.edu/phylip.html]). The MP method was applied to DNA and protein sequences by using DNAPARS and PROTPARS in the Phylip package. The ML method was applied to DNA sequences by using DNAML in the Phylip package; the transition-transversion ratio was optimized at 2.4. The ML method was applied to protein sequences by using PROTML from the MOLPHY package (J. Adachi and M. Hasegawa [http://www.ism.ac.jp/software/ismlib/softother.e.html#molphy]) and the Jones-Taylor-Thornton model of amino acid replacement. These various methods were applied to 1,000 bootstrap replicates generated by SEQBOOT in the Phylip package.
Southern blotting.
Southern blot analysis of viral DNA digested with EcoRI was performed as described previously (34) by using the Bo17 ORF of the BoHV-4 V. test strain as a probe.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the GenBank database. The sequences of the BoHV-4 Bo17 ORF have been determined for the following strains (accession numbers are in parentheses): DN599 (AF465332), LVR140 (AF465330), Movar (AF465331), M40 (AY143158), 108 (AY143155), 130 (AY143156), and Buf. (AY143157). The sequences of the C2GnT-M ORF have been determined for the following members of the Ruminantia (accession numbers are in parentheses): bovine (AF465338), yak (AF465336), African buffalo (AF465337), sheep (AF465335), red deer (AF465333), and giraffe (AF465334). The following previously published sequences were used in this study (accession numbers are in parentheses): the human C2GnT-M ORF (AF102542) (39), the BoHV-4 Bo17 ORF of the V. test strain (AF231105) (35), and the 66-p-347 strain (AF318573) (40).
RESULTS
Bo17 sequence variation among nine BoHV-4 strains.
The goal of the present study was to investigate the origin and evolution of the BoHV-4 Bo17 gene. Since the sequence of Bo17 has been determined for only two strains, V. test (35) and 66-p-347 (40), we first sequenced the Bo17 ORF from seven additional strains in order to estimate the conservation of the gene among BoHV-4 strains. The seven strains were selected among the three groups of BoHV-4 strains that can be defined based on their restriction profiles (Table 1). For the seven strains, the Bo17 gene was amplified by PCR, and the resulting products were sequenced. The same approach was applied to the V. test and 66-p-347 strains, and it confirmed the previously published sequences.
Comparisons of the nine sequences (Table 2) revealed that they fall into two distinct groups, one comprised of the three strains from African buffalo (108, 130, and Buf.) and the other including four strains from taurine cattle (V. test, MOVAR, LVR140, and DN599), one from zebu (M40), and one from bison (66-p-347) (Table 1). The sequence of strain 66-p-347 from bison was identical to the American cattle strain DN599, probably indicating a recent transmission to bison. Two of the three European cattle strains, LVR 140 and MOVAR, were also identical to each other and differed from the third (V. test) at only two nucleotides. The American and European cattle strains differed at 11 to 13 sites, the zebu strain (M40) differed from all cattle strains at 12 to 14 sites, and there were 32 to 45 differences between strains from this group and the African buffalo strains.
TABLE 2.
Bo17 sequence differences among BoHV-4 strainsa
| Strain | No. of amino acid or nucleotide replacements with strain:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 66-p-347 | DN599 | LVR140 | MOVAR | V-test | M40 | 130 | 108 | Buf. | |
| 66-p-347 | 0 | 5 | 5 | 6 | 7 | 12 | 13 | 14 | |
| DN599 | 0 | 5 | 5 | 6 | 7 | 12 | 13 | 14 | |
| LVR140 | 11 | 11 | 0 | 1 | 6 | 13 | 14 | 15 | |
| MOVAR | 11 | 11 | 0 | 1 | 6 | 13 | 14 | 15 | |
| V-test | 13 | 13 | 2 | 2 | 7 | 14 | 15 | 16 | |
| M40 | 13 | 13 | 14 | 14 | 12 | 12 | 13 | 14 | |
| 130 | 35 | 35 | 40 | 40 | 38 | 32 | 1 | 6 | |
| 108 | 40 | 40 | 45 | 45 | 43 | 37 | 9 | 7 | |
| Buf. | 37 | 37 | 42 | 42 | 40 | 34 | 14 | 19 | |
Values above and below the diagonal refer to amino acid and nucleotide sequence differences, respectively. Pairwise sequence comparisons were made across the 426 amino acid residues or 1,280 nucleotides known for all nine sequences.
The extent of nonsynonymous nucleotide substitution, scaled by that of synonymous substitution (Ka/Ks), can be used to examine the constraint on a protein sequence. In comparisons between the groups of BoHV-4 sequences, the Ka/Ks ratio was 0.19. This is similar to that seen among the C2GnT-M sequences of ruminants (see below), indicating that nonsynonymous substitutions have continued to be constrained in the Bo17 gene after the viral gene acquisition event and implying that Bo17 encodes a function that is now important for BoHV-4.
Evolution of the BoHV-4 Bo17 gene.
The Bo17 gene has only been reported in BoHV-4. Although a number of other gammaherpesviruses are known, none are closely related to BoHV-4 (5, 19). Thus, it is not clear how recently an ancestor of BoHV-4 acquired this gene or from what source. Since in many instances herpesviruses appear to have coevolved with their hosts (21), to elucidate the origin of the BoHV-4 Bo17 gene we first sequenced the C2GnT-M gene of B. taurus. The predicted bovine C2GnT-M protein sequence differed from the BoHV-4 sequences by 5 to 6% (depending on the viral strain) of amino acid residues compared to a 16.5% difference between the bovine and human sequences and a 17.5% difference between the human and BoHV-4 sequences. This finding indicates that the BoHV-4 Bo17 gene was acquired from an artiodactyl, but it is unclear how long ago. Using either DNA or protein sequences, simple phylogenetic analyses (such as the least-squares method [10] applied to the Poisson corrected protein sequence distances) suggested that the branch length from the bovine sequence to the common ancestor (CA) of the bovine and viral genes was about 12% of the total distance between the bovine and human genes. Assuming an approximate molecular clock, if the divergence between the lineages leading to artiodactyls and primates occurred 80 to 100 million years ago, the CA of the bovine and viral genes existed 19 to 24 million years ago. That time scale approximates estimates (14) of 20 million years for the CA of Bovinae (e.g., cattle) and Caprinae (e.g., sheep) and 23 million years for the CA of the Bovoidea (Bovinae plus Caprinae) and Cervoidea (e.g., deer). However, the same analysis indicated that the branch length from the CA to BoHV-4 was less than two times that from the CA to bovine gene, a surprising finding given that the viral gene might be expected to evolve much faster than the bovine gene. These observations suggested either that the Bo17 gene was acquired from another lineage within the Bovoidea (i.e., not a direct ancestor of B. taurus), so that much of the branch from the CA to BoHV-4 occurred before the viral gene acquisition, or that unequal rates of evolution were disrupting the analysis.
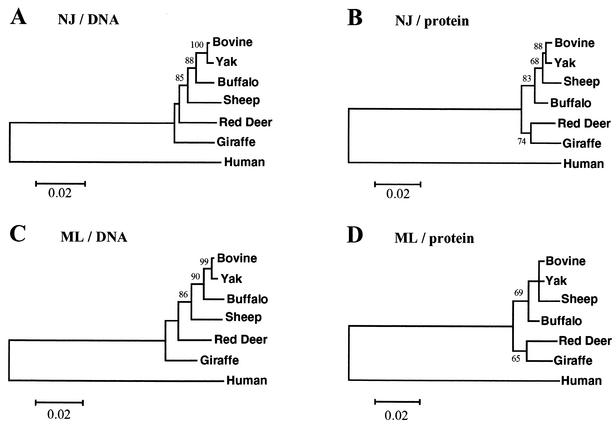
To examine this more closely, we next determined the C2GnT-M sequence from five other species of the Ruminantia selected to represent lineages that diverged from that leading to B. taurus at a range of times in the past. In order of increasing divergence from B. taurus, the chosen species were yak (B. grunniens), African buffalo (S. caffer caffer), sheep (O. aries), red deer (C. elaphus), and giraffe (G. camelopardalis); the various CAs of these species and B. taurus are estimated to have existed between about 1 and 25 million years ago (12, 18). Phylogenetic analyses of the DNA sequences of these ruminant genes, with the human sequence as an out-group, yielded the expected topology with both the NJ and ML methods (Fig. 1A and C). However, the results of the analyses of protein sequences differed (Fig. 1B and D). Both the NJ and ML methods yielded trees in which the buffalo was erroneously placed outside a clade comprising the bovine, yak, and sheep, in which the red deer and giraffe formed a monophyletic clade. This suggests that DNA sequences with their additional information from synonymous changes are more reliable phylogenetic indicators for these data.
FIG. 1.
Phylogenetic relationships among C2GnT-M sequences from ruminants, with the human gene as the out-group, as estimated by NJ (A and B) and ML (C and D) analysis of DNA (A and C) and protein (B and D) sequences. Values on internal branches refer to the percentage of bootstrap replicates in which the branch was found; only values greater than 60% are shown. Horizontal branch lengths are drawn to scale, with the bars indicating 0.02 nucleotide (A and C) or amino acid (B and D) replacements per site.
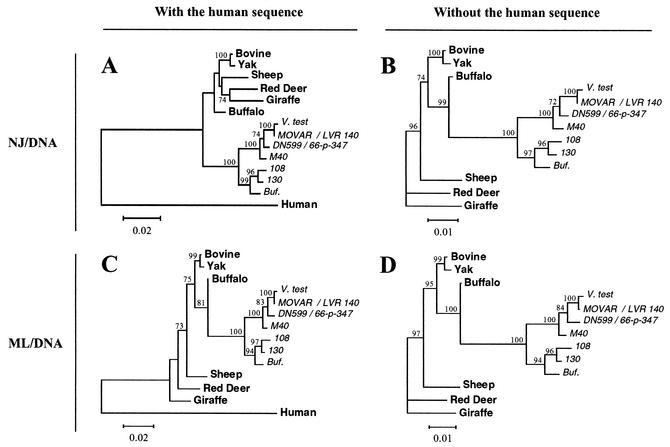
When the BoHV-4 sequences were added to the phylogenetic analyses, they formed a strongly supported monophyletic clade, but the position at which this lineage branched from the mammalian tree varied depending on the methodology used. NJ analysis of DNA sequences (including the human sequence) produced a tree in which the BoHV-4 lineage joined the branch before the CA of the six ruminants (Fig. 2A). However, this result is clearly unreliable because the branching order among the ruminants does not match their known relationships (Fig. 1A and C). Most strikingly, the expected clustering of the buffalo with the bovine and yak (12, 18), which was strongly supported (by 88% of bootstrap replicates) in the analysis without the viral sequences (Fig. 1A), was no longer apparent; instead, the buffalo appeared as an out-group to the other five ruminants (Fig. 2A). In contrast, ML analysis of DNA sequences (including the human sequence) yielded a tree in which the correct relationships among the ruminants were preserved and the BoHV-4 lineage joined the buffalo branch (Fig. 2C). Another obvious difference between the NJ and ML results concerned the distances from the CA of the ruminant and viral sequences to the tips of the branches within the ruminant and viral lineages. In the NJ tree, the viral branches are barely longer than those of the red deer and giraffe (Fig. 2A), whereas in the ML tree, the viral branches extend much further than those of the ruminants (Fig. 2C), as is expected if the viral genes have evolved more rapidly.
FIG. 2.
Phylogenetic relationship of the BoHV-4 Bo17 gene to ruminant C2GnT-M DNA sequences as estimated by NJ (A and B) and ML (C and D) analysis with (A and C) or without (B and D) the human sequence as the out-group. BoHV-4 strains are presented in italics. Values on internal branches refer to the percentage of bootstrap replicates in which the branch was found; only values greater than 60% are shown. The relationship shown in panel A is inferred to be incorrect (see text). Horizontal branch lengths are drawn to scale, with the bars indicating 0.02 (A and C) or 0.01 (B and D) nucleotide replacements per site.
Many methods for phylogenetic reconstruction can be inaccurate when rates of evolution vary substantially among different lineages, with the most-frequent problem being one of long-branch attraction. The present sequences may be susceptible to this problem because the expected faster rate of evolution of the viral genes leads to one long branch while the distance to the human out-group sequence provides a second. When the phylogenetic analyses were repeated excluding the human sequence, and rooted using the giraffe as the out-group, both the NJ (Fig. 2B) and ML (Fig. 2D) methods yielded trees in which the correct ruminant relationships were recovered and the BoHV-4 lineage clustered with the buffalo, as shown in Fig. 2C. Comparison of Fig. 2A and B reveals that, apart from the presence of the human branch in Fig. 2A, and the root imposed by it, the topologies are identical. Rerooting the tree in Fig. 2A with the giraffe as the out-group shows that the human branch has been added to the BoHV-4 lineage, presumably by long-branch attraction.
In the ML analysis including the human sequence (Fig. 2C), the buffalo-BoHV-4 clade was strongly supported (81% of bootstraps), but it is interesting that the other 19% of bootstrap replicates placed the BoHV-4 lineage outside that of the ruminants (as shown in Fig. 2A), indicating that the ML analysis was also at least partly susceptible to the long-branch attraction problem. The MP method is generally thought to be the most sensitive to long-branch attraction, and so it is interesting that MP analysis of DNA sequences (including the human sequence) found the same topology as ML analysis (Fig. 2C). However, earlier MP analyses, when only three BoHV-4 sequences (66-p-347, V. test, and LVR 140) were available, placed the viral clade outside that of the ruminants (as shown in Fig. 2A). Phylogenetic analyses of a protein sequence alignment including the viral sequences are not shown in detail because of the problems in obtaining the correct tree for the ruminants (Fig. 1B and D). However, with respect to the position of the viral clade, the results were the same as for the analyses of DNA sequences (Fig. 2), again supporting the clustering of BoHV-4 with the buffalo gene.
The clustering of the BoHV-4 lineage with the buffalo gene was supported by 99 to 100% of bootstrap replicates when the human sequence was excluded (Fig. 2B and D). This provides very strong evidence that the Bo17 gene was derived by acquisition of the C2GnT-M gene of the African buffalo or its recent ancestor. The acquisition event occurred substantially later than the CA of Syncerus and Bos, which has been estimated at 12 to 14 million years ago (12). There are 8 nucleotide differences (including 5 synonymous differences) shared by the buffalo and BoHV-4 sequences, which are inferred to represent substitutions on the branch leading to their CA. In contrast, there is only a single substitution (a synonymous transversion) unique to the buffalo sequence. This points to the gene acquisition having occurred within the last 2 to 3 million years. Since the BoHV-4 lineage does not derive from an ancestor of B. taurus, that species is inferred to have become infected by BoHV-4 more recently by cross-species transmission.
Rate of nucleotide substitution in herpesviruses.
To estimate the rate of evolution in the BoHV-4 Bo17 gene, we can compare the number of nucleotide substitutions on the branches to the African buffalo and BoHV-4 sequences since their CA. The BoHV-4 sequences are inferred to have undergone 42 to 57 nucleotide substitutions since their CA with the buffalo gene, among which 21 to 33 were synonymous changes. As noted above, there is a single synonymous substitution unique to the African buffalo sequence. Thus, the rate of evolution at synonymous sites in the viral lineage is estimated to have been about 25 to 30 times faster than in the buffalo. This ratio should be taken with caution, because the denominator reflects a single inferred substitution.
For 54 genes, the average number of synonymous substitutions per site between the bovine and caprine genes is 0.08 (E. Jeffs and P. M. Sharp, unpublished data); if those species shared a CA 20 million years ago (14) the rate of synonymous substitution in artiodactyls is estimated as around 2 × 10−9 substitutions per site per year. Then, the rate of evolution of Bo17 in BoHV-4 is estimated to be about 5 × 10−8 to 6 × 10−8 substitutions per site per year.
Distribution of the Bo17 gene among BoHV-4 strains.
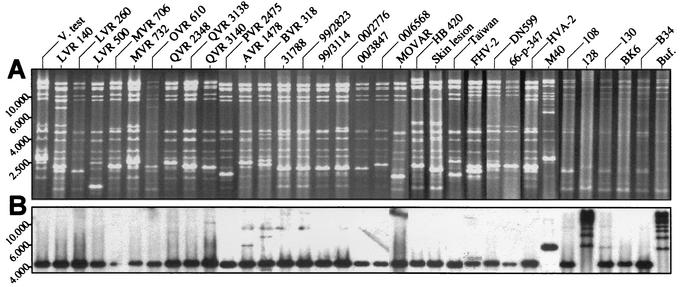
The results presented above reveal that the ancestor of BoHV-4 acquired the Bo17 gene from an ancestor of the African buffalo after the divergence of the Syncerus and Bos lineages. If the gene acquisition was extremely recent, there may be some BoHV-4 strains that do not contain the Bo17 gene. In order to test this hypothesis, 34 BoHV-4 strains (including 6 strains isolated in Kenya from wild African buffalo) (Table 1) isolated throughout the world and from various animal species were analyzed by a combined restriction endonuclease and Southern blotting approach (Fig. 3). Analysis of EcoRI restriction profiles confirmed the identity of the strains and revealed the divergence existing among them as previously reported (see Table 1 for references) (Fig. 3A). However, Southern blot analysis performed with a Bo17 ORF probe revealed that all strains tested contain the Bo17 gene (Fig. 3B). With the exception of three strains, Bo17 was detected in a 4.4-kbp DNA fragment corresponding to the right end of the LUR and the first 200 bp of the first element of the prDNA (4). For the M40 strain, Bo17 was detected in a single fragment of approximately 6.6 kbp due to the loss of the EcoRI site located at the right end of the LUR. For strains 128 and Buf., the blot revealed a ladder profile due to the loss of the EcoRI site in the element of the prDNA and to the random distribution of the number of elements at the ends of BoHV-4 genome (8). Taken together, these results suggest that all BoHV-4 strains possess the Bo17 gene and, thus, that all are the descendants of the virion that acquired this gene from an ancestor of the African buffalo.
FIG. 3.
Distribution of the Bo17 gene among BoHV-4 strains. The DNA of 34 BoHV-4 strains isolated throughout the world (see Table 1) was analyzed by EcoRI restriction (A) and submitted to Southern blot analysis for the detection of the Bo17 gene (B). Marker sizes (in kilobase pairs) are indicated on the left.
The apparent ubiquity of the Bo17 gene among BoHV-4 strains is consistent with the extent of sequence divergence among the nine strains for which sequence data were obtained. On average, the African buffalo strains differ from the Bos strains by a Ks of 0.08 (synonymous substitutions per site). The rate estimated above suggests that the CA of these two groups of BoHV-4 existed around 700,000 years ago. The topology of the phylogenetic tree in Fig. 2C suggests that the gene acquisition was very close to the CA of the BoHV-4 and buffalo sequences, and that was about twice as long ago as the CA of the viral sequences. Indeed, the average Ks between the BoHV-4 and buffalo sequences is similar to that between the two main groups of BoHV-4, suggesting that the gene acquisition occurred about 1.5 million years ago.
DISCUSSION
Many herpesvirus genomes contain a number of homologues of cellular genes that must have been acquired from the host genome at some point in the past. These gene acquisitions have played a major role in the evolution of herpesviruses, but for none of the examples is much known about the nature of the event. It would be interesting to identify the host species that was the source and to know when the acquisition occurred, how the pattern of evolution of the gene may have changed subsequent to its arrival in the viral genome, and what impact this new coding function had on the virus. In many cases these features will remain obscure, simply because the event occurred too long ago. The results reported here, of our investigations into the origin and evolution of the C2GnT-M-encoding Bo17 gene of BoHV-4, make this the best-characterized herpesvirus gene acquisition. Of the examples known, Bo17 appears to be the most-recently acquired gene, making it an ideal model system in which to study this process further.
BoHV-4 is the only virus known to encode a homologue of the C2GnT-M gene. However, prior to this work, the age and source of the Bo17 gene were unknown because no close relatives of BoHV-4 had been characterized and because only one C2GnT-M sequence (that of humans) was known. Phylogenetic analyses of a data set including the Bo17 gene from nine strains of BoHV-4 with the C2GnT-M genes of six ruminant species placed the viral lineage very close to that of the African buffalo, S. caffer, with very strong bootstrap support. This indicates that the Bo17 gene was acquired by a virus infecting a very recent ancestor of buffalo, long after the separation of the Bos and Syncerus lineages, in turn implying that cattle were infected later by cross-species transmission. Consistent with this, BoHV-4 can be isolated from the blood of healthy African buffalo with a high frequency (25%), and almost all (94%) wild African buffalo have antibodies against BoHV-4 (26); such high seroprevalence is rarely observed in the cattle population. It has been proposed that the high prevalence of BoHV-4 in the wild African buffalo population could reflect a selective advantage conferred by the virus to its host (26). In some regions, buffalo and wildebeest (Connochaetes taurinus) compete for grass. Wildebeests carry alcelaphine herpesvirus 1 (AlHV-1), which is lethal for buffalo and other ruminants, including cattle, while apparently harmless to its natural host. Infection of cattle by BoHV-4 has been shown to confer resistance to ulterior infection by AlHV-1, thus conferring an advantage to BoHV-4-infected animals (26) and providing a possible explanation for the initial spread of the virus in cattle.
We estimated the time of acquisition of the Bo17 gene at about 1.5 million years ago. We surveyed 34 BoHV-4 strains isolated from different continents and from different animal species, and we found that they all possess the Bo17 gene (Fig. 3B), suggesting that it has spread to fixation in the viral population. While this may have occurred through random genetic drift, the observation of constraint on nonsynonymous substitutions in the Bo17 gene suggests that the presence of the gene is advantageous and thus that the spread was driven by natural selection. The sequences of the Bo17 gene from nine strains of BoHV-4 fall into two clades, one comprising African buffalo strains and another comprising strains from cattle. The split between these two viral lineages was estimated to have been about 700,000 years ago, clearly predating the domestication of cattle (around 10,000 years ago). This suggests that cattle became infected with BoHV-4 prior to domestication. That interpretation seems problematic because African buffalo are restricted to sub-Saharan Africa, whereas the ancestor of domestic cattle, Bos primigenius, was found in Eurasia and North Africa. However, under different climatic conditions in the past, the ranges of these two species may have overlapped.
Zebu and taurine cattle appear to have been domesticated independently, from different races or subspecies of B. primigenius in Southwest Asia (17). The single BoHV-4 strain from zebu that we examined (M40) fell outside the clade of strains from taurine cattle. The average extent of synonymous substitution between M40 and the taurine strains (0.02 substitutions per site) leads to an estimate of about 200,000 years ago for their divergence time, not dissimilar to the estimate of 275,000 years ago for the common ancestor of zebu and taurine cattle (3). These data are consistent with a relatively ancient transmission of BoHV-4 to the B. primigenius lineage, followed by a host-dependent split between the zebu and taurine viral strains; however, this interpretation should be treated with caution because it is based on a single zebu strain of BoHV-4.
The effect of natural selection on a gene can be gauged by the ratio of nonsynonymous to synonymous substitutions (28). Among the six ruminant C2GnT-M sequences, the ratio averaged 0.20. On the branch before the common ancestor of the BoHV-4 strains, the ratio was 0.35. This elevated ratio could reflect either a relaxation of selective constraint on the protein sequence or adaptively driven amino acid replacements following the transfer of the gene to the viral genome. There are 14 sites at which at least six of the seven different BoHV-4 sequences share an amino acid difference from the ruminants, indicating that replacement occurred on the branch after gene acquisition and pointing to these as candidates for adaptive changes. Among the BoHV-4 strains, the nonsynonymous/synonymous substitution ratio was 0.19, indicating a very similar level of functional constraint to that in mammals. Consistent with this observation, in the V. test strain of BoHV-4, the protein encoded by Bo17 (pBo17) was shown to have conserved all three enzymatic activities exhibited by cellular C2GnT-M, i.e., core 2, core 4, and I branching activities (35); this observation has recently been extended to several other strains (N. Markine-Goriaynoff, unpublished data). Based on studies of cellular C2GnT-M, several hypotheses could be made concerning the role of pBo17 in the biology of BoHV-4 infection. The first is that this enzyme is utilized for the posttranslational modification of structural viral proteins, potentially affecting the tropism of the virion and/or its sensitivity to antibody and/or complement neutralization. A second hypothesis is that pBo17 is involved in the modification of interactions between infected cells and cells of the immune system, protecting the infected cells from the cellular immune response. Consistent with this, several studies have demonstrated that increasing the level of C2GnT-M activity, and that of the resulting core 2 branched oligosaccharides, significantly decreases interactions between the expressing cells and cells of the immune system (11). Experiments to test these hypotheses are in progress. However, the results presented here indicate that acquisition of Bo17 was a relatively recent event in evolution and, clearly, pBo17 function was not essential for viral replication before then. In fact, it was recently demonstrated that a BoHV-4 strain in which Bo17 is deleted can apparently replicate normally in vitro (Markine-Goriaynoff, unpublished data).
By comparison of the divergence of the buffalo C2GnT-M gene and the viral Bo17 gene since their common ancestor, we estimated the basal rate of evolution of BoHV-4, as indicated by the rate of synonymous substitution, to be about 25 to 30 times that in the host genome, or around 5 × 10−8 to 6 × 10−8 substitutions per site per year. Previous estimates of the rate of evolution of herpesviruses have relied on the assumption of host-virus codivergence. The rate of synonymous substitution during the divergence of the two human simplex viruses (human herpesvirus 1 [HHV-1] and HHV-2) has been estimated to be 3 × 10−8 substitutions per site per year (6) by using a divergence time derived from the assumption that alphaherpesviruses have cospeciated with their hosts (20). Similarly, on the assumption that the gammaherpesviruses Epstein Barr virus (HHV-4) and herpesvirus papio (cercopithecine herpesvirus 12) diverged at the same time as their human and Old World monkey hosts, the rate of synonymous substitution in the viral interleukin 10 gene has recently been estimated as about 20 times that of the primate interleukin 10 gene (13). These various estimates of the rate are similar, differing only by a factor of two, which is well within the range of variation of synonymous substitution rates seen among different genes in the comparison of HHV-1 and HHV-2 (6) or among different genes compared between a single pair of mammals (38). Since our rate estimate does not rely on the assumption of codivergence, the consistency with these other estimates provides independent support for the host-virus cospeciation hypothesis.
The difference in the rate of evolution between the viral gene and its cellular homologue provided a striking example of how long-branch attraction can disrupt phylogenetic analyses. Rate differences among lineages may be a common problem in viral evolution, but this most obviously occurs with gene acquisitions where long-branch attraction may obscure the true history of the acquired gene. For example, two recent phylogenetic analyses of ribonucleotide reductase included viral homologues as well as sequences from both prokaryotes and eukaryotes (13, 33). The sequences from herpesviruses formed a clade attached to the ancestral lineage leading to the eukaryotes (represented by animals, fungi, plants, and several protist lineages), even though it seems more likely that the gene was acquired much later from an animal host.
Finally, the present study demonstrates that the Bo17 gene was acquired from the Syncerus lineage rather than from the Bos lineage about 1.5 million years ago and, that after probable gene fixation in the viral population, the virus was transmitted from a recent ancestor of the African buffalo to the ancestors of cattle about 700,000 years ago. The analysis of Bo17 sequences for nine different BoHV-4 strains revealed that strains isolated from cattle and African buffalo fall into two distinct groups. Taking into account the conclusions of the present study and the higher frequency of BoHV-4 infection in African buffalo than in cattle, BoHV-4 isolates from African buffalo should be considered an African buffalo virus rather than a bovine virus. To reinforce this point, and following the rules applied by the International Committee on Taxonomy of Viruses, it might be appropriate to rename the BoHV-4 strains isolated from African buffalo as syncerine herpesvirus 1. Further molecular and biological comparisons between bovine and African buffalo isolates of BoHV-4 are required to assess the extent to which these two closely related virus species have differentiated.
Acknowledgments
This work was supported by a Concerted Action awards program of the French Community of Belgium (ARC 98/03-220). N. Markine-Goriaynoff and A. Vanderplasschen are Research Fellow and Senior Research Associate, respectively, of the Fonds National Belge de la Recherche Scientifique (FNRS).
We thank M. Balligand (University of Liège), C. Charlier (University of Liège), L. Egyed (Veterinary Medical Research Institute, Budapest, Hungary), O. Hanotte (International Livestock Research Institute), P. B. Rossiter (FAO, Nairobi, Kenya), R. M. Rumberia (National Veterinary Research Centre, Muguga, Kenya), and C. Letellier (Cerva, Brussels, Belgium) for providing DNA samples or virus strains.
REFERENCES
- 1.Barahona, H. H., L. V. Meléndez, N. W. King, M. D. Daniel, C. E. O. Fraser, and A. C. Preville. 1973. Herpesvirus aotus type 2: a new viral agent from owl monkeys (Aotus trivirgatus). J. Infect. Dis. 127:171-178. [DOI] [PubMed] [Google Scholar]
- 2.Bartha, A., M. Juhasz, and H. Liebermann. 1966. Isolation of a bovine herpesvirus from calves with respiratory disease and keratoconjunctivitis. Acta Vet. Hung. 16:357-358. [PubMed] [Google Scholar]
- 3.Bradley, D. G., D. E. MacHugh, P. Cunningham, and R. T. Loftus. 1996. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl. Acad. Sci. USA 93:5131-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bublot, M., M. F. Van Bressem, E. Thiry, J. Dubuisson, and P. P. Pastoret. 1990. Bovine herpesvirus 4 genome: cloning, mapping and strain variation analysis. J. Gen Virol. 71:133-142. [DOI] [PubMed] [Google Scholar]
- 5.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 6.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egyed, L., J. P. Kluge, and A. Bartha. 1997. Histological studies of bovine herpesvirus type 4 infection in non-ruminant species. Vet. Microbiol. 57:283-289. [DOI] [PubMed] [Google Scholar]
- 8.Ehlers, B., H.-J. Buhk, and H. Ludwig. 1985. Analysis of bovine cytomegalovirus genome structure: cloning and mapping of the monomeric polyrepetitive DNA unit, and comparison of European and American strains. J. Gen. Virol. 66:55-68. [DOI] [PubMed] [Google Scholar]
- 9.Fabricant, C. G., J. H. Gillespsie, and L. Krook. 1971. Intracellular and extracellular mineral crystal formation induced by viral infection of cell cultures. Infect. Immun. 3:416-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda, M., and S. Tsuboi. 1999. Mucin-type O-glycans and leukosialin. Biochim. Biophys. Acta 1455:205-217. [DOI] [PubMed] [Google Scholar]
- 12.Hassanin, A., and E. J. Douzery. 1999. Evolutionary affinities of the enigmatic saola (Pseudoryx nghetinhensis) in the context of the molecular phylogeny of Bovidae. Proc. R. Soc. Lond. Ser. B 266:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, A. L. 2002. Origin and evolution of viral interleukin-10 and other DNA virus genes with vertebrate homologues. J. Mol. Evol. 54:90-101. [DOI] [PubMed] [Google Scholar]
- 14.Kumar, S., and S. B. Hedges. 1998. A molecular timescale for vertebrate evolution. Nature 392:917-920. [DOI] [PubMed] [Google Scholar]
- 15.Li, W. H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36:96-99. [DOI] [PubMed] [Google Scholar]
- 16.Lin, T. M., G. Y. Shi, S. J. Jiang, C. F. Tsai, B. J. Hwang, C. T. Hsieh, and H. L. Wu. 1999. Persistent infection of bovine herpesvirus type 4 in bovine endothelial cell cultures. Vet. Microbiol. 70:41-53. [DOI] [PubMed] [Google Scholar]
- 17.Loftus, R. T., D. E. MacHugh, D. G. Bradley, P. M. Sharp, and P. Cunningham. 1994. Evidence for two independent domestications of cattle. Proc. Natl. Acad. Sci. USA 91:2757-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthee, C. A., and S. K. Davis. 2001. Molecular insights into the evolution of the family Bovidae: a nuclear DNA perspective. Mol. Biol. Evol. 18:1220-1230. [DOI] [PubMed] [Google Scholar]
- 19.McGeoch, D. J. 2001. Molecular evolution of the gamma-herpesvirinae. Philos. Trans. R. Soc. Lond. Ser. B 356:421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 21.McGeoch, D. J., A. Dolan, and A. C. Ralph. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohanty, S. B., R. C. Hammond, and M. G. Lillie. 1971. A new bovine herpesvirus and its effect on experimentally infected calves. Arch. Gesamte Virusforsch. 34:394-395. [DOI] [PubMed] [Google Scholar]
- 23.Moreno-Lopez, J., M. Goltz, C. Rehbinder, K. V. Valsala, and H. Ludwig. 1989. A bovine herpesvirus (BHV-4) as passenger virus in ethmoidal tumours in Indian cattle. Zentbl. Veterinarmed. Reihe B 36:481-486. [DOI] [PubMed] [Google Scholar]
- 24.Raftery, M., A. Muller, and G. Schonrich. 2000. Herpesvirus homologues of cellular genes. Virus Genes 21:65-75. [PubMed] [Google Scholar]
- 25.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.Rossiter, P. B., I. D. Gumm, D. A. Stagg, P. A. Conrad, S. Mukolwe, F. G. Davies, and H. White. 1989. Isolation of bovine herpesvirus-3 from African buffaloes (Syncerus caffer). Res. Vet. Sci. 46:337-343. [PubMed] [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Sharp, P. M. 1997. In search of molecular darwinism. Nature 385:111-112. [DOI] [PubMed] [Google Scholar]
- 29.Thiry, E., M. Bublot, J. Dubuisson, M. F. Van Bressem, A. S. Lequarre, P. Lomonte, A. Vanderplasschen, and P. P. Pastoret. 1992. Molecular biology of bovine herpesvirus type 4. Vet. Microbiol. 33:79-92. [DOI] [PubMed] [Google Scholar]
- 30.Thiry, E., P.-P. Pastoret, C. Dessy-Doizé, C. Hanzen, C. M. Calberg-Bacq, L. Dagenaix, H. Vindevogel, and F. Ectors. 1981. Réactivation d'un herpèsvirus en culture de cellules testiculaires prélevées chez un taureau atteint d'orchite et d'azoospermie. Ann. Med. Vet. 125:207-214. [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd, W. J., and J. Storz. 1983. Morphogenesis of a cytomegalovirus from an American bison affected with malignant catarrhal fever. J. Gen. Virol. 64:1025-1030. [DOI] [PubMed] [Google Scholar]
- 33.Torrents, E., P. Aloy, I. Gibert, and F. Rodriguez-Trelles. 2002. Ribonucleotide reductases: divergent evolution of an ancient enzyme. J. Mol. Evol. 55:138-152. [DOI] [PubMed] [Google Scholar]
- 34.Vanderplasschen, A., M. Bublot, P. P. Pastoret, and E. Thiry. 1993. Restriction maps of the DNA of cervid herpesvirus 1 and cervid herpesvirus 2, two viruses related to bovine herpesvirus 1. Arch. Virol. 128:379-388. [DOI] [PubMed] [Google Scholar]
- 35.Vanderplasschen, A., N. Markine-Goriaynoff, P. Lomonte, M. Suzuki, N. Hiraoka, J. C. Yeh, F. Bureau, L. Willems, E. Thiry, M. Fukuda, and P. P. Pastoret. 2000. A multipotential beta-1,6-N-acetylglucosaminyl-transferase is encoded by bovine herpesvirus type 4. Proc. Natl. Acad. Sci. USA 97:5756-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Opdenbosch, E., G. Wellemans, and J. Oudewater. 1986. Toevallige isolatie van het boviene herpesvirus 4 uit de long van een schaap. Vlaams Diergeneesk Tijdschr. 55:432. [Google Scholar]
- 37.Wellemans, G., H. Antoine, A. Broes, G. Charlier, and E. Van Opdenbosch. 1983. Isolement d'un virus Herpès chez des bovins atteints de métrite post-partum. Ann. Med. Vet. 127:481-482. [Google Scholar]
- 38.Wolfe, K. H., and P. M. Sharp. 1993. Mammalian gene evolution: nucleotide sequence divergence between mouse and rat. J. Mol. Evol. 37:441-456. [DOI] [PubMed] [Google Scholar]
- 39.Yeh, J. C., E. Ong, and M. Fukuda. 1999. Molecular cloning and expression of a novel beta-1,6-N-acetylglucosaminyltransferase that forms core 2, core 4, and I branches. J. Biol. Chem. 274:3215-3221. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann, W., H. Broll, B. Ehlers, H. J. Buhk, A. Rosenthal, and M. Goltz. 2001. Genome sequence of bovine herpesvirus 4, a bovine Rhadinovirus, and identification of an origin of DNA replication. J. Virol. 75:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]