Abstract
In the context of the Rous sarcoma virus Gag polyprotein, only the nucleocapsid (NC) domain is required to mediate the specificity of genomic RNA packaging. We have previously showed that the Saccharomyces cerevisiae three-hybrid system provides a rapid genetic assay to analyze the RNA and protein components of the avian retroviral RNA-Gag interactions necessary for specific encapsidation. In this study, using both site-directed mutagenesis and in vivo random screening in the yeast three-hybrid binding assay, we have examined the amino acids in NC required for genomic RNA binding. We found that we could delete either of the two Cys-His boxes without greatly abrogating either RNA binding or packaging, although the two Cys-His boxes are likely to be required for efficient viral assembly and release. In contrast, substitutions for the Zn-coordinating residues within the boxes did prevent RNA binding, suggesting changes in the overall conformation of the protein. In the basic region between the two Cys-His boxes, three positively charged residues, as well as basic residues flanking the two boxes, were necessary for both binding and packaging. Our results suggest that the stretches of positively charged residues within NC that need to be in a proper conformation appear to be responsible for selective recognition and binding to the packaging signal (Ψ)-containing RNAs.
Retroviral particles which contain two copies of genomic RNA are assembled through Gag-Gag interactions. During the process of RNA encapsidation, sequences in genomic RNA (termed Ψ) are selectively recognized and specifically bound to domains in the Gag polyprotein. In many retroviruses, such as murine leukemia virus (MuLV), human immunodeficiency virus type 1 (HIV-1), and avian sarcoma leukosis virus (ASLV), packaging sequences are located in the 5′ untranslated leader sequences of the genome. A 160-nucleotide (nt) packaging sequences of ASLV (MΨ) has been identified which confers the efficient packaging of heterologous RNAs into virions, if they are tethered to MΨ RNA (3).
Only the avian retroviral Gag precursor protein is required for viral assembly at the cytoplasmic membrane. Gag is subsequently cleaved by the Gag-encoded protease to yield individual domains; matrix (MA), p2, p10, capsid (CA), nucleocapsid (NC), and protease (PR). Cleavage occurs shortly after or concomitant with viral budding (26). Using deletion analyses and chimeric proteins in which the NC domain of one retrovirus is substituted for the cognate region of another retroviral Gag, the NC domain of the Gag polyprotein has been shown to be solely responsible for specific encapsidation (6, 49). In all Orthoretrovirinae, the NC protein contains one or two copies of the conserved sequence Cys-X2-Cys-X4-His-X4-Cys, which coordinates Zn2+ ion binding (Cys-His zinc finger motif), as well as adjacent basic amino acids flanking the fingers. The zinc finger sequence bears a striking resemblance to those of a variety of eukaryotic transcription factors implicated in recognition of specific DNA sequences (reviewed in reference 17). However, the zinc fingers have not been shown to interact with RNA, whereas the stretches of basic amino acids were found to be a part of the RNA binding motif (45). The property of nucleic acid binding of the Zn-finger NC has been shown to be involved in essential steps of virus replication including reverse transcription, integration, RNA packaging, viral particle assembly, morphogenesis, and infectivity (reviewed in reference 12).
We previously developed a Saccharomyces cerevisiae three-hybrid binding assay to determine the domains in Gag and structural elements in Ψ RNA involved in specific RNA encapsidation (30, 31). Using this assay, we found that the interactions of MΨ RNA with Gag are of high affinity and specificity and showed that there is good agreement between results of the yeast three-hybrid binding assay and those of an in vivo packaging assay. In this study, we analyzed the NC domain in the context of Gag polyprotein using both the site-directed and random mutagenesis in the yeast system to examine the amino acids required for RNA binding. We also examined the abilities of purified recombinant wild-type (wt) and mutant NC proteins to bind to Ψ RNA in vitro and obtained results consistent with those from the in vivo yeast binding assay.
MATERIALS AND METHODS
NC site-directed mutagenesis.
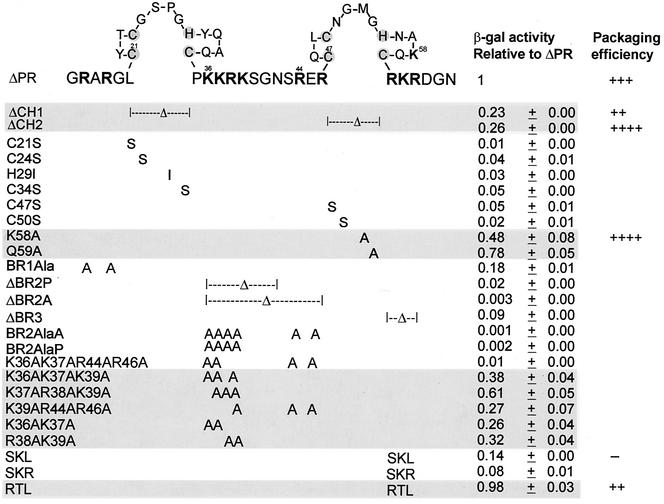
All NC mutations were constructed by two rounds of DNA amplification using PCR technology. All mutations were moved to the pACTIIΔPR plasmid (31) which has all of the domains in the Gag protein, except PR. The ΔCH1 and ΔCH2 mutations lack the first or second Cys-His box (CH box) within the NC domain, respectively (Fig. 1). The basic region deletion mutation ΔBR2A in which the basic region between the two CH boxes was deleted lacks amino acids from Lys36 to Arg46 of NC (Fig. 1). Another basic region deletion mutation, ΔBR2P, lacks amino acids from Lys36 to Gly41.
FIG. 1.
NC site-directed mutations and the resulting β-Gal activities relative to ΔPR. The amino acid sequence of NC is shown using one-letter abbreviations. Amino acid numbering is from the amino terminus of NC. The positively charged residues are indicated in bold type, and the Zn-coordinating residues within the CH boxes are shaded. β-Gal activity relative to that of ΔPR is given as the average ± standard deviation of the values for three to four independent transformants in three assays. The β-Gal activity of cells cotransformed with the RNA hybrid plasmid carrying MΨ sequences and the ΔPR protein hybrid plasmid is about 1,500 U. Mutants with β-Gal activity that was more than 20% of ΔPR are shaded. Packaging efficiencies are summarized from Table 1, Fig. 3 for the SKL mutant, or previous experiments for the RTL mutants (Table 2 in reference 31). Packaging efficiency symbols: ++, the relative packaging efficiency to ΔPR is about 0.8; +++, the relative packaging efficiency to ΔPR is about 1.0; ++++, the relative packaging efficiency to ΔPR is four- to sevenfold more than ΔPR; −, the relative packaging efficiency to ΔPR is about 1% that of ΔPR.
RNA hybrid expression vector, protein hybrid expression vector, and yeast transformation.
We used the pIII/ms2-MΨ RNA hybrid expression vector (31) which carries a 160-nt Rous sarcoma virus (RSV)-PrC strain packaging sequence (MΨ; GenBank accession no. J02342, nt 389 to 548) downstream of the ms2 sequences. To examine the effects of NC mutations on binding to MΨ RNA, mutagenized NC sequences were cloned into the pACTIIΔPR protein hybrid expression vector (31) in which the activation domain of Gal4 is hybridized with the GagΔPR protein. We then introduced mutated plasmids into Saccharomyces cerevisiae L40-coat cells (42) along with the pIII/ms2-MΨ RNA hybrid plasmid as described previously (20). Transformants carrying both the RNA hybrid and protein hybrid plasmids were selected using plates lacking uracil and leucine.
Yeast β-Gal activity assay.
The β-galactosidase (β-Gal) activity of yeast double transformants was quantitatively assayed in a liquid culture by directly measuring the enzyme activity using chlorophenol 5-bromo-4-chloro-3-indolyl-β-d-galactoside (CPRG) as a substrate (4). The filter β-Gal assay was used to qualitatively measure the enzyme activity of yeast transformants after directly transferring colonies to filters. Cells were permeabilized by three cycles of freeze-thaw treatment of filters in a pool of liquid nitrogen, and the filters were placed on another filter that was presoaked in Z-buffer-X-Gal solution (110 mM Na2HPO4, 46 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 330 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal] per ml, 2-mercaptoethanol). Filters were incubated at 30°C, and the appearance of blue β-Gal positive colonies on the filters was monitored.
Cell culture and transfections.
The quail cell line QT6 or QT35 (37) was grown in GM+D+CK (Ham's F10 medium containing 10% tryptose phosphate broth [Difco], 5% calf serum, 1% heat-inactivated chicken serum, and 1% dimethyl sulfoxide). The modified calcium phosphate method (9) was used for DNA transfections on cells seeded in Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum. Stably transfected mass cultures of G418-resistant cells at 0.15 mg/ml were obtained after 2 weeks of selection.
In vivo packaging assay.
Either QT6 or QT35 cultures containing the mutant proviruses were labeled with 250 μCi of [35S]methionine (>1,000 Ci/mmol) (EXPRESS 35S protein labeling mix; NEN Research Products) in 2 ml of serum-free DMEM lacking methionine and cysteine (DMEM-Met-Cys) for 5 h and chased for 18 to 24 h with DMEM-Met-Cys medium supplemented with 10% fetal bovine serum. The supernatants were collected and filtered through a 0.45-μm-pore-size syringe, and virus-like particles were harvested by clearing the supernatant of cell debris by low-speed centrifugation and concentrated by high-speed centrifugation through 20% sucrose cushion. The viral pellet was then resuspended in isotonic buffer as described previously (1). The labeled cells were washed twice with cold isotonic buffer, scraped from plates, and then directly lysed with the lysis buffer (Direct Protect kit; Ambion Inc., Austin, Tex.).
To quantitate virion levels of Gag protein, radioimmunoprecipitation assays (RIPA) were performed with anti-ASLV polyclonal rabbit serum (anti-PrB). Concentrated [35S]methionine-labeled viral particles were incubated in 1.0 ml of antibody buffer (20 mM Tris-HCl [pH 7.4], 50 mM NaCl, 0.5% Nonidet P-40 [NP-40], 0.5% deoxycholic acid [DOC], 0.5% sodium dodecyl sulfate [SDS], 0.5% aprotinin, 1 mM EDTA [pH 8.0]) with 5 μl of anti-PrB antibody and 30 μl of protein A-Sepharose beads (Pharmacia LKB Biotechnology, Inc.). The antigen-antibody complexes were washed twice in RIPA buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1% DOC, 0.1% SDS, 0.5% aprotinin), once in high-salt buffer (10 mM Tris-HCl [pH 7.4], 2 M NaCl, 1% NP-40, 0.5% DOC), and then once more with RIPA buffer. The bound proteins were eluted in SDS sample buffer and loaded onto SDS-10% polyacrylamide gels. The dried gels were scanned directly using a Molecular Dynamics PhosphorImager after overnight exposure. Radioactive bands were quantitated by using the ImageQuant software.
RNase protection assays (RPAs) were performed to measure the amounts of cellular or viral RNAs by using either the antisense neo probe or the antisense gapdh probe as described previously (2, 30). Each probe RNA was in vitro transcribed with a T7 RNA polymerase to produce 32P-labeled RNA. The labeled RNA was purified with phenol-chloroform and passed through a G-25 column (Amersham Pharmacia) to remove unincorporated 32P. The probes were ethanol precipitated and used for hybridization with cellular and viral RNAs. RPAs were performed by the method specified for the Direct Protect kit. Reaction mixtures were run on a 5% polyacrylamide gel, and radioactively labeled protected bands were quantitated, in machine units, by using ImageQuant software.
Packaging efficiencies for neo RNA were determined by calculating the amount of neo RNA in virions normalized to the level of neo RNA in the cells (as measured by RPA). This calculated level of RNA was then further normalized by the number of virus particles as measured by the amount of Gag protein in the pelletable particles using RIPA.
Random mutagenesis of NC.
PCR random mutagenesis was based on manganese-induced misincorporation of nucleotides by AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.) (21). The PCR was modified to use an increased concentration of magnesium to stabilize noncomplementary pairs, unequal amounts of nucleotides, a higher concentration of DNA polymerase, and an increased number of PCR cycles to decrease the fidelity of PCR amplification (28, 32). To make changes in the coding sequences of the 266-nt nucleocapsid DNA, we designed three PCR protocols employing different combinations of the modifications described above. The first protocol used a 100-μl volume of mutagenesis buffer containing 1× PCR buffer II (10 mM Tris-HCl [pH 8.3], 50 mM KCl [Perkin-Elmer]), 7 mM MgCl2, 200 μM (each) dCTP and dTTP, 20 μM (each) dATP and dGTP, 10 pmol of each primer, and 7.5 U of AmpliTaq DNA polymerase. The second PCR protocol adds 0.5 mM MnCl2 to the buffer of the first PCR protocol. The third mutagenesis PCR protocol has 0.5 mM MnCl2 in reaction buffer containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM dTTP, 20 μM (each) dATP, dCTP, and dGTP, 10 pmol of each primer, and 7.5 U of AmpliTaq DNA polymerase.
A library of RSV NC mutant sequences from each reaction was digested with AscI and EcoRI, which cleave the restriction enzyme sites encoded by the primers, and ligated with the AscI-EcoRI fragment of the pACTIIΔPR-Asc plasmid. This plasmid was derived from the hybrid protein expression vector pACIIΔPR (31) into which the unique AscI restriction site was engineered at the 5′ end of the NC sequence for directional cloning. The ligated DNAs were cotransformed into the yeast strain L40-coat (42) along with the pIII/ms2-MΨ RNA hybrid expression vector (31).
In addition to the PCR-based random mutagenesis, the second protocol used a mutator strain of Escherichia coli. To obtain randomly mutagenized DNA pools, the pACTIIΔPR-Asc protein hybrid expression plasmid was transformed into a DNA repair deficient strain Epicurian Coli XL1-Red (Stratagene). The lawn of transformant colonies was then scraped from plates, and the mutant library DNAs were extracted, digested, and used for ligation and transformation, as described above for PCR mutagenesis.
Expression and purification of recombinant NC proteins.
A T7 polymerase-based expression system was used for high-level expression of recombinant NC proteins. 5′ six-His-tagged wt and a mutant version (NC-SKL) of NC sequences were PCR amplified and subcloned into the sequence downstream of T7 promoter of the expression vector, and recombinant plasmids were transformed into a BL21(DE3) E. coli strain. Recombinant proteins were extracted and partially purified by Ni-nitrilotriacetic acid affinity chromatography under denaturing conditions including 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris-Cl (pH 8.0). The proteins were eluted by reducing the pH to 4.5 and refolded by step dialysis. Refolded proteins were then once more dialyzed against the optimized buffer (20 mM HEPES [pH 7.5], 10% glycerol, 100 mM NaCl, 0.1 mM ZnCl2, 100 mM Arg HCl, 5 mM 2-mercaptoethanol) for further purification by both ion-exchange and gel filtration fast-performance liquid chromatographies.
In vitro binding assay.
Either a 160-nt sequence of RSV-PrC strain MΨ (GenBank accession no. J02342, nt 389 to 548), antisense MΨ, or an 82-nt packaging signal (μΨ; GenBank accession no. J02342, nt 389 to 470) (2) was inserted into the MluI site of pASY161, a pCMVneo derivative (3). Each recombinant plasmid was linearized with XbaI and used as a template for in vitro transcription with a T7 RNA polymerase to produce 32P-labeled RNA. The labeled RNA was purified with phenol-chloroform and passed through a G-25 column (Amersham Pharmacia) to remove unincorporated 32P. Ψ RNA (100 nM) was incubated with 20 U of RNase inhibitor (Promega) and various amounts of either recombinant wt NC or NC-SKL proteins that were resuspended in a buffer containing 20 mM HEPES (pH 7.5), 10% glycerol, 100 mM NaCl, 0.1 mM ZnCl2, 100 mM Arg HCl, and 5 mM 2-mercaptoethanol for 1 h at room temperature. The samples were loaded onto a 10% 0.5× Tris-borate-EDTA (TBE) native polyacrylamide gel, which ran at 5 W for 2.5 h. The gel was dried and exposed to a phosphorimager screen.
RESULTS
One CH box is sufficient for RNA binding and efficient packaging.
To define domains of avian retroviral NC involved in genomic RNA binding, and hence packaging, we have used the yeast three-hybrid binding assay. The site-directed mutations were constructed in the NC sequences of the pACTIIΔPR plasmid in which the activation domain of Gal4 is fused to a Gag molecule containing all of the domains, except PR. We found that ΔPR allows efficient packaging of specific RNA and leads to a higher level of sensitivity than wt Gag in the three-hybrid binding assay (31). The NC mutant DNAs were cotransformed with the pIII/ms2-MΨ RNA hybrid expression plasmid into a yeast strain (L40-coat), and the effects of mutations on binding to MΨ RNA were assayed by measuring enzymatic activity of the reporter gene, lacZ. Both deletions and alanine substitutions were made in the NC domain (Fig. 1). We also constructed single substitutions for the Zn-coordinating residues of the CH boxes. The β-Gal activity relative to ΔPR for each mutant is given in Fig. 1. All of the mutants stably expressed equivalent amounts of the mutant proteins, as detected by immunoprecipitation assays (data not shown). We previously found that a level of β-Gal activity which was ≤20% of that of the wt most often led to a large reduction in RNA packaging (30, 31). Thus, any mutant which has a binding efficiency of more than 0.2 is likely to be packaging competent, whereas one which has an efficiency of less than 0.2 is likely to be packaging deficient. We found that deletion of either of the CH boxes gives ambiguous binding efficiency values of 0.23 and 0.26. In contrast, a single substitution for some of the Zn-coordinating residues (C21S, C24S, H29I, C34S, C47S, and C50S) within the boxes leads to a 20- to 50-fold reduction in relative β-Gal activity, whereas strains with mutations in the non-Zn-coordinating residues (K58A and Q59A) showed levels of β-Gal activity comparable to that of ΔPR. This suggests that while the substitutions might disrupt the overall conformation of the protein, the deletions had less of an effect.
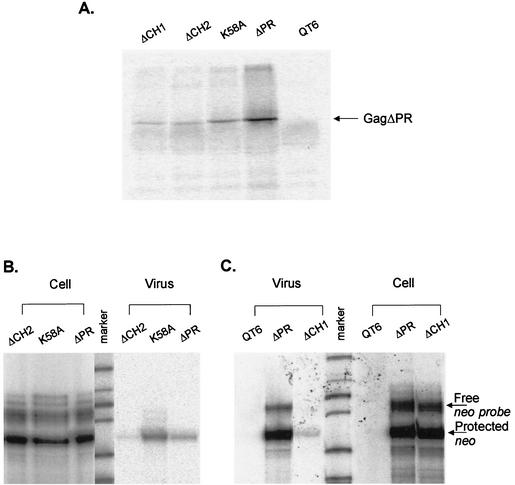
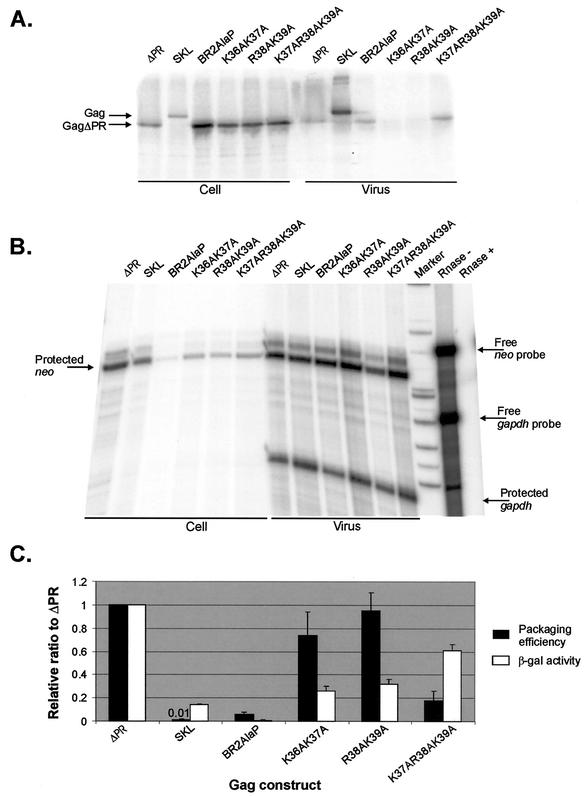
Several CH box mutations were subcloned into the packaging vector pASY165 (31) which was then transfected into QT6 cells to measure the packaging efficiency in vivo, as described in Materials and Methods. Deletion of either of the CH boxes greatly reduced the relative efficiency of particle assembly and release by about 30-fold, as measured by RIPA (Fig. 2A and Table 1). A mutation of Lys58 to Ala also led to somewhat decreased production of viral Gag proteins (fivefold reduction relative to ΔPR). As the pASY165 packaging vector contains neo RNA in place of src in the full-length RSV RNA with the intact packaging sequences at the 5′ end of the genome, we were able to examine the efficiency of packaging the viral RNA into particles by using RPA to measure the amount of viral neo RNA. Since each transfected cells produced somewhat different levels of genomic viral RNA in the cells, we also measured the level of cellular neo RNA to normalize the level of viral RNA (Fig. 2B and C). To quantitate the packaging efficiency of viral RNA into particles, the normalized level of viral RNA was corrected by the number of virus particles, as measured by the level of Gag protein in the pelletable particles. The packaging efficiency of each mutant relative to ΔPR is shown in Table 1. Deletion of either CH boxes greatly reduced the efficiency of particle assembly and release, and thus increased the apparent ratio of RNA to Gag protein in virions. The deletion of either of the boxes did not affect the ability of the Gag polyprotein to efficiently package its own genomic RNA, suggesting that one CH box is sufficient for RNA binding and packaging but that the two zinc finger motifs are required for efficient viral assembly and release. Not surprisingly, the K58A mutant, which had a β-Gal activity of 0.48 relative to ΔPR, showed a high packaging efficiency.
FIG. 2.
In vivo packaging assays of the NC mutants. (A) RIPA analysis of pelleted virus-like particles collected from supernatants to quantitate the number of viral particles released to the medium. QT6 cells represent untransfected cells. (B) RPA to measure the amount of neo RNA in cells transfected with either ΔCH2, K58A, or ΔPR mutation or supernatant virions, using an antisense neo RNA as the riboprobe. (C) RPA to measure the effect of the ΔCH1 mutation on the ability of Gag polyprotein to package viral RNA into virions. The amount of neo RNA was measured in transfected cells and virions.
TABLE 1.
Relative packaging efficiencies of NC mutants to ΔPR
| Gag constructa | Viral Gagb | Ratio of viral to cellular RNAc | Relative packaging efficiencyd |
|---|---|---|---|
| ΔCH1 | 0.033 ± 0.01 | 0.026 ± 0.01 | 0.78 ± 0.08 |
| ΔCH2 | 0.033 ± 0.01 | 0.164 ± 0.04 | 3.89 ± 0.95 |
| K58A | 0.21 ± 0.02 | 1.485 ± 0.11 | 6.59 ± 0.47 |
| ΔPR | 1 | 1 | 1 |
NC mutations were made in Gag polyproteins lacking the PR domain.
RIPA were performed three times, and a representative gel is shown in Fig. 2A. Gels were scanned by a Molecular Dynamics PhosphorImager, and radioactive bands were quantitated, in machine units, by using ImageQuant software. The averages ± standard deviations are shown.
RPAs were performed three times (Fig. 2B and C), and quantitation was done by PhosphorImager analysis. The averages ± standard deviations are indicated.
The relative packaging efficiencies to ΔPR were summarized from Fig. 2. The averages ± standard deviations were calculated from three independent experiments.
In RSV, there are three regions of clustered basic residues surrounding the two CH boxes. The first basic region has an ambiguous role in RNA binding, since the β-Gal activity of a mutant (BR1Ala) relative to ΔPR is close to 0.2 (Fig. 1). In the region between the two boxes, when four or more of the six positively charged residues were changed to alanine, binding diminished greatly. However, mutations of three or two did not greatly affect binding, regardless of the position. This indicates that three positively charged residues between the CH boxes are necessary for MΨ RNA binding.
To examine whether the results of yeast three-hybrid binding assays correlated well with those of in vivo packaging assays, several mutations in the basic region between the CH boxes were subcloned into the packaging vector pASY165 and transfected into QT35 quail cells. Packaging efficiencies were then determined, as described in Materials and Methods. Mutation of two positively charged residues to alanine (Fig. 3A, K36AK37A or R38AK39A) greatly decreased the efficiencies of both particle assembly and release into media. However, the levels of packaged viral neo RNA were about 10-fold more than that of a quadruple mutant (Fig. 3B, BR2AlaP) which released a number of viral particles comparable to that of ΔPR (Fig. 3A). The packaging efficiency of each mutant was calculated relative to ΔPR as described in Materials and Methods and compared with the relative β-Gal activity in the yeast three-hybrid binding assays (Fig. 3C). Mutants with mutations of two or three amino acids efficiently packaged viral genomic RNA at levels 20- to 100-fold higher than that of the SKL mutant (Fig. 1), whereas four mutations (BR2AlaP) showed a negligible level of RNA packaging (fivefold more than SKL). These results indicated a good correlation between the two assays, suggesting that three positively charged residues between the CH boxes are sufficient for MΨ RNA binding and packaging.
FIG. 3.
(A) RIPA to measure the amount of Gag protein in cells transfected with various Gag constructs and to determine the number of virus particles released to the medium. All Gag polyproteins lack the PR domain at the C terminus except SKL, which has a point mutation at the active site in PR. (B) RPA to measure the amount of neo RNA in virions and the amounts of neo RNA and cellular gapdh RNA in transfected cells. The Rnase + lane contains a mixture of the neo probe RNA and gapdh probe RNA treated with RNase cocktail (RNase A and T1). The Rnase − lane was not treated with RNase cocktail. (C) Comparison of the β-Gal activity measured in the yeast three-hybrid binding assays and the packaging efficiency determined in vivo. The packaging efficiency was calculated as follows: the amount of neo RNA in virions (as measured by RPA [shown in panel B]) was normalized to the amount of cellular neo RNA, relative to the level of cellular gapdh RNA (as measured by RPA of whole-cell lysates [shown in panel B]). This calculated neo RNA was then normalized to the number of virions (as measured by RIPA [shown in panel A]). Each experiment was done three times, and the means ± standard deviations (error bars) are shown. Both the β-Gal activity and packaging efficiency were normalized to those for ΔPR. The relative packaging efficiency of the SKL mutant is shown above the bar. The β-Gal activities were summarized from Fig. 1.
In the distal region, the mutation of RKR to either SKL or SKR greatly reduced binding, whereas mutation of RKR to RTL restored the binding (Fig. 1). Previously we have shown that the RTL mutant packages the viral genomic RNA as efficiently as the wt (31), whereas the SKL mutant is defective in RNA encapsidation. These findings suggest that only the first basic residue is required for binding and efficient packaging.
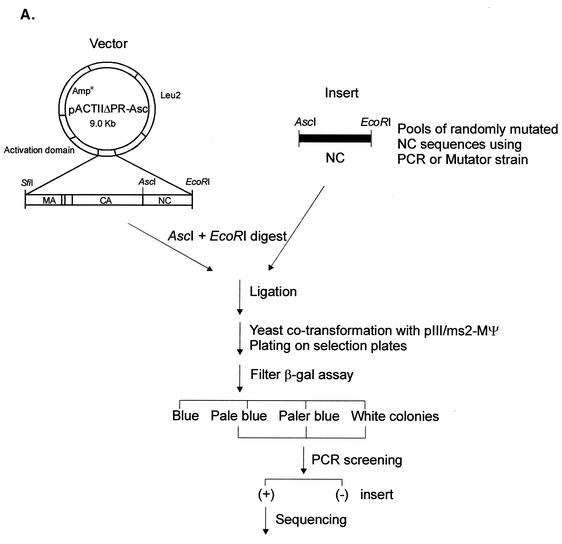
In vivo screening of random NC mutants reveals additional amino acids important for RNA binding.
To examine a large collection of mutations in RSV NC, we randomly mutagenized the NC sequences by using either PCR or an E. coli DNA repair-deficient mutator strain, as described in Materials and Methods. The pools of randomly mutated NC sequences were swapped for the wt NC sequences of the activation domain hybrid plasmid and then transformed into yeast cells to screen large number of mutants (Fig. 4A). First, yeast cells were stably transformed with the pIII/ms2-MΨ RNA hybrid plasmid, and then the mutant library DNA plasmids were introduced into these MΨ+ yeast cells. After 3 days of incubation, transformant colonies were directly transferred to filters, and enzymatic activity was assayed by the filter β-Gal assay. Of 600 colonies screened in four β-Gal screens, 49 colonies were found to be pale blue or paler blue than the wt cells carrying the ΔPR plasmid. All these candidates were amplified by PCR to examine whether NC sequences were present in the protein hybrid expression plasmid. Thirty-nine colonies contained NC inserts, and these inserts were sequenced to locate mutations in the NC sequences. About half of them had either one, two, or three amino acid changes in the NC sequences (9 single mutations, 3 double mutations, and 10 triple mutations). The other 17 clones had multiple mutations (6 mutants with four mutations and 11 mutants with five or more mutations). In our PCR-based random mutagenesis, we used concentrations of dATP and dGTP which were 10 times lower than dCTP and dTTP. However, this did not lead to a bias in the mutational changes, since of the 180 mutations analyzed, 54 (30%) had misincorporation of C or T nucleotide instead of A or G, 49% had misincorporation of A or G, and the remaining 21% had neutral changes.
FIG. 4.
(A) Strategy for in vivo screening of random NC mutants using the yeast three-hybrid system. Pools of randomly mutated NC sequences by using either PCR or a mutator strain were swapped for the wt NC sequences of the pACTIIΔPR-Asc protein hybrid plasmid. The ligated DNAs were used to cotransform yeast cells along with the pIII/MS2-MΨ RNA hybrid plasmid and plated onto plates lacking Ura and Leu to select both plasmids. β-Gal activity of each transformant was qualitatively measured by filter β-Gal assays. Colonies with low β-Gal activities were screened for the NC inserts using PCR. The positive (+) clones were sequenced to locate mutations in the NC sequences. (B) Mutational changes resulting from random mutagenesis and in vivo screening. The amino acid sequence of NC is shown at the top, with the numbering starting from the amino terminus of NC. The two CH boxes are underlined, and the Zn-coordinating residues within the CH boxes are shaded. The mutational changes predicted to significantly lower the β-Gal activity are shown in bold type. The asterisk in line 9 represents a mutational change from glutamine to arginine at residue 33 of NC. A stop mutation at Gln69 (line 12) led to the truncation of NC at the C terminus. The relative β-Gal activities indicated are the averages from three independent assays. The mutant with low β-Gal activity (less than 20% of that of ΔPR) are indicated by shading. (C) Additional site-directed mutants. ΔN has residues 1 to 13 of NC deleted; ΔC lacks all of the amino acids following position 68. The G52W mutant which had β-Gal activity of less than 0.2 is shaded.
The informative mutations are shown in Fig. 4B, which gives the results of a liquid β-Gal assay relative to the β-Gal activity of the ΔPR protein. Many mutations were found to map to the amino and carboxy termini of the NC protein. Four mutants (Fig. 4B, lines 1 to 4) had a single amino acid change in the region upstream of the proximal CH box and showed wt levels of binding. This implies that the N-terminal region flanking the proximal CH box is dispensable for RNA binding. This result was confirmed by constructing a site-directed N-terminal truncation mutant, which had a β-Gal activity as high as that of ΔPR (Fig. 4C, ΔN). We also deleted the C-terminal 21 amino acids from NC, and this protein behaved like the wt protein in binding to MΨ RNA (Fig. 4C, line 2, ΔC). Thus, only the central portion of NC, including both the CH boxes and the basic regions surrounding the boxes, contains major determinants for specific binding to MΨ RNA.
In most cases, the residues involved in RNA binding were located within the CH boxes. This is not unexpected, because in the surrounding basic residues, multiple changes would be required for the binding-negative phenotype, given our results from the site-directed mutagenesis studies.
A change from tyrosine at residue 22 to isoleucine (Fig. 4B, line 5) decreased β-Gal activity to 3% that of ΔPR. In many retroviruses, an amino acid containing an aromatic ring such as Tyr, Phe, and Trp is conserved at this position, suggesting that a bulky residue might be required for proper conformation of the zinc finger motif. We isolated three mutants having an amino acid change at glutamine 33, and their β-Gal activities relative to ΔPR ranged from 0.08 to 0.14 (lines 6, 8, and 9). Either leucine or arginine was substituted at this position, but the arginine mutant (shown with an asterisk in Fig. 4B, line 9) had a second mutation at Cys47 which is predicted to decrease binding. In some instances of multiple mutations, a substitution for the Zn-coordinating residues (CCHC) within the CH boxes was simultaneously isolated with other mutations (lines 9 and 10). Thus, it is possible that the lower β-Gal activity of these mutants might be a function of the mutations in the CCHC residues rather than the other changes. A triple mutant (line 10) had two mutations in the distal CH box; one is a change of glutamine 48 to histidine and the other is a change of cysteine 60 to tyrosine. To examine the effect of the histidine mutation, we constructed a single mutant, Q48H (Fig. 4C). This single mutant bound to MΨ RNA as efficiently as ΔPR. Thus, an additional Zn-coordinating residue within the box did not prevent the CCHC residues from Zn2+ ion binding.
A triple mutant (Fig. 4B, line 11) which contained a mutation of glycine 52 to tryptophan and two mutations at the C terminus of the NC domain impaired binding. To test whether the Gly52 mutation alone prevents binding, we engineered a single G52W mutation (Fig. 4C), which displayed the same binding affinity as the triple mutant. In all Orthoretrovirinae, without exception, the histidine residue within the CH boxes is preceded by glycine. A mutation of glycine 54 to valine was obtained (Fig. 4B, line 7) which completely abolished binding, suggesting steric hindrance at this position. To examine the role of glycine at the corresponding position within the proximal CH box (residue 28), we changed glycine 28 to either leucine or isoleucine, but these changes prevented stable expression of the proteins in yeast cells (data not shown). The mutational change of arginine 61 to glycine (line 12) led to the reduction of β-Gal activity, confirming the importance of the first basic residue downstream of the distal CH box for RNA binding, as also shown in mutants SKL and SKR (Fig. 1). Taken together, these results show the importance of the Zn-coordinating residues and some conserved residues within the CH boxes in RNA binding.
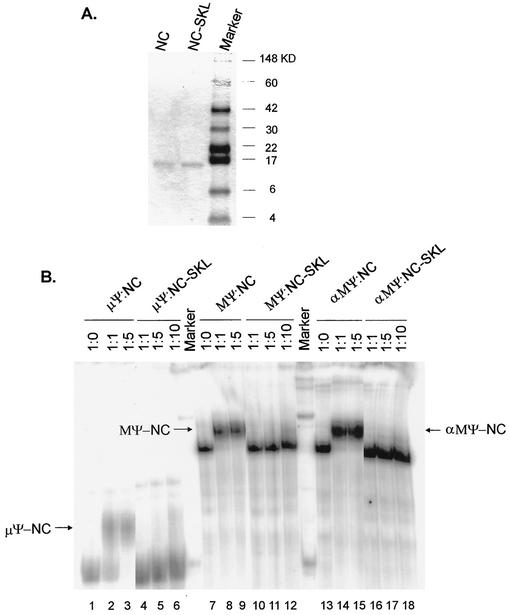
Recombinant NC protein binds to ΨRNA in vitro, but the mutant NC-SKL protein does not.
Since the NC domain in the context of Gag polyprotein specifically bound to Ψ RNA in the in vivo yeast three-hybrid binding assay (31), we examined the abilities of purified NC proteins to bind to Ψ RNA in vitro. Recombinant wt and mutant SKL (Fig. 1) NC proteins were synthesized in E. coli using a T7 RNA polymerase expression system and purified them as described in Materials and Methods. wt NC and NC-SKL were bound to 32P-labeled Ψ RNA (either μΨ, an 82-nt minimal packaging sequence [3], MΨ, or antisense MΨ) in a buffer containing 0.1 mM ZnCl2 (see Materials and Methods) and electrophoresed on native gels to examine the mobility shift (Fig. 5). We found that μΨ, MΨ, and αMΨ shifted to slower-migrating bands (Fig. 5B, lanes 1 to 3, 7 to 9, and 13 to 15) at a RNA/NC molar ratio of 1:1 or 1:5, suggesting the nonspecific NC binding to Ψ RNAs. The property of a rather nonspecific RNA binding was predicted from our earlier observation that yeast cells transformed with a plasmid expressing NC alone are sick, forming tiny colonies, probably due to the nonspecific binding to yeast cellular RNAs (data not shown), and consistent with findings of other laboratories (27, 29). However, the mutant NC-SKL protein did not cause any shift with any RNA (lanes 4 to 6, 10 to 12, and 16 to 18), consistent with the inability to bind RNA in the yeast three-hybrid binding assay.
FIG. 5.
Binding of Ψ RNA to the NC proteins. (A) Purification of NC proteins. wt NC and a mutant version of NC (NC-SKL) were expressed, purified by fast-performance liquid chromatography, electrophoresed on a SDS-10% polyacrylamide gel, and stained with Coomassie brilliant blue. The positions of molecular mass markers (in kilodaltons) are shown to the right of the gel. (B) In vitro-transcribed RNA was not mixed or mixed with NC or NC-SKL proteins at the indicated molar ratios. The band shifts are indicated by arrows. μΨ, an 82-nt minimal packaging sequence; MΨ, a 160-nt packaging sequence; αMΨ, antisense MΨ RNA.
DISCUSSION
In this study, we sought to determine the important residues in NC, in the context of the Gag polyprotein, which are involved in Ψ RNA binding and packaging. We found that substitutions for the Zn-coordinating residues within the CH boxes abolished binding. However, deletion of either CH box had no effect on binding, although this greatly reduced the efficiency of particle assembly and release. Three regions of clustered basic amino acids surrounding the two CH boxes were found to play important roles in RNA binding.
NC proteins are highly basic and can bind to either DNA or RNA with a preference for single strands (reviewed in references 10 and 19). The properties of nucleic acid binding, annealing, and strand transfer activities of NC are involved in many stages of virus replication (reviewed in reference 12). These include the processes of primer tRNA annealing to the primer binding site, of reverse transcription, of retroviral RNA dimerization and packaging, and of viral particle assembly. NC also functions in virus morphogenesis and infectivity. All retroviral NC proteins except those of spumavirus include one or two copies of the CH boxes and positively charged residues clustered around the boxes. In RSV and HIV NC, one of the assembly domains for viral particles overlaps the two Cys-His zinc finger motifs and the flanking basic residues. This domain is required for the formation of viral particles with proper density (7). It has been previously shown that deletion of either of the CH boxes leads to inefficient RNA packaging and that deletion of the proximal box has a greater effect on RNA packaging than the distal one, suggesting that the two boxes are distinct and not functionally equivalent (23, 35). Our results confirmed this, as we found that the mutation of the proximal box led to about sixfold-less viral RNA packaging than mutation of the distal box. However, we found that mutants lacking either of the boxes also showed a great reduction in particle assembly. This is also true for a mutant with both boxes deleted (31). Because of both the lower particle assembly and packaging, these mutants give an apparent wt RNA packaging efficiency. Previously Meric et al. (35) found that deletion of either of the CH boxes produced particles containing the same level of Gag proteins as that for the wt. These researchers used immunoblotting, which is not as quantitative as the immunoprecipitation assay that we used, which could explain the different results.
Not surprisingly, we found that point mutations at the strictly conserved amino acids (Cys21, Cys24, His 29, Cys34, Cys47, and Cys50) completely abolished RNA binding, but those at the nonconserved residues (Lys58 and Gln59) within the CH box did not impair either RNA binding or packaging, consistent with previous studies using point mutations within the CH boxes of RSV, MuLV, and HIV-1 (16, 25, 34, 35). Although NC proteins isolated from mature virus particles have been shown to lack tightly bound Zn2+ ions (25), the nuclear magnetic resonance (NMR) structure of HIV-1 NC demonstrates that the Zn finger structure is stabilized by the coordination of the CCHC residues with Zn (43). Thus, substitutions for Zn-coordinating residues probably alter the NC structure and lead to the defect in RNA binding. However, the precise deletion of either of the CH boxes may not have a gross effect on the overall structure of the NC protein and so do not affect the ability of mutant NC protein to bind to Ψ RNA. In contrast to RSV, MuLV, and HIV-1 NC Zn-finger mutants that produced virus with no genomic RNA, simian immunodeficiency virus (SIV) (Mne) NC mutants with substitutions at Zn-coordinating residues were found to package viral RNA levels comparable to or greater than those found in the wt virus, suggesting that the NC domain of SIV (Mne) is not as sensitive to alteration with respect to genomic RNA packaging as are HIV-1, RSV, and MuLV (22).
Many studies have been undertaken to characterize roles of the clustered positively charged residues surrounding the two CH boxes for RNA packaging (11, 18, 39-41). These results indicated that certain positively charged residues are more critical for RNA binding and packaging than others and that multiple substitutions have greater effects. Our systematic alanine substitutions in the basic region linking the two CH boxes suggests that three positively charged residues are essential for both RNA binding and packaging, but the precise residues are not important. Removal of positively charged residues either reduces the affinity of NC for the negatively charged RNA or causes conformational changes in the protein. The former explanation is more likely, since substitutions for more than three basic residues, regardless of their positions, abolished binding. Many retroviral NCs, including that of HIV-1, contain several positively charged amino acids within the CH boxes. NMR structural studies of HIV-1 (13) showed that certain basic residues within the box interact with nucleotides in one of the stem-loops of the HIV-1 packaging region, and alanine substitutions at these residues reduced the efficiency of RNA packaging (40). However, unlike other retroviruses, RSV NC has only a single lysine residue within the two boxes (Lys58), and in contrast to the HIV-1 results, we found that its mutation to alanine led to both wt binding and RNA packaging as efficiently as ΔPR.
Truncation of the first 15 amino acids from the amino terminus of NC did not alter RNA binding. This is consistent with works of others (15, 46), in which a shorter version of HIV-1 NC with the N-terminal 12 residues deleted did not greatly affect RNA binding activity. We also found that deletion of the carboxy terminus, which left only eight amino acids downstream of the distal CH box, has no effect on RNA binding. This indicated that the minimal active domain of NC is a 53-residue peptide, which might form the globular structure of NC necessary for RNA binding during the process of specific encapsidation. The NMR structures of MuLV and HIV-1 NC reveals a central globular domain containing the zinc finger(s) with flexible amino- and carboxy-terminal domains (14, 36).
Most retroviral NC proteins contain at least one aromatic amino acid within the zinc finger motifs (Tyr in RSV and MuLV; Phe in HIV-1). We isolated a single mutation at Tyr22 within the proximal CH box which completely abrogated binding. In other retroviruses such as MuLV and HIV-1, mutations of this highly conserved residue to amino acids without aromatic rings greatly reduced both RNA binding and packaging (11, 16, 24, 34, 46). The NMR structure of HIV-1 NC indicates that this bulky residue is at the surface and involved in hydrophobic interactions with RNA (13, 44). Thus, the bulky tyrosine residue in RSV NC might be structurally important for proper conformation of the zinc finger motif and/or serve as a recognition element for Ψ RNA, as in HIV-1 NC.
Overall, it appears that trans-acting determinants for RNA packaging reside in the clustered basic residues surrounding the CH boxes, which is required for the recognition and binding to Ψ RNA. The property of binding is highly likely to be the electrostatic interactions between the basic residues and the acidic phosphodiester backbone of the nucleic acids. However, both the basic residues itself and the specific conformation of NC that is mainly contributed by the neighboring zinc finger motifs are essential.
NC also plays important roles for assembly of viral particles (5). Although there is evidence supporting a direct role for NC in establishing Gag-Gag interactions (33, 48), a more plausible mechanism is that NC indirectly influences the Gag-Gag interactions mediated by RNA binding. RNA seems to work as a scaffold upon which Gag proteins tightly condense (8, 38, 47). Yu et al. (47) reported that while deletion of the positively charged residues between the two CH boxes abrogated assembly of RSV Gag particles in vitro, multiple alanine substitutions for cysteine and histidine residues within the proximal box did not affect the efficiency of RSV Gag particles assembled in vitro. This result in combination with our results suggests that there might be two distinct structural requirements of NC for processes of retroviral assembly. In this hypothesis, during genomic RNA encapsidation, clustered basic residues in NC with the proper conformation recognize and bind to a high-affinity binding site (Ψ) in the cell where Gag and genomic RNA concentrations are low, but the clustered positively charged residues can mediate low-affinity binding to any RNA and Gag-Gag interactions at the cytoplasmic membrane during the process of viral assembly.
Acknowledgments
We thank Michael Emerman for helpful comments on the manuscript.
This work was supported in part by a grant from the National Cancer Institute (CA 18282) to M.L.L. E.-G.L. was supported in part by an NIH postdoctoral training grant (T32 CA09229).
REFERENCES
- 1.Aronoff, R., and M. L. Linial. 1991. Specificity of retroviral RNA packaging. J. Virol. 65:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banks, J. D., and M. L. Linial. 2000. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J. Virol. 74:456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banks, J. D., A. Yeo, K. Green, F. Cepeda, and M. L. Linial. 1998. A minimal avian retroviral packaging sequence has a complex structure. J. Virol. 72:6190-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel, P. L., J. A. Roecklein, D. SenGupta, and S. Fields. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72-77. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 67:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz, R. D., A. Ohagen, S. Hoglund, and S. P. Goff. 1995. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J. Virol. 69:6445-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowzard, J. B., R. P. Bennett, N. K. Krisina, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin, J. M. 1985. Genome structure, p. 17-74. In R. Weiss, N. Teich, H. Varmus, and J. Coffin (ed.), RNA tumor viruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Dannull, J., A. Surovoy, G. Jung, and K. Moelling. 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 13:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darlix, J.-L., M. Lapadat-Tapolsky, H. de Rocquigny, and B. P. Roques. 1995. First glimpses at structure-function relationship of the nucleocapsid protein of retroviruses. J. Mol. Biol. 254:523-537. [DOI] [PubMed] [Google Scholar]
- 13.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 14.Demene, H., N. Jullian, N. Morellet, H. Rocquigny, F. Cornille, B. Maigret, and B. Roques. 1994. Three-dimensional 1H NMR structure of the nucleocapsid protein NCp10 of MoMuLV. J. Biomol. NMR 4:153-170. [DOI] [PubMed] [Google Scholar]
- 15.de Rocquigny, H., C. Gabus, A. Vincent, M. C. Fournie-Zaluski, B. Roques, and J. L. Darlix. 1992. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. USA 89:6472-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupraz, P., S. Oertle, C. Meric, P. Damay, and P. F. Spahr. 1990. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J. Virol. 64:4978-4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans, R. M., and S. M. Hollenberg. 1988. Zinc fingers: gilt by association. Cell 52:1-3. [DOI] [PubMed] [Google Scholar]
- 18.Fu, X., R. A. Katz, A. M. Skalka, and J. Leis. 1988. Site-directed mutagenesis of the avian retrovirus nucleocapsid protein, pp12. J. Biol. Chem. 263:2140-2145. [PubMed] [Google Scholar]
- 19.Gelfand, C. A., Q. Wang, S. Randall, and J. E. Jentoft. 1993. Interactions of avian myeloblastosis virus nucleocapsid protein with nucleic acids. J. Biol. Chem. 268:18450-18456. [PubMed] [Google Scholar]
- 20.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 21.Goodman, M. F., S. Keener, and S. Guidotti. 1983. On the enzymatic basis for mutagenesis by manganese. J. Biol. Chem. 258:3469-3475. [PubMed] [Google Scholar]
- 22.Gorelick, R. J., R. E. Beveniste, T. D. Gagliardi, T. A. Wiltrout, L. K. Busch, W. J. Bosche, L. V. Coren, J. D. Lifson, P. J. Bradley, L. E. Henderson, and L. O. Arthur. 2002. Nucleocapsid protein zinc-finger mutants of simian immunodeficiency virus strain Mne produce virions that are replication defective in vitro and in vivo. Virology 253:259-270. [DOI] [PubMed] [Google Scholar]
- 23.Gorelick, R. J., D. J. Chabot, A. Rein, L. E. Henderson, and L. O. Arthur. 1993. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 67:4027-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jentoft, J. E., L. M. Smith, X. D. Fu, M. Johnson, and J. Leis. 1988. Conserved cysteine and histidine residues of the avian myeloblastosis virus nucleocapsid protein are essential for viral replication but are not “zinc-binding fingers.” Proc. Natl. Acad. Sci. USA 85:7094-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan, A. H., M. Manchester, and R. Swanstrom. 1994. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximal efficiency. J. Virol. 68:6782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karpel, R. L., L. E. Henderson, and S. Oroszlan. 1987. Interactions of retroviral structural proteins with single-stranded nucleic acids. J. Biol. Chem. 262:4961-4967. [PubMed] [Google Scholar]
- 28.Keohavong, P., and W. G. Thilly. 1989. Fidelity of DNA polymerase in DNA amplification. Proc. Natl. Acad. Sci. USA 86:9253-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan, R., and D. P. Giedroc. 1994. Nucleic acid binding properties of recombinant Zn2 HIV-1 nucleocapsid protein are modulated by COOH-terminal processing. J. Biol. Chem. 269:22538-22546. [PubMed] [Google Scholar]
- 30.Lee, E.-G., and M. L. Linial. 2000. Yeast three-hybrid screening of Rous sarcoma virus mutants with a randomly mutagenized minimal packaging signal reveals regions important for Gag interactions. J. Virol. 74:9167-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, E.-G., A. Yeo, B. Kraemer, M. Wickens, and M. L. Linial. 1999. The Gag domains for avian retroviral RNA encapsidation determined using two independent methods. J. Virol. 73:6282-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin-Goerke, J. L., D. J. Robbins, and J. D. Burczak. 1997. PCR-based random mutagenesis using manganese and reduced dNTP concentration. BioTechniques 23:409-412. [DOI] [PubMed] [Google Scholar]
- 33.McDermott, J., L. Farrell, R. Ross, and E. Barklis. 1996. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J. Virol. 70:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meric, C., and S. P. Goff. 1989. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J. Virol. 63:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meric, C., E. Gouilloud, and P.-F. Spahr. 1988. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J. Virol. 62:3328-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morellet, N., H. de Rocquigny, Y. Mely, N. Jullian, H. Demene, M. Ottmann, D. Gerard, J.-L. Darlix, M.-C. Fournie-Zaluski, and B. Roques. 1994. Conformational behavior of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H-NMR. J. Mol. Biol. 235:287-301. [DOI] [PubMed] [Google Scholar]
- 37.Moscovici, C., M. G. Moscovici, H. Jiminez, M. M. C. Lai, M. J. Hayman, and P. K. Vogt. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11:95-103. [DOI] [PubMed] [Google Scholar]
- 38.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ottmann, M., C. Gabus, and J. L. Darlix. 1995. The central globular domain of the nucleocapsid protein of human immunodeficiency virus type 1 is critical for virion structure and infectivity. J. Virol. 69:1778-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poon, D. T., J. Wu, and A. Aldovini. 1996. Charged amino acid residues of human immunodeficiency virus type 1 nucleocapsid p7 protein involved in RNA packaging and infectivity. J. Virol. 70:6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmalzbauer, E., B. Strack, J. Dannull, S. Guehmann, and K. Moelling. 1996. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J. Virol. 70:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sengupta, D. J., B. L. Zhang, B. Kraemer, P. Pochart, S. Fields, and M. Wickens. 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA 93:8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Summers, M. F., L. E. Henderson, M. R. Chance, J. W. Bess, T. L. South, P. R. Blake, I. Sagi, G. Perez-Alvarado, R. C. Sowder, and D. R. Hare. 1992. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1:563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Summers, M. F., T. L. South, and D. R. Hare. 1990. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry 29:329-340. [DOI] [PubMed] [Google Scholar]
- 45.Tan, R., and A. D. Frankel. 1998. A novel glutamine-RNA interaction identified by screening libraries in mammalian cells. Biochemistry 95:4247-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urbaneja, M. A., B. P. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. R. Casas-Finet. 1999. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 287:59-75. [DOI] [PubMed] [Google Scholar]
- 47.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y. Q., and E. Barklis. 1995. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J. Virol. 69:5716-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]