Abstract
Although combinatorial antibody libraries have solved the problem of access to large immunological repertoires, efficient production of these complex molecules remains a problem. Here we demonstrate the efficient expression of a unique large single-chain (lsc) antibody in the chloroplast of the unicellular, green alga, Chlamydomonas reinhardtii. We achieved high levels of protein accumulation by synthesizing the lsc gene in chloroplast codon bias and by driving expression of the chimeric gene using either of two C. reinhardtii chloroplast promoters and 5′ and 3′ RNA elements. This lsc antibody, directed against glycoprotein D of the herpes simplex virus, is produced in a soluble form by the alga and assembles into higher order complexes in vivo. Aside from dimerization by disulfide bond formation, the antibody undergoes no detectable posttranslational modification. We further demonstrate that accumulation of the antibody can be modulated by the specific growth regime used to culture the alga, and by the choice of 5′ and 3′ elements used to drive expression of the antibody gene. These results demonstrate the utility of alga as an expression platform for recombinant proteins, and describe a new type of single chain antibody containing the entire heavy chain protein, including the Fc domain.
Currently, there are a number of heterologous protein expression systems available for the production of recombinant proteins, and each of these systems offers distinct advantages in terms of protein yield and ease of manipulation and cost of operation (1). mAbs are produced primarily by culture of transgenic mammalian cells in fermentation facilities. Because of high capital costs and the inherent complexity of mammalian production systems, monoclonal antibody production capacity will fall substantially short of requirements over the next 5 years (2).
As a consequence of the projected shortfall in mAb production via mammalian cell culture, alternative, cost-effective, means to produce mAbs will be required to maintain the present pace of therapeutic protein development. Yeast and bacterial systems, although more economical in terms of media components, have several shortcomings, including an inability to efficiently produce properly folded functional molecules, as well as poor yields of more complex proteins. In addition to traditional fermentation, several groups have sought to exploit the productivity of terrestrial plants for mAb production (3–5). In such systems, the plant itself becomes the bioreactor, with the antibody deposited into leaf or seed tissue. Although plants afford an economy of scale unprecedented in the biotechnology industry (one can plant thousands of acres of corn, for example), there are several inherent drawbacks to this approach as well. First, the length of time required from the initial transformation event to having usable (mg to gram) quantities of recombinant protein on hand, can be as long as 3 years for species such as corn. A second concern surrounding the expression of human therapeutics in food crops is the potential for gene flow (via pollen) to surrounding crops (6), as occurred between transgenic corn expressing Bacillus thuringiensis insecticidal proteins and native landraces (7). These concerns raise the possibility that regulatory agencies will prohibit the open cultivation of transgenic food plants (like corn, rice, and soybeans) expressing human therapeutics.
Only a few attempts have been made to engineer chloroplasts for the expression of therapeutic proteins (8), although in some instances quite high levels of recombinant protein expression have been achieved in this organelle (9–12). There have been even fewer reports on the generation of transgenic algae for the expression of recombinant proteins, even though green algae have served as a model organism for understanding everything from the mechanisms of light and nutrient regulated gene expression to the assembly and function of the photosynthetic apparatus (13). In previous work, we demonstrated that by optimizing codon usage of a GFP reporter gene to reflect the codon bias of the Chlamydomonas reinhardtii chloroplast genome, we were able to increase GFP accumulation by ≈80-fold, to 0.5% of soluble protein (14).
In this work, we show that human monoclonal antibodies can be expressed in transgenic algae chloroplasts. We engineered a large single-chain (lsc) antibody gene in C. reinhardtii chloroplast codon bias, and used the C. reinhardtii chloroplast atpA or rbcL promoters and 5′ untranslated regions to drive expression. This antibody is directed against herpes simplex virus (HSV) glycoprotein D (15), and contains the entire IgA heavy chain protein fused to the variable region of the light chain by a flexible linker peptide. The lsc antibody accumulates as a soluble protein in transgenic chloroplasts, and binds herpes virus proteins, as determined by ELISA assays. This lsc antibody assembles into higher order structures (dimers), in vivo, and contains no obvious posttranslational modifications, aside from the disulfide bonds associated with dimerization. These results demonstrate the utility of algae as an expression platform for complex recombinant proteins.
Experimental Procedures
C. reinhardtii Strains, Transformation, and Growth Conditions.
All transformations were carried out on C. reinhardtii strain 137c (mt+) as described in ref. 14. Cultivation of C. reinhardtii transformants for expression of HSV8-lsc was carried out in TAP medium (16) at 23°C under illumination and cell density as described.
Plasmid Construction.
All DNA and RNA manipulations were carried out essentially as described in refs. 17 and 18. The coding region of the HSV8-lsc gene was synthesized de novo according to the method of ref. 19 and as described in ref. 14. The resulting 1,926-bp PCR product was cloned into plasmid pCR2.1 TOPO (Invitrogen) according to the manufacturers protocol. The atpA and rbcL promoters and 5′ UTR and the rbcL 3′ UTR were generated via PCR and described in ref. 14.
Southern and Northern Blots.
Southern blots and 32P labeling of DNA for use as probes were carried out as described in ref. 17. Radioactive probes used on Southern blots included the 2.2-kb BamHI/PstI fragment of p322 (probe 5′ p322), the 2.0-kb BamHI/XhoI fragment of p322 (probe 3′ p322) and the 1,926-bp NdeI/XbaI fragments from HSV8-lsc. Additional radioactive probes used in Northern blot analysis included the psbA cDNA. Northern and Southern blots were visualized by using a Packard Cyclone Storage Phosphor System equipped with optiquant software.
Protein Expression, Western Blotting, and ELISA.
For Western blot analysis proteins were isolated from C. reinhardtii as described in ref. 14. Flag affinity-purified C. reinhardtii HSV8-lsc were isolated in Tris-buffered saline (25 mM Tris, pH 7.4/150 mM NaCl) containing Complete protease inhibitor tablets (Roche Diagnostics) and PMSF at 1 mM final concentration. Extracts were purified by using anti-Flag M2 agarose beads (Sigma) according to the manufacturer's protocol. ELISAs were carried out in volumes of 100 μl in 96-well microtiter plates (Costar) coated with 100 μl of HSV proteins. Samples for use in ELISA were diluted in blocking buffer comprised of PBS (137 mM NaCl/2.7 mM KCl/1.8 mM K2HPO4/10 mM Na2HPO4, pH 7.4) and 5% nonfat dry milk. Incubations were carried out for 8 h at 4°C with rocking. Plates were then rinsed with PBS plus 0.5% Tween 20 three times, then incubated with anti-Flag antibody (Sigma) for 8 h at 4°C. Plates were again rinsed three times and incubated with alkaline phosphatase conjugated goat-anti-mouse antibody (Santa Cruz Biotechnology) for 8 h at 4°C. Plates were once again rinsed three times with PBS plus 0.5% Tween 20 and developed with 100 μl of p-nitrophenyl phosphate (Sigma). Reactions were terminated by the addition of 50 μl 3 N NaOH. Protein concentrations were determined by using BioRad's protein assay reagent. Western blots were carried out as described in ref. 18, by using a murine anti-Flag primary antibody (Sigma) and an alkaline phosphatase conjugated goat anti-mouse secondary antibody (Santa Cruz Biotechnology).
Results
De Novo Synthesis of a lsc Antibody Gene in C. reinhardtii Chloroplast Codon Bias.
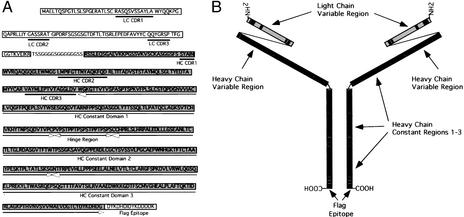
To develop robust expression of recombinant antibodies in the C. reinhardtii chloroplast, we synthesized a single chain antibody gene by using codons optimized to reflect abundantly translated C. reinhardtii chloroplast mRNAs. The antibody we engineered was derived from a human antibody library displayed on phage, and identified by panning with herpes simplex virus proteins (15). This antibody, termed HSV8, was previously shown to bind the viral surface antigen glycoprotein D (20), and both Fab or IgG1 versions of this antibody act as efficient neutralizing antibodies, in vivo and in vitro (15, 20). As simple single-chain Fv antibodies can be made in bacterial or yeast systems, we attempted the synthesis of a more complex antibody in chloroplast, but one that could still be translated from a single mRNA. We designed a single chain antibody containing the entire heavy chain region, fused to the variable region of the light chain gene via a flexible linker peptide. The primary amino acid sequence of this unique, lsc protein is shown in Fig. 1A, and a model of the assembled (dimerized) molecule is shown in Fig. 1B.
Figure 1.
Synthesis of a lsc antibody gene optimized for C. reinhardtii chloroplast expression. (A) The amino acid sequence is indicated by single-letter code and the heavy chain region is boxed and shaded with specific domains indicated by open arrows. The light chain variable region is boxed but unshaded. The complementarity-determining regions (CDR) are indicated by heavy boxes, and the Flag tag is indicated under the amino acid sequence. The linker region is unboxed. (B) Schematic diagram of the lsc protein as a dimer; the heavy chain is gray, the CDRs are black, and the linker and hinge region are indicated by a thin line. Cysteine residues capable of forming disulfide bonds are indicated as a line between the heavy chain constant regions, and the flag epitope is indicated.
Construction of a Chimeric C. reinhardtii Chloroplast lsc Antibody Gene.
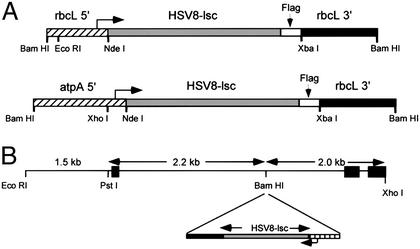
To generate transgenic chloroplast expressing the recombinant antibody, we constructed chimeric genes containing either the atpA or rbcL promoter and 5′ UTRs fused to the codon optimized HSV8-lsc coding region, followed by the rbcL 3′ UTR (Fig. 2A). Integration of genes into the chloroplast genome occurs by homologous recombination, and requires sequence homology between the transformation vector and the chloroplast genome (21). We used the C. reinhardtii chloroplast transformation vector p322 (14). As diagrammed in Fig. 2B, the chimeric antibody genes were ligated into the BamHI site of p322 to create plasmid p322/atpA-HSV8 and plasmid p322/rbcL-HSV8. The p322/HSV8 constructs were cotransformed into C. reinhardtii chloroplasts by means of particle bombardment (21), along with plasmid p228, containing a 16S ribosomal gene conferring spectinomycin resistance.
Figure 2.
Restriction maps of HSV8-lsc genes for expression in C. reinhardtii chloroplasts. (A) Relevant restriction sites delineating the rbcL 5′ UTR (BamHI/NdeI), the HSV8 coding region and flag tag (NdeI/XbaI), and the rbcL 3′ UTR (XbaI/BamHI), as well as relevant restriction sites of the atpA 5′ UTR (BamHI/NdeI), the HSV8 coding region and flag tag (NdeI/XbaI), and the rbcL 3′ UTR (XbaI/BamHI) are shown. (B) Restriction map showing the site of integration of the HSV8-lsc genes into plasmid p322 for integration into the C. reinhardtii chloroplast genome. p322 DNA includes the 5.7-kb region from EcoRI to XhoI in the C. reinhardtii chloroplast genome corresponding to position 44,877–50,577 (www.biology.duke.edu/chlamy_genome/chloro.html). Double-headed arrows indicate regions corresponding to the probes used in the Southern blot analysis. Black boxes indicate, from left to right, psbA exon 5, and the 5S and a small portion of the 23S ribosomal RNA genes, respectively.
Southern Blot Analysis of HSV8-lsc Transgenic Chloroplast.
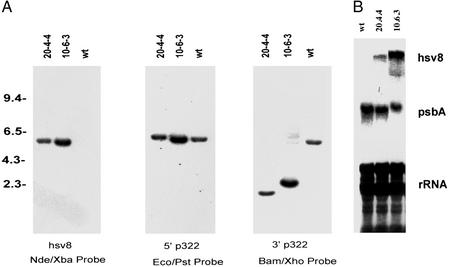
Primary transformants were selected on media containing spectinomycin and screened by Southern blot analysis for HSV8 gene integration. HSV8 positive transformants were taken through additional rounds of selection to isolate homoplasmic lines in which all copies of the chloroplast genome contained the introduced HSV8-lsc gene. Two homoplasmic transformants were selected, one 10-6-3, containing the atpA promoter driving HSV8-lsc and the other, 20-4-4, containing the rbcL promoter driving HSV8-lsc. Genomic DNA from WT and the two HSV8-lsc transformants was digested with EcoRI and XhoI, separated on agarose gels, and subjected to Southern blot analysis. As shown in Fig. 3A, hybridization with a 32P-labeled NdeI/XbaI fragment of the HSV8 coding region identified a 6.0-kb band in both the atpA-HSV8 and rbcL-HSV8 transgenic strains, whereas no detectable band was observed in the WT lane, as expected. When these same blots were hybridized with a 32P-labeled 1.5-kb EcoRI to PstI fragment from the 5′ end of p322, a 5.7-kb fragment was visualized in the WT sample, whereas a slightly larger 6.0-kb fragment was identified in the two transgenic strains. Hybridization with a 32P-labeled BamHI/XhoI fragment from the 3′ end of p322, resulted in the visualization of 2.5 and 2.0 kb in 10-6-3 and 20-4-4, respectively, whereas the WT strain again showed a band of 5.7 kb. These data demonstrate that the HSV8 gene had correctly integrated into the p322 silent site of the chloroplast genome, and that all copies of the chloroplast genome contained the HSV8 gene.
Figure 3.
Southern and Northern blot analysis of HSV8-lsc in C. reinhardtii chloroplast transformants. (A) C. reinhardtii DNAs were prepared as described in experimental procedures, digested with EcoRI and XhoI, and subjected to Southern blot analysis. Filters were hybridized with the radioactive probes indicated by the double-headed arrows in Fig. 2B. (B) Detection of chloroplast-expressed HSV8-lsc mRNA in transgenic C. reinhardtii strains. Total RNA isolated from untransformed (WT), atpA HSV8-lsc transformed (10-6-3), and rbcL transformed (20-4-4) strains was separated on denaturing agarose gels and blotted to nylon membrane. The membranes were either stained with methylene blue (Bottom) or hybridized with a psbA cDNA probe (Middle) or a hsv8-specific probe (Top).
Accumulation of HSV8-lsc mRNA in Transgenic Strains.
To determine whether the HSV8 genes were transcribed in transgenic C. reinhardtii chloroplasts, Northern blot analysis of total RNA was used. Ten micrograms of total RNA from WT and the two transgenic lines was separated on denaturing agarose gels and blotted to nylon membrane. Duplicate filters were stained with methylene blue and hybridized with either a 32P-labeled psbA cDNA probe or an HSV8-specific probe. As shown in Fig. 3B, ribosomal RNA and psbA mRNA accumulate to similar levels in WT and each of the transgenic strains, demonstrating that equal amounts of RNA were loaded, and that introduction of the transgene does not alter endogenous mRNA accumulation. Hybridization with an HSV8-specific probe showed that strains 10-6-3 and 20-4-4 accumulate HSV8-lsc mRNA of the correct size, whereas no HSV8 signal is detected in the WT lane, as expected.
Analysis of HSV8-lsc Protein Accumulation in Transgenic C. reinhardtii Chloroplasts.
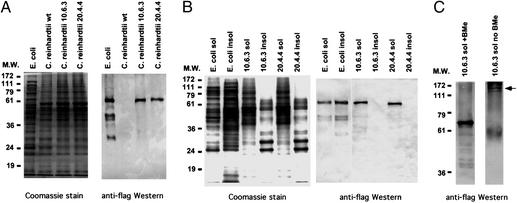
To determine whether HSV8-lsc protein accumulated in the transgenic lines, HSV8-lsc antibody levels were measured by Western blot analysis using an anti-flag antibody. Twenty micrograms of total protein from an Escherichia coli strain expressing HSV8-lsc from a pET vector, and 20 μg of total protein from C. reinhardtii WT and the two transgenic lines, was separated by SDS/PAGE and either stained with Coomassie blue (Fig. 4A Left), or subjected to Western blot analysis with anti-Flag antisera (Fig. 4A Right). For bacterial expression, the NdeI/BamHI fragment of codon optimized HSV8-lsc gene was ligated into a pET vector, and expression was induced by addition of isopropyl β-d-thiogalactoside. The Coomassie- stained gel indicates that equal amounts of protein were loaded in each lane, and that overall protein accumulation is normal in the transgenic lines. Western blot analysis of the same samples using an anti-Flag antibody showed a robust signal of the correct molecular weight in both of the HSV8-lsc transgenic strains and E. coli, but no signal in the C. reinhardtii WT lane, as expected.
Figure 4.
Expression of HSV8-lsc proteins in bacteria and chloroplast. (A) Twenty micrograms of crude protein from E. coli, WT C. reinhardtii, and transgenic lines 10-6-3 and 20-4-4 were separated by SDS/PAGE and either stained with Coomassie blue (Left) or blotted to nitrocellulose membrane and decorated with an anti-flag antibody (Right). (B) Proteins from E. coli and C. reinhardtii expressing the Hsv8-lsc antibody were separated into soluble and insoluble pellets by centrifugation. Twenty micrograms of protein were either stained with Coomassie blue (Left) or blotted to nitrocellulose membrane and decorated with an anti-flag antibody (Right). (C) Soluble proteins from C. reinhardtii transgenic line 10-6-3 were either treated with (+Bme) or without (no Bme) reducing agent before separation on SDS/PAGE. Proteins were blotted to nitrocellulose membrane and decorated with anti-flag antibody.
Characterization of HSV8-lsc Antibodies Expressed in E. coli and Chloroplast.
To ascertain whether the HSV8-lsc that accumulated in C. reinhardtii chloroplast was functional, we characterized the chloroplast-expressed protein along with that of the bacterial-expressed HSV8-lsc. HSV8-lsc transgenic bacteria and algae were resuspended in Tris-buffered saline, and the cells lysed by sonication. Soluble proteins were separated from insoluble proteins by centrifugation. Equal amounts of protein from the soluble fractions and from the insoluble pellets were separated by SDS/PAGE, and HSV8-lsc proteins visualized by Western blot analysis. As shown in Fig. 4B, ≈60% of the HSV8-lsc produced in bacteria partitioned to the insoluble fraction, whereas the HSV8-lsc produced in chloroplast was found exclusively in the soluble fraction.
To determine whether chloroplast-expressed antibodies contained any posttranslational modifications we first examined the antibodies by SDS/PAGE and Western blot analysis on reducing and nonreducing gels. Under nonreducing conditions any disulfide bonds formed between the two heavy chain moieties of the antibody should remain intact allowing the antibody to migrate as a larger species. As shown in Fig. 4C (arrowhead), under nonreducing conditions chloroplast-expressed HSV8-lsc runs as a much larger protein of ≈140 kDa, the size expected of a dimer. Treatment with BMe, to reduce disulfide bonds, results in the migration of the chloroplast HSV8-lsc proteins at the predicted molecular mass of the monomer at 68 kDa. To ascertain whether any other posttranslational modifications might be present in the chloroplast-expressed proteins, we characterized the bacterial- and chloroplast-expressed proteins by mass spectrometry. The mass spectra of peptide fragments from both the E. coli- and chloroplast-expressed protein have an almost identical pattern, indicating that no additional modifications are made to the chloroplast protein (data not shown).
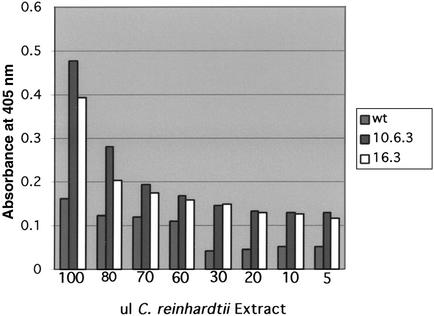
Finally, to determine whether the HSV8-lsc accumulating in the transgenic chloroplast was functional, we examined the ability of chloroplast-expressed HSV8-lsc to bind HSV8 proteins. HSV8-lsc was purified from transgenic chloroplast by using an anti-flag affinity resin. As shown in Fig. 5, the chloroplast produced antibody recognized HSV8 proteins in ELISA assays in a robust manner.
Figure 5.
Characterization of HSV8-lsc binding to HSV8 viral protein via ELISA. Affinity-purified HSV8-lsc from the transgenic C. reinhardtii strains (10-6-3 and 16-3) were screened in an ELISA assay against HSV proteins prepared from virus-infected cells. A total of 100, 80, 70, 60, 30, 20, 10, or 5 μl of Flag-purified hsv8-lsc were incubated in microtiter plates coated with a constant amount of viral protein. Protein concentrations in these affinity-purified extracts were 13 ng/μl, of which ≈10% was HSV8-lsc as judged by Coomassie staining. Equal volumes of WT C. reinhardtii proteins were used as a negative control (concentration = 1 μg/μl).
Modulation of HSV8-lsc Accumulation in Transgenic Algae.
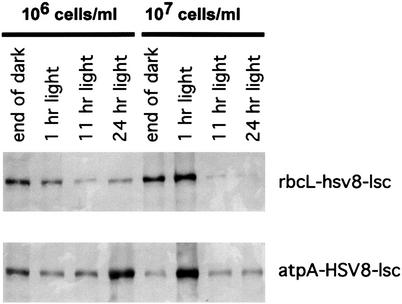
To determine whether we could modulate the expression of HSV8-lsc in C. reinhardtii chloroplast, we examined the effect of different growth regimes on the accumulation of antibody in the two transgenic strains, 10-6-3 and 20-4-4. Cultures of each strain were maintained at 106 or 107 cells per ml and grown either in a 12/12 light/dark cycle (5,000 lux) or under continuous light (5,000 lux). Cells were harvested by centrifugation and 20 μg of soluble protein was resolved on SDS/PAGE, and HSV8-lsc visualized by Western blotting with anti Flag antibody. As shown in Fig. 6, accumulation of HSV8-lsc varies considerably depending on the growth conditions. Expression under the control of the rbcL promoter/5′ UTR in strain 20-4-4, shows a marked increase in the accumulation of antibody at the end of the dark phase or immediately after entering the light phase, regardless of the cell density. Alternatively, The atpA promoter/5′ UTR in strain 10-6-3 drives fairly constant levels of HSV8-lsc production at 106 cells per ml in a light/dark cycle, yet shows a tremendous increase in lsc accumulation on entering the light phase when cells are cultured at 107 cells per ml. When grown under continuous light, both strains exhibited higher accumulation at 106 cells per ml than at 107 cells per ml. These data clearly show that accumulation of HSV8-lsc in chloroplast of C. reinhardtii can be optimized, dependent on the light regime used to culture the cells, the phase in the cycle at which cells are harvested, and the promoter/UTR used to drive expression.
Figure 6.
Effect of growth condition on accumulation of HSV8-lsc in C. reinhardtii. Before harvest, C. reinhardtii transgenic lines 10-6-3 and 20-4-4 were maintained at either 1 × 106 cells per ml or 1 × 107 cells per ml. Cultures were grown either under continuous illumination or under a 12-h light/12-h dark cycle. Total soluble protein (20 μg) was separated by SDS/PAGE, blotted to nitrocellulose membrane, and decorated with anti-Flag antibody.
Discussion
We have expressed a human monoclonal antibody in the chloroplast of green algae. High levels of recombinant protein expression were achieved by optimizing the codon usage within the antibody coding sequence to reflect the codon usage of abundant chloroplast proteins, and by driving expression of the chimeric gene using the chloroplast atpA or rbcL promoters and 5′ UTRs. This lsc antibody contains the entire IgA heavy chain fused to the variable region of the light chain by a flexible linker, and accumulates as a fully soluble protein in chloroplast. The antibody was directed against glycoprotein D of HSV, and the alga-expressed antibody binds to herpes proteins as determined through ELISA. This lsc antibody contains the Fc portion of the heavy chain, which is the site normally involved in intermolecular disulfide bond formation leading to dimerization of the antibody. The chloroplast-expressed antibody assembles into higher order complexes that are susceptible to reduction by BMe, suggesting that the chloroplast-expressed antibody is forming dimers, in vivo. Formation of disulfide bonds in recombinant proteins expressed in chloroplast has previously been shown for human somatotropin, expressed in tobacco chloroplast (8), and was somewhat expected given the presence of protein disulfide isomerase in algal chloroplasts (22). This lsc antibody also contains putative sites for N-linked glycosylation. Chloroplast encoded proteins are not known to be glycosylated and indeed, we see no evidence for glycosylation of our chloroplast-expressed antibody based on mass spectral analysis.
The transgenic strains we have generated show differential accumulation of antibody depending on the promoter used to drive expression, as well as the cell density and light conditions under which they are cultured. The reasons for these large fluctuations in antibody accumulation likely arise from a variety of factors including stability and translational competence of the chimeric mRNAs, and turnover of the antibody protein. These data demonstrate that antibody accumulation can be positively impacted by growth conditions and suggest that high levels of antibody accumulation (>1% of soluble protein) should be achievable in alga, simply by identifying optimal growth conditions compatable with specific promoter and UTR combinations.
Recombinant proteins can be produced in a variety of protein expression systems. Complex therapeutic proteins, like mAbs, are primarily produced by culture of transgenic mammalian cells. Costs for mAb production in cultured mammalian cells averages approximately $150 per gram for raw materials, whereas in plant systems mAb production has been estimated to cost $0.05 per gram (1). Costs for production of mAbs in algal systems are expected to rival those in terrestrial plants, given that media costs for algae are quite reasonable ($0.002 per liter). In addition, algae can be grown in continuous culture, and their growth medium can be recycled (S.E.F., unpublished observation).
Aside from the tremendous cost advantage of producing mAbs in algae, there are a number of specific attributes that make alga ideal candidates for recombinant protein production. First, transgenic algae can be generated quickly, requiring only a few weeks between the generation of initial transformants and their scale up to production volumes. Second, both the chloroplast and nuclear genome of algae can be genetically transformed, opening the possibility of producing a variety of transgenic proteins in a single organism, a requirement if we are to produce multimeric protein complexes, like secretory antibodies. In addition, algae have the ability to be grown on scales ranging from a few milliliters to 500,000 liters in a cost-effective manner. These attributes, and the fact that green algae fall into the GRAS (generally regarded as safe) category, make C. reinhardtii a particularly attractive alternative to other systems for the expression of recombinant proteins. Finally, although this work specifically addresses the production of antibodies in algae, this system should be amenable to the production of virtually any recombinant protein.
Acknowledgments
We thank Drs. Jeff Kelly and Kim Janda for review of the manuscript, Paul Haynes for help with the mass spectrometry analysis of the antibodies, Anna Coragliotti, Jason Schultz, Michelle Hartmann, and Ryan Henry for expert technical assistance, Maria Jose Gonzales for help with ELISA, and Pietro Sanna for his generous gift of HSV extracts. This work was supported by funds from Sea Grant, the National Institutes of Health, and Syngenta Corporation (to S.P.M.).
Abbreviation
- lsc
large single-chain
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY191459).
References
- 1.Dove A. Nat Biotechnol. 2002;20:777–779. doi: 10.1038/nbt0802-777. [DOI] [PubMed] [Google Scholar]
- 2.Motmans K, Bouche B. Antibodies: The Next Generation. New York: Auerbach Grayson; 2000. [Google Scholar]
- 3.Hiatt A, Cafferky R, Bowdish K. Nature. 1989;342:76–78. doi: 10.1038/342076a0. [DOI] [PubMed] [Google Scholar]
- 4.Ma J, Lehner T, Stabila P, Fux C I, Hiatt A. Eur J Immunol. 1994;24:131–138. doi: 10.1002/eji.1830240120. [DOI] [PubMed] [Google Scholar]
- 5.Ma J, Hiatt A, Hein M, Vine N D, Wang F, Satabila P, van Dollweerd C, Mostov K, Lehner T. Science. 1995;268:716–719. doi: 10.1126/science.7732380. [DOI] [PubMed] [Google Scholar]
- 6.Ellstrand N C. Plant Physiol. 2001;125:1543–1545. doi: 10.1104/pp.125.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quist D, Chapela I. Nature. 2001;414:541–543. doi: 10.1038/35107068. [DOI] [PubMed] [Google Scholar]
- 8.Staub J M, Garcia B, Graves J, Hajdukiewicz P T, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll J A, Spatola L, et al. Nat Biotechnol. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- 9.Kota M, Daniell H, Varma S, Garczynski S F, Gould F, Moar W J. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sidrov V A, Kasten D, Pang S Z, Hajdukiewwicz P T, Staub J M, Nehra N S. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- 11.Ruf S, Herrmann M, Berger I, Carrer I J, Bock R. Nat Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- 12.Heifetz P B. Biochimie. 2000;82:655–666. doi: 10.1016/s0300-9084(00)00608-8. [DOI] [PubMed] [Google Scholar]
- 13.Harris E. The Chlamydomonas Sourcebook. New York: Academic; 1989. [Google Scholar]
- 14.Franklin S E, Ngo B, Effuet E, Mayfield S P. Plant J. 2002;30:733–744. doi: 10.1046/j.1365-313x.2002.01319.x. [DOI] [PubMed] [Google Scholar]
- 15.Burioni R, Williamson R A, Sanna P P, Bloom F E, Burton D R. Proc Natl Acad Sci USA. 1994;91:355–359. doi: 10.1073/pnas.91.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorman D S, Levine R P. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatas T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Cohen A, Yohn C B, Bruick R K, Mayfield S P. Methods Enzymol. 1998;297:192–208. [Google Scholar]
- 19.Stemmer W, Crameri A, Ha K, Brennan T, Heyneker H. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 20.De Logu A, Williamson R A, Rozenshteyn R, Ramiro-Ibanez F, Simpson C D, Burton D R, Sanna P P. J Clin Microbiol. 1998;36:3198–3204. doi: 10.1128/jcm.36.11.3198-3204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boynton J E, Gillham N W, Harris E H, Hosler J P, Johnson A M, Jones A R, Randolph-Anderson B L, Robertson D, Klein T M, Shark K B, Sanford J C. Science. 1988;240:1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Mayfield S P. Science. 1997;278:1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]