Abstract
The reversible phosphorylation of tyrosine residues is an important mechanism for modulating biological processes such as cellular signaling, differentiation, and growth, and if deregulated, can result in various types of cancer. Therefore, an understanding of these dynamic cellular processes at the molecular level requires the ability to assess changes in the sites of tyrosine phosphorylation across numerous proteins simultaneously as well as over time. Here we describe a sensitive approach based on multidimensional liquid chromatography/mass spectrometry that enables the rapid identification of numerous sites of tyrosine phosphorylation on a number of different proteins from human whole cell lysates. We used this methodology to follow changes in tyrosine phosphorylation patterns that occur over time during either the activation of human T cells or the inhibition of the oncogenic BCR-ABL fusion product in chronic myelogenous leukemia cells in response to treatment with STI571 (Gleevec). Together, these experiments rapidly identified 64 unique sites of tyrosine phosphorylation on 32 different proteins. Half of these sites have been documented in the literature, validating the merits of our approach, whereas motif analysis suggests that a number of the undocumented sites are also potentially involved in biological pathways. This methodology should enable the rapid generation of new insights into signaling pathways as they occur in states of health and disease.
Many cellular processes are directly controlled through the reversible phosphorylation of protein tyrosine residues. These regulatory functions are ultimately affected through the coordinated phosphorylation of numerous tyrosine residues across multiple proteins over time. Clearly, there are benefits to individually characterizing specific components of a particular pathway, such as identifying a site of phosphorylation on a given protein, the kinase responsible for the modification, or the identity of subsequently interacting proteins. Ultimately, though, a thorough understanding of these signaling pathways at the molecular level requires the wide-scale, simultaneous evaluation of these phosphorylation events as they occur over time.
To date, two-dimensional gel electrophoresis (2D-GE) remains the most common methodology for assessing wide-scale changes in phosphorylation (1). However, this methodology is relatively slow, and suffers from a number of well documented operational limitations. For example, 2D-GE has been shown to be poorly suited for the direct detection and analysis of medium to low abundance proteins from whole cell lysates, a particular concern in the case of regulatory proteins such as kinases, which often exist at very low copy numbers per cell (2). Even with the improved dynamic range afforded by multiple 2D-GE runs of prefractionated samples, the individual gel-isolated proteins still require further characterization by using methods such as two-dimensional tryptic phosphopeptide mapping (3), Edman degradation (4), or precursor ion scanning MS (5).
Several recent publications have described alternative approaches for assessing changes in phosphorylation patterns based primarily on MS methodologies (6–10). However, because only ≈0.1% of all protein phosphorylations occur on tyrosine residues (11), these important modifications are difficult to assess by these methods that either preferentially detect (7) or specifically target (9) the far more numerous sites of serine and threonine phosphorylation. Therefore, by the strategic application of phosphotyrosine immunoprecipitation (12) in tandem with methyl esterification and immobilized metal affinity chromatography of tryptic peptides, we have developed an effective method for the wide-scale determination of sites of tyrosine phosphorylation. This procedure can be used to rapidly gain insights into various cellular regulatory pathways.
Materials and Methods
Cell Culture, Stimulation, and Immunoprecipitation.
Jurkat clone E6-1, and Lck-deficient Jurkat clone J.CaM1.6 were obtained from American Type Culture Collection. The 32Dp210 and K562 cells were a kind gift from Brian Druker at Oregon Health Sciences University (Portland, OR). All cell lines were grown in RPMI medium 1640 supplemented with 10% FBS, 2 mM l-glutamine, 100 μg/ml streptomycin sulfate, and 100 units/ml penicillin G (all from Invitrogen) in a 5.0% CO2 incubator at 37°C. The 32Dp210 cells were grown in the presence of 20% WEHI-3B conditioned media (13). After treatment, cells were resuspended for 5 min at 4°C in 1× lysis buffer [20 μg/ml aprotinin/20 μg/ml leupeptin/50 mM Tris, pH 7.5/100 mM NaCl/1% Nonidet P-40/10% glycerol/1 mM Perfabloc/2 mM Na3VO4 (tyrosine phosphatase inhibitor)/1 mM EDTA/10 mM β-glycerophosphate (Sigma)].
Anti-CD3/CD4 (OKT3/OKT4) antibody treatments were done as described (14). Briefly, Jurkat cells were treated at 1 × 108 cells/ml in PBS with 2.5 μg/ml each of OKT3 and OKT4 (Ortho Biotech, Raritan, NJ) for 10 min at 4°C. Cells were then treated with 22 μg/ml goat anti-mouse IgG (Jackson ImmunoResearch) at 37°C for the times listed in Table 1 and lysed by the addition of 5× lysis buffer. STI571 (Novartis, Basel) was freshly prepared as 1 mM stock in PBS. The 32Dp210 or K562 cells were treated at a final concentration of 1 μM STI571 at 37°C followed by lysis with 1× lysis buffer. Lysates from all experiments were centrifuged at 12,000 × g for 15 min at 4°C. Monoclonal anti-phosphotyrosine agarose (Sigma) was added at 150 μl of resin per 1 × 109 cells for 4 h at 4°C with stirring. Beads were washed three times with 1× lysis buffer and three times with 20 mM Tris, pH 7.4/120 mM NaCl. Proteins were eluted with 8 M urea/100 mM NH4CO3 for 5 min at 96°C followed by filtration through a poly(vinylidene difluoride) (PVDF) 0.2-μm filter. Lysates were diluted to 4 M urea and subjected to overnight tryptic digestion with 2.5 μg of trypsin per 1 × 109 cells at 37°C.
Table 1.
Phosphorylated peptides from anti-CD3/CD4-stimulated Jurkat cells
| Protein | NCBI GI no. | Peptide sequence | PO4 site | Known PO4 site (ref.) | Top scansite hits (24) | 0 min | 0.5 min | 1 min | 2 min | 5 min |
|---|---|---|---|---|---|---|---|---|---|---|
| Jurkat | ||||||||||
| CAS-L | 5453680 | TGHGYVpYEYPSR | Y166 | J-0.32, Y-0.25, JJ-0.27 | x | |||||
| CBL | 115855 | IKPSSpSANAIpYSLAAR | S669, Y674 | Y674 (39) | S669:None,Y674:B-2.2 | x | ||||
| CD3δ | 4502669 | NDQVpYQPLR | Y149 | S,X-1.7,GG-2.8 | x | x | ||||
| CD3δ | 4502669 | DRDDAQpYSHLGGNWAR | Y160 | S,X-3.4,GG-4.9 | x | x | x | x | x | |
| CD3ɛ | 4502671 | DLpYSGLNQR | Y199 | Y199 (40) | S,X-0.36,GG-0.52 | x | x | x | x | x |
| CD3ζ | 115997 | NPQEGLpYNELQK | Y110 | Y110 (29) | T,N-0.22,X-0.02,II-0.83 | x | ||||
| CD3ζ | 115997 | RKNPQEGLpYNELQK | Y110 | Y110 (29) | T,N-0.22,X-0.02,II-0.83 | x | x | |||
| CD3ζ | 115997 | MAEApYSEIGMK | Y122 | Y122 (29) | T,P-1.6,Y-1.9,JJ-1.9 | x | x | |||
| CD3ζ | 115997 | GHDGLpYQGLSTATK | Y141 | Y141 (29) | U,X-0.24,Y-3.6,GG-0.76 | x | x | x | x | x |
| CD3ζ | 115997 | REEpYDVLDKR | Y83 | Y83 (29) | S,X-0.36,GG-1.5,II-1.9 | x | ||||
| CD5 | 7656965 | pSHAENPTASHVDNE pYSQPPRNpSR | S439, Y453, S460 | Y453 (41) | Y453:J-0.96,W-0.66,Z-0.47 S460:None,S439:None | x | ||||
| GADS | 4758476 | RHpTDPVQLQAAGR | T262 | A-1.0,D-4.7,EE-0.65 | x | x | x | |||
| HS1 | 4885405 | GFGGQpYGIQK | Y198 | None | x | x | ||||
| LIM lipoma | 5031887 | YYEGYpYAAGPGYGGR | Y301 | B-3.3,W-2.1 | x | |||||
| PYK2 | 4758976 | YIEDEDpYpYKASVTR | Y579,Y580 | Y579,Y580 (42) | Y579:L-1.6,O-1.4,Q-2.3 Y580:None | x | ||||
| ZAP-70 | 1177033 | RIDTLNSDGpYTPEPAR | Y292 | Y292 (20) | None | x | x | x | ||
| ZAP-70 | 1177044 | PMPMDTSVpYESPpYSDPE ELKDK | Y315,Y319 | Y315, Y319 (21) | Y315:C-0.30,W-0.12,Z-0.08 Y319:C-1.9,W-2.1,Z-1.6 | x | x | |||
| ZAP-70 | 1177044 | ALGADDSpYpYTAR | Y492,Y493 | Y492, Y493 (20) | Y492:None,Y493:None | x | ||||
| ZAP-70 | 1177044 | ALGADDSYpYTAR | Y493 | Y493 (20) | None | x | x | x |
Times are after anti-mouse IgG treatment (T cell receptor crosslinking). NA, time points not analyzed in the experiments. Scansite motifs are represented by a letter indicating the motif followed by a percentile score for that motif. A, 14-3-3 mode 1 site; B, ABL kinase; C, ABL SH2; D, AKT kinase; E, ATM kinase; F, casein kinase 2; G, Cdc2 kinase; H, Cdk5 kinase; I, Clk2 kinase; J, Crk SH2; K, DNA PK; L, EGFR kinase; M, Erk1 kinase; N, FGFR kinase; O, FGFR SH2; P, Fyn SH2; Q, Grb2 SH2; R, GSK3 kinase; S, ITAM 1; T, ITAM 2; U, ITAM 3; V, ltk kinase; W, ltk SH2; X, Lck kinase; Y, Lck SH2; Z, Nck SH2; AA, p85 SH2; BB, PDGFR kinase; CC, PDZ class II; DD, PLCg N-terminal SH2; EE, protein kinase A; FF, Shc PTB; GG, Shc SH2; HH, SHIP SH2; II, Src kinase; JJ, Src SH2.
Methyl Esterification of Tryptic Peptides.
Tryptic peptides were methyl esterified as described (7), with modifications. Tryptic digests of 1–2 × 109 cell equivalents of immunoprecipitated protein were loaded onto C18 peptide macrotrap cartridges (Michrom Bioresources, Auburn, CA) and washed with 1 ml of 0.1% acetic acid. Peptides were eluted with 70% acetonitrile containing 0.1% acetic acid, dried in a Speed Vac plus (Thermo Savant, Holbrook, NY), and treated at room temperature for 2 h with 2 N d3-methanolic DCl. This reagent was prepared by adding 120 μl of acetyl chloride to 0.75 ml of d3-methyl d-alcohol (Cambridge Isotope Laboratories, Cambridge, MA) with stirring. d3-Methanolic DCl was removed in a Speed Vac, and the sample was reconstituted in 100 μl of 0.1% acetic acid.
Immobilized Metal Affinity Chromatography (IMAC).
IMAC was performed as described (15). Briefly, methyl esterified tryptic peptides were loaded onto iron-activated IMAC columns (360 μm OD × 100 μm ID fused silica packed with 8-cm POROS 20 MC from Perceptive Biosystems, Framingham, MA). The column was washed with 20 μl of a 25:74:1 acetonitrile/water/acetic acid mixture containing 100 mM NaCl followed by 10 μl of 0.1% acetic acid. Enriched phosphopeptides were eluted to microcapillary columns (360 μm OD × 75 μm ID) containing 6 cm of 5- to 15-μm C18 resin (Waters, Milford, MA) by using 5 μl of 50 mM potassium phosphate (pH 9.0) and rinsed with 0.1% acetic acid.
Nanoflow Reversed-Phase HPLC/μESI/MS.
Methylated phosphopeptides were analyzed by nanoflow reversed-phase HPLC/μESI/MS with an LCQ ion trap mass spectrometer using a system modified from that described (16). Microcapillary columns loaded with methylated phosphopeptides were connected to analytical columns (360 μm OD × 50 μm ID fused silica packed with 8 cm of 5-μm C18 particles) with integrated electrospray ionization emitter tips (≈5-μm diameter). Peptides were eluted into the mass spectrometer with an HPLC gradient (0–70% B in 30 min; A = water with 0.1 M acetic acid, B = acetonitrile with 0.1 M acetic acid). Initially, the flow rate was set to 60 nl/min. When peptides began to elute, the flow rate was lowered to ≈10 nl/min by lowering the flow rate delivered by the HPLC such that chromatographic peaks were ≈1 min wide (full width half maximum). The electrospray voltage (1.6 kV) was applied to the HPLC mobile phase before flow splitting. The mass spectrometer was programmed to record continuous cycles composed of one MS scan followed by MS/MS scans of the five most abundant ions in each MS scan. The dynamic exclusion option was selected with a repeat count of 1, and an exclusion duration of 1 min.
Database Analysis.
MS/MS spectra were matched to amino acid sequences in the National Center for Biotechnology Information nonredundant protein database by using the SEQUEST algorithm (17). Search parameters specified a differential modification of +80 Da to serine, threonine, and tyrosine residues and a static modification of +17 Da to glutamic acid and aspartic acid residues and the C terminus of each peptide. All reported SEQUEST assignments were verified by manual interpretation of spectra.
Results
The goal of this work was the development of a methodology that would greatly enhance the speed of identifying both the nature and temporal ordering of tyrosine phosphorylation events that occur in signaling cascades. To this end, total Jurkat cell lysates from various time points or different treatment regimens were first immunoprecipitated with an anti-phosphotyrosine antibody (clone PT66) to reduce the background of serine and threonine phosphorylation, and the immunoprecipitated proteins were digested with trypsin. Although we found the PT66 antibody to enrich a large subset of tyrosine phosphorylated proteins, other anti-phosphotyrosine antibodies such as 4G10 could be used independently or in combination to generate a different collection of phosphorylated proteins. Immunoprecipitation of undigested proteins rather than tryptic peptides results in the additional identification of serine and threonine phosphorylation sites on tyrosine phosphorylated proteins and their binding partners. Despite the sample simplification afforded by this initial affinity selection, the direct analysis of the resulting tryptic digests by reversed-phase HPLC/MS resulted in the identification of only a very limited number of sites of tyrosine phosphorylation (data not shown). Most identified peptides were not phosphorylated and were derived from known tyrosine kinase substrates or abundant cytoskeletal proteins. Because of the difficulties of detecting phosphorylated peptides, the tryptic mixtures were further enriched by using a combination of methyl esterification and immobilized metal affinity chromatography (6). The resulting samples were then analyzed by using reversed phase HPLC and tandem MS methods, enabling the unambiguous assignment of numerous sites of tyrosine phosphorylation (Fig. 1). To demonstrate the power of this experimental methodology, we investigated the temporal organization of phosphorylation events in a highly studied signaling cascade, as well as the changes that occur in another signaling pathway after chemical perturbation.
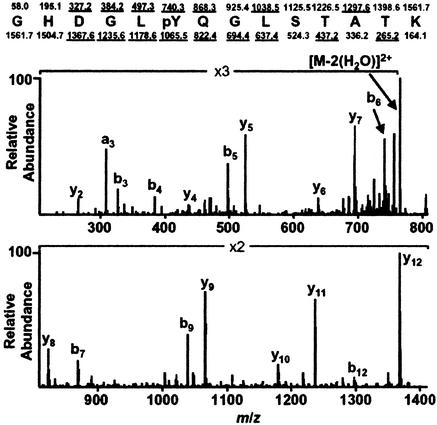
Figure 1.
MS/MS spectrum of the CD3ζ-derived peptide GHDGLpYQGLSTATK. Predicted nominal masses of type b and type y ions appear above and below the sequence, respectively. Those observed are underlined. Note that the mass difference between y8 and y9, and also b5 and b6, is 243 Da, corresponding to a phosphotyrosine residue.
T cell receptor (TCR) signaling is an important process in the activation of T cells, and many of the proteins involved have been identified (18). We therefore decided to test the overall suitability of our approach by independently measuring tyrosine phosphorylation events in this system with respect to time, and comparing our results with those documented in the literature. Thus, Jurkat cells were treated with antibodies that bind to the CD3 and CD4 receptors followed by crosslinking with anti-mouse IgG. This stimulation mimics TCR ligation and is known to induce cascades of tyrosine phosphorylation (18, 19). Subsequent analysis of fractions soluble in Nonidet P-40 was expected to yield data from readily soluble phosphorylated proteins involved in T cell signaling. The synthetic peptide LIEDNEpYTAR (5 pmol) was added to each sample before immunoprecipitation as an external control. This peptide, derived from the activation loop of several tyrosine kinases including Lck, contains four carboxylic acid functional groups and serves as a control for immunoprecipitation, methylation, and HPLC/MS. A signal for LIEDNEpYTAR was not detected in extracts of Jurkat cells treated with anti-CD3, CD4 antibodies (probably because of the inclusion of this protein into Nonidet P-40 insoluble lipid rafts) until the synthetic peptide was added. The use of this standard precludes detection of LIEDNEpYTAR generated from digests of cellular proteins; however, a different standard may be chosen to profile this phosphorylation. Approximately 5 × 108 cell equivalents of soluble protein from each time point were processed and analyzed (Table 1 and http://phosphopeptide.com). As expected, overall protein tyrosine phosphorylation levels increased on antibody stimulation (Fig. 2). The LIEDNEpYTAR control was detected in every sample with chromatographic peak areas all within a factor of four, suggesting consistent recovery of phosphopeptides from each of the cell lysates.
Figure 2.
Time course of chromatographic peak areas of phosphopeptides normalized to LIEDNEpYTAR peak areas. CD3ζ Tyr-141 (♦), CD3δ Tyr-160 (■), CD3ɛ Tyr-199 (●), CD3ζ Tyr-122(+), CD3δ Tyr-149 (−), ZAP-70 Tyr-315,319 (▴), and ZAP-70 Tyr-493 (*). Profiles shown here are representative of all observed.
Fig. 3 illustrates the data obtained for a representative peptide derived from the protein tyrosine kinase ZAP-70, the phosphorylation of which was clearly induced within 1 min of T cell receptor ligation. All of the known in vivo tyrosine phosphorylation sites of ZAP-70 were detected (Tyr-292, Tyr-315, Tyr-319, Tyr-492, and Tyr-493) (20, 21). In total, 16 known sites of tyrosine phosphorylation and their relative temporal organization were rapidly identified. Furthermore, tyrosine phosphorylation was effectively absent in the Lck-depleted mutant cell line J.CaM1.6 on CD3/CD4 stimulation (data not shown).
Figure 3.
ZAP-70 Tyr-315,319 phosphorylation in CD3/CD4-stimulated Jurkat cells compared with LIEDNEpYTAR phosphorylation. Total ion chromatogram (TIC) recorded during MS analysis of phosphorylated peptides from unstimulated Jurkat cells (a) and Jurkat cells stimulated with crosslinked anti-CD3 and anti-CD4 antibodies for 1 min (b). Selected-ion chromatogram (SIC) or plot of the ion current vs. time for m/z 990 corresponding to the (M + 3H)3+ of the phosphopeptide ZAP-70 Tyr-315,319 for unstimulated (c) and 1 min stimulated (d) Jurkat cells. SIC for m/z 742.6 corresponding to the (M + 4H)4+ of the phosphopeptide ZAP-70 Tyr-315,319 for unstimulated (e) and 1 min stimulated (f) Jurkat cells. SIC for m/z 686 corresponding to the (M + 2H)2+ of the phosphopeptide LIEDNEpYTAR for unstimulated (g) and 1 min stimulated (h) Jurkat cells.
We next investigated changes in the sites of tyrosine phosphorylation triggered by chemical inhibition of the BCR-ABL signaling pathway. Constitutive activity of the fusion tyrosine kinase BCR-ABL is the primary cause of chronic myeloid leukemia (CML) (22). The inhibition of pathways activated by BCR-ABL was investigated by treatment of the BCR-ABL containing cell lines 32Dp210 (13) (murine myeloid cell line with human BCR-ABL) and K562 (human CML) with the BCR-ABL inhibitor STI571 (Gleevec) (23). Phosphorylation of both the Grb-2 binding site Tyr-177 on BCR and the autophosphorylation site Tyr-393 (Tyr-793 on BCR-ABL fusion protein) were inhibited by STI571 (Table 2 and http://phosphopeptide.com/). Both sites of phosphorylation have been reported to be directly implicated in the function of BCR-ABL (22).
Table 2.
Phosphorylated peptides from BCR/ABL-expressing cells treated with STI571
| Protein | NCBI GI no. | Peptide sequence | PO4 site | Known PO4 site (ref.) | Top scansite hits (24) | 0 hr | 1.5 hr | 3 hr |
|---|---|---|---|---|---|---|---|---|
| 32d p210 | ||||||||
| ABL | 125135 | LMTGDTpYTAHAGAK | Y393 | Y393 (43) | None | x | NA | |
| ABL | 125135 | LMTGDTpYpTAHAGAK | Y393, T394 | Y393 (43) | Y393:None,Y394:None | x | NA | |
| ABL | 125135 | LpYVSSESR | Y185 | None | x | x | NA | |
| ABL | 125135 | pYSLTVAVK | Y264 | Y264 (44) | None | x | NA | |
| ABL | 125135 | NKPTVpYGVSPNYDK | Y226 | Y226 (43) | B-3.3,X-3.4,HH-2.5 | x | NA | |
| ABL | 125135 | VpYELMR | Y469 | AA-0.25,GG-2.8,II-3.3 | x | NA | ||
| BCR | 11038641 | HQDGLPYIDDpSPSSpSPHLSSK | S459, S463 | S459:M-4.0,R-0.56,S463:G-1.3,H-1.6,R-0.32 | x | NA | ||
| CAS-L | 8567380 | LpYQVPNSQAASR | Y91 | B-1.5,C-1.0,J-0.56 | x | x | NA | |
| CAS-L | 8567380 | DVpYDVPPSHSTQGVpYDIPPS SVK | Y176, Y188 | Y176:C-0.14,J-0.01,Z-0.04 Y188:C-0.20,J-0.01,Z-0.03 | x | NA | ||
| CBL | 115857 | IKPSSSANAIpYSLAAR | Y655 | Y655 (39) | B-2.2 | x | x | NA |
| CBL | 115857 | LPPGEQGEpSEEDTEpYMTPTS RPVGVQKPEPK | S675, Y681 | Y681 (45) (Y700) | S675:E-3.6,F-0.40,K-1.8 Y681:J-0.08,W-0.04,Z-0.06 | x | x | NA |
| DYRK1A | 6015042 | IYQpYIQSR | Y321 | Y321 (46) | None | x | NA | |
| SHEP1 | 7305329 | AGESpYTHIR | Y273 | None | x | NA | ||
| SHP-2 | 6755228 | VpYENVGLMQQQR | Y584 | Y584 (47) | O-0.55,Y-1.9,JJ-1.9 | x | NA | |
| k562 | ||||||||
| ABL | 125135 | LMTGDTpYTAHAGAK | Y393 | Y393 (43) | None | x | x | |
| ABL | 125135 | LMTGDTpYpTAHAGAK | Y393, T394 | Y393 (43) | Y393:None,Y394:None | x | x | |
| ABL | 125135 | LpYVSSESR | Y185 | None | x | x | ||
| ABL | 125135 | LGGGQpYGEVYEGVWK | Y253 | N-4.0,P-4.0,X-0.20 | x | |||
| ABL | 125135 | LGGGQYGEVpYEGVWK | Y257 | X-0.32,Y-0.65,II-0.83 | x | |||
| Activated p21cdc42Hs | 18553914 | pYATPQVIQAPGPR | Y827 | J-4.6,W-3.0 | x | |||
| Activated p21cdc42Hs | 423137 | VSSTHpYpYLLPERPSYLER | Y912, Y913 | Y912:BB-1.2,GG-2.1,HH-0.04 Y913:C-3.9,J-3.4,DD-1.3 | x | |||
| BCR | 11038641 | PFpYVNVEFHHER | Y177 | Y177 (30) | N-2.7,Q-0.03,BB-3.2 | x | ||
| BCR | 11038641 | HQDGLPYIDDpSPSSpSPHLSSK | S459, S463 | S459:M-4.0,R-0.56,S463:G-1.3,H-1.6,R-0.32 | x | x | ||
| BCR | 11038641 | RLpTWPR | T310 | A-2.4,D-1.9,EE-0.55 | x | x | ||
| BCR | 11038641 | NSLETLLpYKPVDR | Y644 | None | x | |||
| BCR | 11038641 | LASQLGVpYR | Y591 | None | x | |||
| CBL | 115855 | IKPSSpSANAIpYSLAAR | S669, Y674 | Y674 (39) | Y674:B-2.2,S669:None | x | ||
| CBL | 115855 | IKPSSSANAIpYSLAAR | Y674 | Y674 (39) | B-2.2 | x | ||
| Cortactin | 4885205 | LPSSPVpYEDAASFK | Y421 | Y421 (48) | O-1.6,Y-1.3,AA-2.0 | x | ||
| DNABP1 | 4557447 | ELEEIpYMLPR | Y1068 | B-0.37,W-0.66,DD-1.3 | x | |||
| e3b1 | 2245671 | HNSTTSSTSSGGpYR | Y305 | None | x | |||
| GAB1 | 4503851 | SYpSHDVLPK | S266 | D-0.82,E-3.6,I-4.2 | x | |||
| GAB1 | 4503851 | APSASVDSSLpYNLPR | Y259 | C-2.8,J-0.76,Z-2.1 | x | x | x | |
| GAB2 | 18105042 | HNTEFRDSTpYDLPR | Y266 | C-0.65,J-0.56,Z-0.22 | x | |||
| GAGE-3 | 4503881 | STpYYWPRPR | Y10 | B-0.59,C-1.3,J-2.9 | x | |||
| p56dok | 4503359 | GQEGEpYAVPFDAVAR | Y299 | B-0.04,W-0.66,X-0.67 | x | |||
| p62dok | 4503357 | SHNSALpYSQVQK | Y449 | P-3.0 | x | |||
| plakophilin 4 | 4505843 | NNYALNTTATpYAEPpYRPIQYR | Y478, Y482 | Y478:B-0.02,C-0.52,W-1.0,Y482:None | x | |||
| SHC | 284403 | ELFDDPSpYVNVQNLDK | Y317 | Y317 (49) | L-0.47,Q-0.01,BB-0.33 | x | ||
| SHIP-2 | 4755142 | TLSEVDpYAPAGPAR | Y1135 | W-4.3 | x | x | x | |
| SHIP-2 | 4755142 | GLPSDpYGR(PLSFPPPR) | Y1162 | Y1162 (39) | B-0.60,J-0.96,C-2.8 | x | x | |
| SHIP-2 | 4755142 | DTpYAWHK | Y671 | None | x | |||
| SHIP-2 | 4755142 | LpYEWISIDKDEAGAK | Y886 | P-1.1,W-1.5,GG-0.52 | x | |||
| SHIP-2 | 4755142 | NSFNNPApYYVLEGVPHQLLPP EPPpSPAR | Y986, S994 | Y986 (50) | Y986:L-0.20,N-1.7,FF-0.19,S994:None | x | ||
| SYK | 1174527 | EALPMDTEVpYESPpYADPEEIR PK | Y348, Y352 | Y348, Y352 (51) | Y348:C-0.65,W-0.07,Z-0.22 Y352:C-1.9,W-0.18,BB-3.2 | x |
Times are after STI571 treatment. Scansite motifs are described in Table 1. NA, time points not analyzed in the experiments.
In addition, nearly 30 other previously undescribed sites of tyrosine phosphorylation were identified (Table 2). To better understand the nature of these previously undescribed sites, motif analysis using the Scansite tool was performed (http://scansite.mit.edu/) (24). When this method was used, scores of <0.4% were considered most significant based on the examination of known phosphorylation sites, though all predictions require evaluation in the biological context of the system studied (24). Higher scores may be due to either divergences from prototypical motifs or novel motifs not contained in Scansite.
Discussion
The rapid simultaneous identification of numerous changes in tyrosine phosphorylation on different proteins that were already documented as relevant to the T cell signaling pathway validates the potential power of this methodology. Interestingly, despite the large volume of published work on T cell signaling, these experiments also identified unreported sites of tyrosine phosphorylation such as Tyr-166 of Cas-L, which contains a strong consensus site for binding to the Lck SH2 domain. Cas-L is known to bind to Lck kinase in CD3-stimulated T cells, but the site of interaction has not been defined (25); phosphorylation of a Lck SH2 site on Cas-L suggests that Lck could bind this site. Another novel phosphorylation site was observed at Tyr-198 of HS1. HS1 is known to be tyrosine phosphorylated as a consequence of anti-CD3 stimulation, possibly through the action of Syk (26, 27), but again the site of phosphorylation was not previously identified. Up-regulation of ZAP-70 catalytic activity results from phosphorylation of Tyr-493 by Lck and autophosphorylation of Tyr-319 after tandem interactions of the ZAP-70 SH2 domains with immunoreceptor tyrosine-based activation motifs (ITAMs) on CD3ζ (21, 28). Consistent with the reported sequential phosphorylation of ZAP-70 and CD3ζ, we observed maximal phosphorylation of Tyr-493 and Tyr-319 of ZAP-70 to occur 1 min after maximal CD3ζ phosphorylation (Fig. 2). Induced phosphorylation of CD3ζ, CD3δ, and CD3ɛ was consistent with the expected phosphorylation of these ITAM regions (29). The absence of tyrosine phosphorylation in the Lck-depleted mutant cell line J.CaM1.6 on CD3/CD4 stimulation verifies the predicted importance of this kinase in T cell signaling (18).
Application of this methodology to assess changes in tyrosine phosphorylation on treatment of chronic myelogenous leukemia cells with STI571 resulted in the identification of both sites of phosphorylation reported to be directly implicated in the function of BCR-ABL, again demonstrating the potential power of this methodology. In addition, nearly 30 other previously undescribed sites of tyrosine phosphorylation were identified. Among the recently identified phosphorylation sites found were Tyr-469 and Tyr-257 on ABL. The Scansite algorithm identified Tyr-177 as a strong candidate for binding to the Grb2-SH2 domain (0.03 percentile). In fact, Tyr-177 is known to bind this domain, demonstrating the potential of scansite to discern such interactions (30).
Although the changes in phosphorylation patterns reported on STI571 treatment have focused primarily on the inhibition of phosphorylation, it should be noted that numerous cases of induced modification were also observed. The inhibition of BCR-ABL kinase activity by STI571 is known to restore expression of the transcriptional regulator C/EBPα, which leads to expression of genes related to cellular differentiation such as the granulocyte-colony stimulating factor (G-CSF) receptor (31, 32). STI571 alone is known to induce differentiation of K562 cells as illustrated by CD11b and hemoglobin up-regulation (31). Exogenous expression of G-CSF receptor in K562 cells and addition of G-CSF also leads to up-regulation of CD11b (33). Treatment of K562 cells with STI571 for 3 h induced the tyrosine phosphorylation of G-CSF signaling pathway members including Syk, cortactin, and SHC (34–36) (Table 2), suggesting that inhibition of BCR-ABL allows restoration of a functional G-CSF signaling cascade.
To our knowledge, the rapid determination of comparable numbers of tyrosine phosphorylation sites across multiple proteins in a single HPLC/MS experiment has not previously been reported. Although these experiments revealed more than 60 sites of phosphorylation on many proteins, we did not detect all expected phosphorylation sites. For example, we did not detect phosphorylation of SLP-76, LAT, or PAG. This could be explained by the fact that these proteins reside in Nonidet P-40 insoluble lipid rafts (37, 38), or because certain tryptic phosphopeptides were not amenable to HPLC/MS because of low abundance, unsuitable size, or poor ionization efficiency. Increased numbers of tyrosine phosphorylated peptides could likely be identified with our method by solublizing proteins with different detergents, analyzing phosphopeptide pools from larger numbers of cells, digesting proteins with enzymes other than trypsin (i.e., glu-c or chymotrypsin), or by analyzing phosphopeptide pools using two-dimensional HPLC/MS. These experiments can be performed without any knowledge of the expected identity and functions of the various relevant proteins by leveraging the availability of genomic sequence information. Subsequently, more directed approaches can be used to elucidate the biological relevance of these newly discovered sites. In addition, this method enables the examination of changes in the sites of phosphorylation over time, helping to clarify the temporal orchestration of protein tyrosine phosphorylation within signaling pathways. Finally, profiling of tyrosine kinase inhibitors such as STI571 should provide a deeper understanding of the downstream effects of these agents on a variety of signaling pathways.
Acknowledgments
We thank Dr. Brian Druker for providing the BCR-ABL-expressing cell lines and Dr. Badry Bursulaya for modeling the novel phosphorylation sites on the ABL/STI571 crystal structure. Dr. Achim Brinker was supported by a fellowship of the Humboldt Foundation.
References
- 1.Sickmann A, Meyer H E. Proteomics. 2001;1:200–206. doi: 10.1002/1615-9861(200102)1:2<200::AID-PROT200>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 2.Gygi S P, Corthals G L, Zhang Y, Rochon Y, Aebersold R. Proc Natl Acad Sci USA. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle W J, van der Geer P, Hunter T. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- 4.Aebersold R H, Leavitt J, Saavedra R A, Hood L E, Kent S B. Proc Natl Acad Sci USA. 1987;84:6970–6974. doi: 10.1073/pnas.84.20.6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr S A, Huddleston M J, Annan R S. Anal Biochem. 1996;239:180–192. doi: 10.1006/abio.1996.0313. [DOI] [PubMed] [Google Scholar]
- 6.Posewitz M C, Tempst P. Anal Chem. 1999;71:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 7.Ficarro S B, McCleland M L, Stukenberg P T, Burke D J, Ross M M, Shabanowitz J, Hunt D F, White F M. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Watts J, Aebersold R. Nat Biotechnol. 2001;19:375–378. doi: 10.1038/86777. [DOI] [PubMed] [Google Scholar]
- 9.Goshe M, Conrads T, Panisko E, Angell N, Veenstra T, Smith R. Anal Chem. 2001;73:2578–2586. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- 10.Oda Y, Nagasu T, Chait B. Nat Biotechnol. 2001;19:379–382. doi: 10.1038/86783. [DOI] [PubMed] [Google Scholar]
- 11.Cooper J A, Sefton B M, Hunter T. Methods Enzymol. 1983;99:387–402. doi: 10.1016/0076-6879(83)99075-4. [DOI] [PubMed] [Google Scholar]
- 12.Pandey A, Podtelejnikov A V, Blagoev B, Bustelo X R, Mann M, Lodish H F. Proc Natl Acad Sci USA. 2000;97:179–184. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heaney C, Kolibaba K, Bhat A, Oda T, Ohno S, Fanning S, Druker B J. Blood. 1997;89:297–306. [PubMed] [Google Scholar]
- 14.Sauer K, Liou J, Singh S B, Yablonski D, Weiss A, Perlmutter R M. J Biol Chem. 2001;276:45207–45216. doi: 10.1074/jbc.M106811200. [DOI] [PubMed] [Google Scholar]
- 15.Zarling A L, Ficarro S B, White F M, Shabanowitz J, Hunt D F, Engelhard V H. J Exp Med. 2000;192:1755–1762. doi: 10.1084/jem.192.12.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin S E, Shabanowitz J, Hunt D F, Marto J A. Anal Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 17.Eng J, McCormack A, Yates J R. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 18.Kane L P, Lin J, Weiss A. Curr Opin Immunol. 2000;12:242–249. doi: 10.1016/s0952-7915(00)00083-2. [DOI] [PubMed] [Google Scholar]
- 19.June C H, Fletcher M C, Ledbetter J A, Samelson L E. J Immunol. 1990;144:1591–1599. [PubMed] [Google Scholar]
- 20.Watts J D, Affolter M, Krebs D L, Wange R L, Samelson L E, Aebersold R. J Biol Chem. 1994;269:29520–29529. [PubMed] [Google Scholar]
- 21.Di Bartolo V, Mege D, Germain V, Pelosi M, Dufour E, Michel F, Magistrelli G, Isacchi A, Acuto O. J Biol Chem. 1999;274:6285–6294. doi: 10.1074/jbc.274.10.6285. [DOI] [PubMed] [Google Scholar]
- 22.Chopra R, Pu Q Q, Elefanty A G. Blood Rev. 1999;13:211–229. doi: 10.1054/blre.1999.0119. [DOI] [PubMed] [Google Scholar]
- 23.Schindler T, Bornmann W, Pellicena P, Miller W T, Clarkson B, Kuriyan J. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 24.Yaffe M B, Leparc G G, Lai J, Obata T, Volinia S, Cantley L C. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 25.Kanda H, Mimura T, Hamasaki K, Yamamoto K, Yazaki Y, Hirai H, Nojima Y. Immunology. 1999;97:56–61. doi: 10.1046/j.1365-2567.1999.00753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchcroft J E, Slavik J M, Lin H, Watanabe T, Bierer B E. J Immunol. 1998;161:4506–4512. [PubMed] [Google Scholar]
- 27.Ruzzene M, Brunati A M, Marin O, Donella-Deana A, Pinna L A. Biochemistry. 1996;35:5327–5332. doi: 10.1021/bi9528614. [DOI] [PubMed] [Google Scholar]
- 28.Chan A C, Dalton M, Johnson R, Kong G H, Wang T, Thoma R, Kurosaki T. EMBO J. 1995;14:2499–2508. doi: 10.1002/j.1460-2075.1995.tb07247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kersh E N, Shaw A S, Allen P M. Science. 1998;281:572–575. doi: 10.1126/science.281.5376.572. [DOI] [PubMed] [Google Scholar]
- 30.Ma G, Lu D, Wu Y, Liu J, Arlinghaus R B. Oncogene. 1997;14:2367–2372. doi: 10.1038/sj.onc.1201053. [DOI] [PubMed] [Google Scholar]
- 31.Fang G, Kim C N, Perkins C L, Ramadevi N, Winton E, Wittmann S, Bhalla K N. Blood. 2000;96:2246–2253. [PubMed] [Google Scholar]
- 32.Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K, Iervolino A, Condorelli F, Gambacorti-Passerini C, Caligiuri M A, Calabretta B. Nat Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- 33.el-Sonbaty S S, Watanabe M, Hochito K, Yamaguchi K, Matsuda I, Tsuchiya H. Int J Hematol. 1995;61:61–68. doi: 10.1016/0925-5710(95)00352-s. [DOI] [PubMed] [Google Scholar]
- 34.Maruyama S, Kurosaki T, Sada K, Yamanashi Y, Yamamoto T, Yamamura H. J Biol Chem. 1996;271:6631–6635. doi: 10.1074/jbc.271.12.6631. [DOI] [PubMed] [Google Scholar]
- 35.Grishin A, Sinha S, Roginskaya V, Boyer M J, Gomez-Cambronero J, Zuo S, Kurosaki T, Romero G, Corey S J. Oncogene. 2000;19:97–105. doi: 10.1038/sj.onc.1203254. [DOI] [PubMed] [Google Scholar]
- 36.Corey S J, Burkhardt A L, Bolen J B, Geahlen R L, Tkatch L S, Tweardy D J. Proc Natl Acad Sci USA. 1994;91:4683–4687. doi: 10.1073/pnas.91.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cinek T, Horejsi V. J Immunol. 1992;149:2262–2270. [PubMed] [Google Scholar]
- 38.Zhang W, Trible R P, Samelson L E. Immunity. 1998;9:239–246. doi: 10.1016/s1074-7613(00)80606-8. [DOI] [PubMed] [Google Scholar]
- 39.Steen H, Kuster B, Fernandez M, Pandey A, Mann M. J Biol Chem. 2002;277:1031–1039. doi: 10.1074/jbc.M109992200. [DOI] [PubMed] [Google Scholar]
- 40.de Aos I, Metzger M H, Exley M, Dahl C E, Misra S, Zheng D, Varticovski L, Terhorst C, Sancho J. J Biol Chem. 1997;272:25310–25318. doi: 10.1074/jbc.272.40.25310. [DOI] [PubMed] [Google Scholar]
- 41.Vila J M, Gimferrer I, Padilla O, Arman M, Places L, Simarro M, Vives J, Lozano F. Eur J Immunol. 2001;31:1191–1198. doi: 10.1002/1521-4141(200104)31:4<1191::aid-immu1191>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 42.Schlaepfer D D, Hauck C R, Sieg D J. Prog Biophys Mol Biol. 1999;71:435–478. doi: 10.1016/s0079-6107(98)00052-2. [DOI] [PubMed] [Google Scholar]
- 43.Brasher B B, Van Etten R A. J Biol Chem. 2000;275:35631–35637. doi: 10.1074/jbc.M005401200. [DOI] [PubMed] [Google Scholar]
- 44.Konopka J B, Witte O N. Mol Cell Biol. 1985;5:3116–3123. doi: 10.1128/mcb.5.11.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andoniou C E, Thien C B, Langdon W Y. Oncogene. 1996;12:1981–1989. [PubMed] [Google Scholar]
- 46.Himpel S, Panzer P, Eirmbter K, Czajkowska H, Sayed M, Packman L C, Blundell T, Kentrup H, Grotzinger J, Joost H G, Becker W. Biochem J. 2001;359:497–505. doi: 10.1042/0264-6021:3590497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogel W, Ullrich A. Cell Growth Differ. 1996;7:1589–1597. [PubMed] [Google Scholar]
- 48.Huang C, Liu J, Haudenschild C C, Zhan X. J Biol Chem. 1998;273:25770–25776. doi: 10.1074/jbc.273.40.25770. [DOI] [PubMed] [Google Scholar]
- 49.van der Geer P, Wiley S, Lai V K, Olivier J P, Gish G D, Stephens R, Kaplan D, Shoelson S, Pawson T. Curr Biol. 1995;5:404–412. doi: 10.1016/s0960-9822(95)00081-9. [DOI] [PubMed] [Google Scholar]
- 50.Taylor V, Wong M, Brandts C, Reilly L, Dean N M, Cowsert L M, Moodie S, Stokoe D. Mol Cell Biol. 2000;20:6860–6871. doi: 10.1128/mcb.20.18.6860-6871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furlong M T, Mahrenholz A M, Kim K H, Ashendel C L, Harrison M L, Geahlen R L. Biochim Biophys Acta. 1997;1355:177–190. doi: 10.1016/s0167-4889(96)00131-0. [DOI] [PubMed] [Google Scholar]