Abstract
Autosomal dominant dyskeratosis congenita (DKC), as well as aplastic anemia, has been linked to mutations in the RNA component of telomerase, the ribonucleoprotein responsible for telomere maintenance. Here we examine the effect of the DKC mutations on the structure and stability of human telomerase RNA pseudoknot and CR7 domains by using NMR and thermal melting. The CR7 domain point mutation decreases stability and alters a conserved secondary structure thought to be involved in human telomerase RNA accumulation in vivo. We find that pseudoknot constructs containing the conserved elements of the pseudoknot domain are in equilibrium with a hairpin conformation. The solution structure of the wild-type hairpin reveals that it forms a continuous helix containing a novel run of three consecutive U⋅U and a U⋅C base pairs closed by a pentaloop. The six base pairs unique to the hairpin conformation are phylogenetically conserved in mammals, suggesting that this conformation is also functionally important. The DKC mutation in the pseudoknot domain results in a shift in the equilibrium toward the hairpin form, primarily due to destabilization of the pseudoknot. Our results provide insight into the effect of these mutations on telomerase structure and suggest that the catalytic cycle of telomerase involves a delicate interplay between RNA conformational states, alteration of which leads to the disease state.
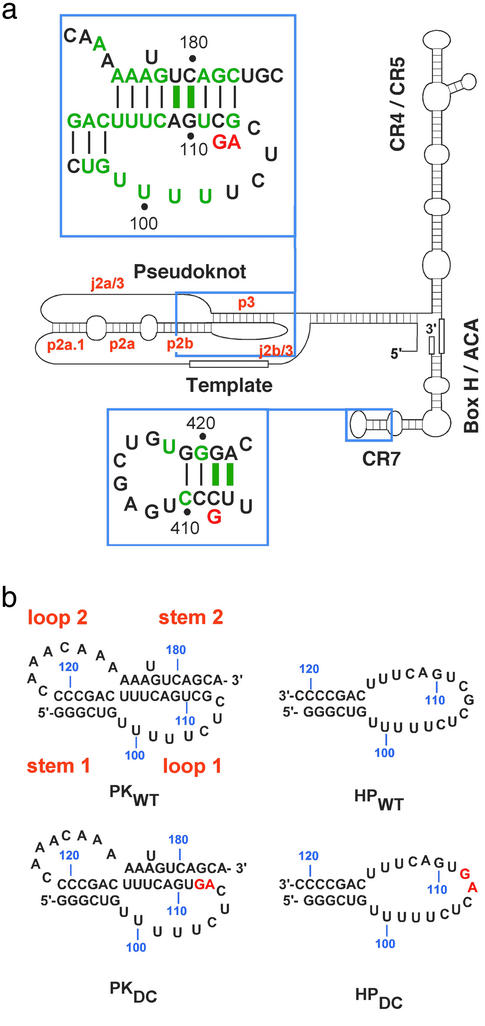
Dyskeratosis congenita (DKC) is a rare inherited multisystemic disorder characterized by abnormal skin pigmentation, leukoplakia, and nail dystrophy (1). The known phenotypic hallmarks of DKC are defects in highly proliferative tissues that could ensue from a telomere maintenance disorder (2, 3). The leading causes of premature mortality due to DKC are progressive bone marrow failure, pulmonary disease, and malignancy. X-linked, autosomal recessive, and autosomal dominant inheritance patterns have been observed for DKC, and telomere lengths in patients with DKC are found to be significantly reduced in all three forms of inheritance (4). Similarly, some patients with ideopathic aplastic anemia also have shorter telomeres than age-matched controls (5). Although the X-linked form of DKC is characterized by mutations in the gene encoding the protein dyskerin (6), a component of the telomerase ribonucleoprotein (7), the autosomal dominant form of DKC has been linked to mutations in the gene encoding human telomerase RNA (hTR). The presence of telomerase RNA mutations and their segregation with the disease were confirmed in three families with DKC inheritance (8). One mutation results in deletion of the H/ACA and CR7 domains (9) required for nucleolar localization, 3′-end processing, and RNA stability (7, 10–12), whereas the second is a point mutation (C408G) in the CR7 domain (Fig. 1a). The third mutation is a two-base substitution in the essential pseudoknot domain (9), which is required for activity (13, 14) and is involved in the binding of the protein catalytic subunit (10) (Fig. 1a). Some patients with aplastic anemia are also heterozygous for mutations in hTR, one of which occurs in the same region of the pseudoknot as the DKC mutation (15).
Figure 1.
Human telomerase RNA and conserved subdomains. (a) Secondary structure representation based on the phylogenetic analysis of the vertebrate telomerase RNA gene from 32 species (9). The conserved domains are labeled and regions containing DKC mutations are boxed and shown in the larger insets. Green letters and bars indicate sequences and base pairs, respectively, that are 100% conserved. DKC mutations are indicated in red below the wild-type sequence. (b) RNA oligonucleotide sequences derived from the pseudoknot domain used in this work. Stem and loop designations are indicated in orange.
Here we report structural and thermodynamic studies of wild-type and mutant human telomerase subdomains that provide, to our knowledge, the first 3D structural insight into the molecular basis of a disease caused by a defect in RNA. This work further reveals the presence of a phylogenetically conserved hairpin in equilibrium with the pseudoknot in the telomerase pseudoknot domain, which is stabilized by a unique uridine helix. We propose that interconversion between the hairpin and pseudoknot conformations is functionally important, and that the pseudoknot DKC mutation affects this interconversion.
Materials and Methods
RNA Synthesis and Purification.
Unlabeled, uniformly 13C,15N-labeled, and 13C,15N-U-labeled RNA oligonucleotides were prepared by in vitro transcription and purified as previously described (16). RNAs were annealed under dilute conditions (1–10 μM) in NMR buffer (10 mM sodium phosphate, pH 6.3/200 mM KCl/50 μM EDTA/0.2% NaN3) and concentrated by ultrafiltration to ≈1.0 mM.
NMR Spectroscopy.
All NMR spectra were recorded on Bruker (Billerica, MA) DRX 500 and 600 MHz spectrometers. Exchangeable proton spectra were measured in 95% H2O/5% 2H2O at 278 K, and nonexchangeable proton spectra were measured in 100% 2H2O at 293 K. Total correlation spectroscopy (TOCSY) and exchangeable and nonexchangeable NOESY experiments were performed on all RNA constructs. For HPWT and PKWT, exchangeable protons and nitrogens in the Watson–Crick base pairs were assigned by using NOESY (100-ms mixing time), 1H-15N heteronuclear multiple quantum correlation (HMQC), and 15N correlated Carr–Purcell–Meiboom–Gill-NOESY spectra (17). For HPWT, confirmation of the assignments and assignment of the U⋅U base-pair exchangeable protons was achieved by using 2D H5(C5C4N)H (18), H(CCN)H-TOCSY, and NOESY (300 ms) (19). Nonexchangeable protons and carbons were assigned by using NOESY spectra (50, 100, 150, 200, 250, and 300 ms), homonuclear TOCSY, 1H-13C CT-heteronuclear sequential quantum correlation (HSQC), 3D MQ-H(C)N(C)H, 3D HCCH-COSY, 3D HCCH-TOCSY, and 3D NOESY-HMQC experiments (17). Additional assignments were made by using 2D 13C and 12C-edited NOESY spectra (unpublished pulse sequences) on the 13C,15N-U labeled HPWT. Hydrogen bonding patterns were confirmed by using JNN-HNN-COSY (20). Hydrogen bonding of the U⋅U base pairs was determined by the detection of a 3hJN3C′2 coupling for U100⋅U114 by using the long range JNC′ H(N)CO (21) and C4/C2 carbonyl chemical shifts for U99⋅U115 and U101⋅U113. The U102⋅C112 base-pairing scheme is supported by absence of a U102 imino proton, upfield-shifted C112 amino protons, A-form helical nuclear Overhauser effect (NOE) pattern, and C112 N4 amino to U102 H5 NOE. 3JH3′P and 3JCP were measured by using 31P spin echo difference CT-HSQCs to determine β and ɛ torsion angles (22). Residual dipolar couplings were measured for 1JHN and 1JHC in C12E6/hexanol by using CT-CE-HSQC (23). Spectra were processed and analyzed by using Bruker XWINNMR 2.6 and AURELIA 3.108.
Structure Calculations.
Interproton distances from 2D NOESY with various mixing times and 3D NOESY HMQC spectra were generated as previously described (22) except for the classification of semiquantitative NOEs, which were as follows: strong (1.8–3.5 Å), medium (2.5–4.5 Å), weak (3.5–5.5 Å), and very weak (4.5–6.5 Å). In total, 266 intranucleotide and 318 internucleotide distance restraints were generated. One hundred seventeen dihedral angle constraints for α, β, δ, and ɛ backbone angles were obtained (22), and the γ dihedral angles for 20 nucleotides involved in base pairs were constrained to the A-form value (54 ± 30°) (17). Hydrogen bonding distance restraints were used for the eight Watson–Crick, three U⋅U, and C⋅U base pairs, consistent with experimental data. The structures of HPWT were calculated by using x-plor 3.851 starting from an extended RNA conformation. The folding and refinement stages followed standard x-plor protocols. Structures with no experimental restraint violations were then subjected to refinement against 24 residual dipolar couplings. The protocol involved slow cooling from 1,000 to 100 K in 18 cycles of molecular dynamics corresponding to a total of 18 ps. During this stage, the force constant for dipolar couplings was slowly increased from 0.001 to 0.2 kcal⋅mol−1Hz−2. Estimates of axial and rhombic components of the alignment tensor were based on the results of order matrix analysis (24). The experimentally derived residual dipolar couplings together with the lowest energy structure from the structure calculation employing NOE and dihedral restraints were used as input information. Calculations were performed separately for the stem and loop regions of the molecule, and the results indicated that the magnitude and asymmetry of the alignment tensor are described by Da = −39 Hz and R = 0.1. The subsequently performed grid search yielded Da = −39.75 Hz and R = 0.125. The force constants used in the refinement stage of structure calculation were 50 kcal⋅mol−1⋅Å−2, 200 kcal⋅mol−1⋅deg−2, and 0.2 kcal⋅mol−1⋅Hz−2 for NOEs, dihedral angles, and residual dipolar couplings, respectively. The correlation coefficients for the back-calculated residual dipolar couplings (RDCs) increased from 0.54 to 0.97 after inclusion of the experimental RDCs in the structure calculations (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Structural statistics for the 18 lowest energy structures are included in Table 2. Helical parameters were calculated by using curves (25) with a linear helical axis, except for the parameters Zp and C1′-C1′ distance, which were calculated by using 3DNA (26) (Fig. 6, which is published as supporting information on the PNAS web site).
Table 2.
Structural statistics for HPWT
| Experimental data used for structure calculations | |
| NOE-derived distance restraints | |
| Intranucleotide NOEs | 266 |
| Internucleotide NOEs | 318 |
| Hydrogen bond for paired residues | 29 |
| Dihedral restraints | 117 |
| Residual dipolar couplings | |
| 1DN-H (Hz) | 9 |
| 1DC-H (Hz) | 15 |
| Additional restraints | |
| Base-pair planarity restraints | 12 |
| rms deviation from experimental restraints* | |
| Distance restraints, ņ | 0.036 ± 0.001 |
| Dihedral restraints, °‡ | 0.073 ± 0.003 |
| Dipolar couplings, Hz | 1.03 ± 0.06 |
| Deviations from idealized geometry | |
| Bonds, Å | 0.0048 ± 0.0001 |
| Angles, ° | 1.01 ± 0.01 |
| Impropers, ° | 0.48 ± 0.01 |
| Overall rms deviation, Å (residue: 94–120)* | |
| From mean structure | 0.59 |
| Mean pairwise | 0.74 ± 0.29 |
Averaged over accepted structures.
No violations >0.2 Å.
No violations >5°.
Thermal Denaturation.
Thermal melting experiments were collected on a Varian Cary 1 scanning spectrophotometer and absorption recorded as a function of temperature and [KCl] at both 260 and 280 nm simultaneously. RNA samples (2–4 μM) were prepared, heated to 65°C for 10 min, slow-cooled, and equilibrated at 2.0°C for 30 min. The temperature was increased at 0.3°C per min from 2 to 98°C with data collected at 0.3°C increments. Melting profiles were obtained by taking the first derivative of the absorbance with respect to temperature (∂A/∂T) and subjected to nonlinear least-squares parameter estimation of tm,i (melting temperature), ΔHi (van't Hoff enthalpy), A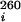 and A
and A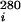 (∂A/∂T amplitude at 260 and 280 nm) for each ith transition on application of a sequential two-state unfolding model by using the tmelt program (27).
(∂A/∂T amplitude at 260 and 280 nm) for each ith transition on application of a sequential two-state unfolding model by using the tmelt program (27).
Results and Discussion
The Wild-Type Pseudoknot Is in Equilibrium with a 5′ Hairpin.
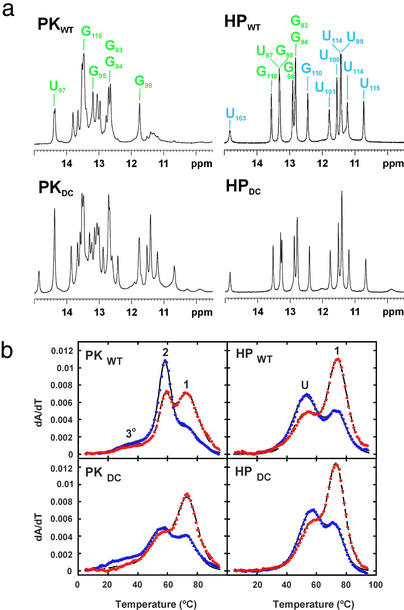
RNA constructs were designed and NMR and optical melting experiments were performed on the wild-type (PKWT) and DKC (PKDC) pseudoknot sequences (Fig. 1b) to characterize the effects of these mutations on structure and thermodynamic stability. The model pseudoknots contain all phylogenetically conserved nucleotides in the p2b (stem 1) and p3 (stem 2) helices, the exact human sequence for the j2b/3 loop (loop 1), and all of the highly conserved nucleotides in the j2a/3 loop (loop 2) (Fig. 1). Assignment of the imino proton resonances of the PKWT construct by using multidimensional heteronuclear NMR spectroscopy indicated formation of all of the base pairs expected for the pseudoknot stems (Fig. 2a), consistent with formation of a hairpin-type pseudoknot.
Figure 2.
The pseudoknot–stem 1 hairpin equilibrium at 200 mM KCl. (a) 1D imino proton NMR spectra of PKWT, PKDC, HPWT, and HPDC. The small resonances at low field in the PKWT spectrum arise from the stem 1 hairpin conformation. The imino proton resonances from the six predicted Watson–Crick base pairs are labeled in green with the additional iminos in HPWT labeled in blue. (b) UV melting profiles, with every 10th data point obtained at 260 and 280 nm shown as blue and red circles, respectively. The calculated fits to the data are indicated by solid (260-nm) and dashed (280-nm) lines. The pseudoknot profiles display two intense transitions with different 260-/280-nm ratios and a much less intense transition at lower temperature arising from tertiary structure. The transitions in PKWT are labeled corresponding to the identified unfolding events. Tertiary structure (3°) unfolds first, followed by stem 2 (2) and then stem 1 (1). The transitions in HPWT are labeled corresponding to unfolding of the four pyrimidine base pairs and top two Watson–Crick base pairs (U) followed by stem 1 (1).
The thermodynamic parameters obtained from the optical melting data (Fig. 2b) for PKWT confirm the formation of two helical stems with the correct base-pair compositions (Fig. 1b), although the stability of the PKWT pseudoknot is 3.5 kcal⋅mol−1 greater than expected from the secondary structure predictions (Table 1). This additional stability was assigned to a tertiary interaction between stem 1 and loop 2 by using variant pseudoknot sequences that abolish this interaction (data not shown). This interaction involves 170CAAA173, the only conserved nucleotides in the j2a/3 loop. Analysis of NOESY and 1H-15N HMQC spectra for PKWT confirmed the presence of tertiary contacts in the minor groove of stem 1 (data not shown).
Table 1.
Thermodynamic parameters obtained at 1 M KCl
| RNA | Observed
|
Predicted
|
|
|---|---|---|---|
| −ΔHtotal | −ΔGtotal | −ΔGtotal | |
| PKWT | 164.7 | 17.8 | 14.2 |
| PKDC | 124.0 | 11.2 | 8.2 |
| HPWT | 92.0 | 9.8 | 9.3*, 10.1† |
| HPDC | 99.0 | 10.5 | 9.3*, 10.1† |
| CR7WT | 88.9 | 9.7 | 11.2 |
| CR7DC | 87.0 | 5.3 | 7.1 |
ΔH is model-dependent van't Hoff enthalpy. ΔH and ΔG are reported in kcal mol−1. ΔG is calculated at 37°C and compared to nearest-neighbor predictions for stacking (35), pseudoknot loop penalties (36), tandem mismatches, and internal loop free energies (37).
Calculated for the expected hairpin with a six-Watson–Crick base-pair stem and a 17-nucleotide loop.
Calculated based on the observed base pairing, with the pyrimidine tract stability estimated from tandem mismatch data.
The pseudoknot-stem 1 hairpin equilibrium for WT and DC is illustrated below. 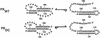
Although the pseudoknot is the predominant conformation, the presence of additional imino proton resonances and exchange crosspeaks in NOESY spectra indicated a minor conformation (≈5%) in equilibrium with PKWT. Hairpin-type pseudoknots typically unfold either via a stem 1 (5′) or stem 2 (3′) hairpin intermediate, depending on the stability of the component hairpins. Given the base-paired regions observed upstream of p2b in the pseudoknot domain, the GC-rich content of stem 1, and the conserved bulged nucleotide in stem 2, the most likely candidate for the alternate conformation is the stem 1 hairpin. To confirm this prediction, we also studied RNA constructs containing the sequence for the wild-type (HPWT) and mutant (HPDC) stem 1 hairpins (Fig. 1b). Comparison of the HPWT and PKWT spectra (Fig. 2a) showed that the additional resonances in the imino region of the pseudoknot correspond to hairpin peaks, identifying the stem 1 hairpin as the alternate conformation. These results are consistent with the unfolding pathway for PKWT determined by analysis of the melting profiles of variant pseudoknots with altered stem compositions (data not shown), which indicates that stem 1 unfolds last.
The DKC Mutation Shifts the Equilibrium Toward the Hairpin Conformation.
NMR spectra of the mutant pseudoknot, PKDC, containing the two consecutive base changes in stem 2, also reveal the presence of two conformations in equilibrium, but in this case, their populations are approximately equal. Comparison of the NMR spectra of PKDC with a stem 1 hairpin containing the GC→AG mutation, HPDC, shows that the alternative conformation in PKDC also corresponds to the stem 1 hairpin (Fig. 2a). Furthermore, the imino proton resonances of HPDC and HPWT have identical chemical shifts, indicating that the mutant and wild-type hairpins have identical base pairs (Fig. 2a), which was confirmed by analysis of the NOESY spectra. The resonances in PKDC that do not arise from the hairpin conformation correspond almost completely to those from PKWT. Therefore, except for disruption of base pairing in stem 2, the wild-type and mutant pseudoknot conformations are also the same. Significantly, however, the stem 1 hairpin conformation is much more highly populated for the mutant sequence (≈50%) than for the wild type (≈5%).
The thermodynamic parameters obtained from optical melting data for PKDC are consistent with the formation of two helical stems and the tertiary interaction (Fig. 2b, Table 1). Although the PKDC mutant pseudoknot is also stabilized by this tertiary interaction, PKDC is less stable than the PKWT pseudoknot, consistent with disruption of the two closing base pairs in stem 2 (Table 1). The large shift in the equilibrium of the mutant pseudoknot toward the stem 1hairpin conformation was unexpected based on the predicted stability of a 17-nucleotide loop with a 6-bp stem.
The Stem 1 Hairpin Structure Is Stabilized by a Unique Uridine Helix.
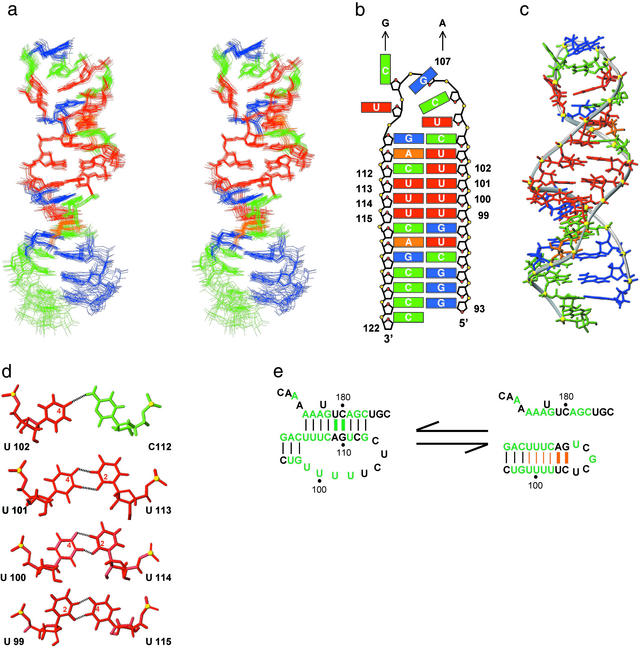
In addition to the six imino protons expected for the predicted Watson–Crick base pairs, the imino proton spectra of HPWT also contains eight additional imino proton resonances (Fig. 2a), indicating that it is significantly more structured than predicted. The solution structure of the stem 1 hairpin (Fig. 3a) shows that it has a well-defined structure (Table 2) in which the loop nucleotides pair to form three consecutive U⋅U base pairs stacked on the Watson–Crick stem followed by a U⋅C pair and two additional Watson–Crick base pairs (Fig. 3 b and c). Thus, the hairpin forms a continuous 12-bp helix capped by a structured pentaloop. The run of four consecutive U⋅U and U⋅C base pairs (Fig. 3d) is, to our knowledge, unprecedented in nucleic acid structures observed to date. A search of the Non-Canonical Base Pair and NDB databases found no structures with more than two consecutive U⋅U (28, 29) or U⋅U/U⋅C base pairs. Each U⋅U base pair has two imino to carbonyl hydrogen bonds. In the first U⋅U base pair, which is stacked on the Watson–Crick G98-C116, the O2 carbonyl of U99 hydrogen bonds with the N3 of U115, whereas the following U100⋅U115 and U101⋅U114 base pairs have hydrogen bonds between the O4 carbonyl of the 5′ base and the N3 imino proton of the 3′ base (Fig. 3d). The U⋅C base pair has a U102 O4 to C112 N4H hydrogen bond and a potential water-mediated hydrogen bond between U102 N3H and C112 N3 (30). There is an abrupt reduction in the C1′-C1′ distance (≈8 Å) for the first two U⋅U base pairs relative to the preceding Watson–Crick base pairs, similar to that observed for the two consecutive U⋅U base pairs in the crystal structure of the UUCG duplex (28). This is followed by a gradual increase in helix width for the third U101⋅U133 base pair progressing toward A-form helical parameters for the U102⋅C112 base pair on the 3′ side of the uridine tract. Overall, the hairpin stem has an extended A-RNA structure with the helical parameters of inclination and X-displacement for the pyrimidine tract intermediate between canonical A- and B-form helices.
Figure 3.
Solution structure of the stem 1 hairpin. (a) Stereoview of the superposition of the 18 lowest energy structures. Nucleotides are colored red (U), green (C), orange (A), and blue (G). (b) Schematic representation of the HPWT structure. DKC mutations are indicated by arrows. (c) Lowest energy structure, with a minor groove view of the central uridine helix and a major groove view of the ordered pentaloop. (d) Hydrogen bonding pattern of the three consecutive U⋅U and the U⋅C base pairs. Hydrogen bonds are indicated by dotted lines and the carbonyl carbons involved in hydrogen bonds are identified by number inside the pyrimidine ring. (e) Sequence conservation of nucleotides that form the hairpin helix. Green letters and bars indicate sequences and base pairs, respectively, that are 100% conserved among vertebrates (9). Base pairs conserved in the hairpin are indicated in orange with thick bars representing the two base pairs, which are conserved to covary among the mammalian sequences.
The run of five Us in loop 1 of the pseudoknot was expected to form an unpaired internal loop, which would destabilize rather than stabilize hairpin formation relative to the pseudoknot. Instead, all of these Us are base paired in the hairpin conformation. Thus, the loop 1 uridine sequence plays a surprisingly important role in the pseudoknot–hairpin equilibrium.
The Mutant Hairpin Forms the Same Structure as the Wild-Type Hairpin.
The DKC mutation occurs in the third and fourth bases of the pentaloop in the hairpin conformation (Fig. 3b). The only large (>0.5-ppm) chemical shift differences observed for HPDC occur in the pentaloop with much smaller shifts occurring in the top three base pairs of the stem (data not shown). The optical melting data (Fig. 2b) for both HPWT and HPDC showed two transitions and a higher stability than predicted for a Watson–Crick 6-bp stem (Table 1), with HPDC slightly more stable than HPWT. The optical melting profiles were consistent with the second transition resulting from the predicted 6-bp stem and the first transition from the additional structure in the hairpin loop region. The thermodynamic and NMR data indicate that the DKC mutation does not alter the unique stem structure observed for the wild-type hairpin. The slight increase in stability of HPDC vs. HPWT can be accounted for by a small change in the stacking interactions in the structured pentaloop. The HPWT structure explains the higher thermal stability observed for both the wild-type and DKC hairpins.
A Proposed Molecular Switch in the Pseudoknot Domain.
Formation of the pseudoknot appears to be required for catalytic activity, because substitutions or insertions which disrupt stem 2 of the pseudoknot abolish human telomerase activity in vitro (13, 14). However, the nucleotides that form the uridine helix in the hairpin are 100% conserved in vertebrates, although sequence conservation of U99→U102 is not required for pseudoknot base pairing or topology. Further inspection of the mammalian telomerase sequences (9) revealed that the two Watson–Crick base pairs at the top of the helix covary to maintain canonical base pairing (Fig. 3e). This previously undetected phylogenetic conservation suggests that the extended stem 1 hairpin conformation is also functionally important, because these base pairs are not formed in the pseudoknot conformation. In vivo chemical probing of human telomerase RNA structure has indicated the presence of an alternative conformation to the putative pseudoknot (31), which we propose is the stem 1 hairpin. A pseudoknot (32) conformational equilibrium has been proposed for the Tetrahymena telomerase RNA (33), and a pseudoknot–hairpin equilibrium also acts as a molecular switch to control translation of the ribosomal protein S15 mRNA (34).
In the wild-type sequence, the greater stability difference between PKWT and HPWT results in the fully folded pseudoknot being the predominant conformation under physiological conditions for the model RNAs. In contrast, the higher stability of HPDC combined with the large destabilization of the mutant pseudoknot, PKDC, results in a shift in the equilibrium to favor the hairpin conformation under physiological conditions. Although the proportions of pseudoknot and hairpin in the intact ribonucleoprotein cannot be predicted from these studies, it is clear that different population distributions would occur for the wild-type and DKC sequences. We propose that it is this change in the relative energy required to switch between the two conformations that accounts for the defect in telomerase activity in the DKC mutation.
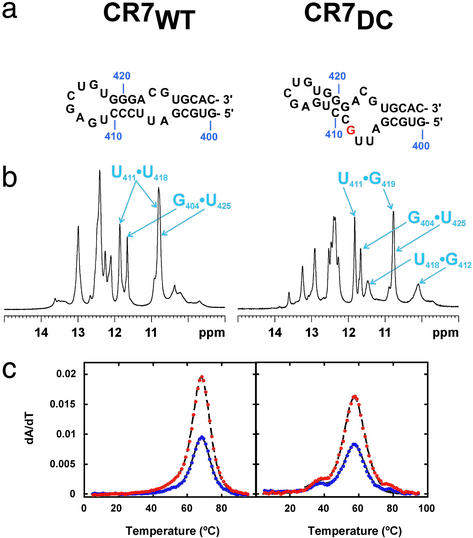
The CR7 Mutation Changes the Secondary Structure and Stability.
The model CR7 domain hairpin contains the exact human sequence for the conserved stem and loop regions (402–429) (Fig. 4a). The DKC mutation in the CR7 domain (C408G) involves disruption of the phylogenetically conserved (9) C408-G421 base pair (Fig. 1a). The wild-type (CR7WT) and to a greater extent the mutant (CR7DC) constructs also exhibit crosspeaks in 2D NMR spectra consistent with multiple conformational states. The predominant secondary structures of CR7WT and CR7DC (Fig. 4a) were determined on the basis of exchangeable proton resonance assignments obtained from NOESY spectra. In CR7WT, the base-pairing scheme is consistent with the phylogenetically predicted secondary structure as the predominant conformation and includes G404⋅U425 wobble and U418⋅U411 base pairs (Fig. 4b). Analysis of CR7DC spectra indicated that it contained the same lower stem base pairs but exhibited significant differences for the upper stem including two consecutive G⋅U base pairs (G419⋅U411 and G412⋅U418) (Fig. 4 a and b). These base pairs can form only if the mutated G shifts into the internal loop, altering the upper stem base pairing and decreasing the size of the terminal hairpin loop (Fig. 4a). Thermodynamic parameters determined for CR7WT and CR7DC (Fig. 4c) demonstrate that, whereas CR7 constructs exhibit more enthalpy than expected, most likely due to base stacking in the loops, CR7DC shows a significant decrease in the melting temperature of both unfolding transitions, with the observed decrease in ΔG consistent with the altered base pairing observed by NMR (Table 1).
Figure 4.
Effect of CR7 mutation on secondary structure and stability. (a) CR7WT and CR7DC RNA constructs shown with secondary structures determined by NMR. The CR7DC stems are oriented to emphasize the change in base pairing that shifts the mutated nucleotide into the internal loop. (b) Imino proton spectra with resonances arising from U⋅U and G⋅U base pairs labeled. (c) UV melting profiles with data displayed as described in Fig. 2B. CR7WT unfolds in two tightly coupled transitions above 65°C. In CR7DC, the two unfolding transitions are uncoupled and shifted to lower temperature.
In vivo experiments with human and mouse telomerase RNAs have shown that alteration (411UGAG414→ACUC) (10) or deletion (Δ412–418) (10, 11) of the CR7 terminal hairpin loop abolishes both accumulation of telomerase RNA and telomerase activity. The upper stem and hairpin loop nucleotides of the CR7 domain (except for C415) are ≥90% conserved among mammals (9) and are less susceptible to chemical cleavage in vivo than in the absence of proteins in vitro (31). Taken together, these observations suggest that this conserved region of the CR7 domain is a binding site for proteins required for nucleolar localization and/or human telomerase RNA stability. Our results demonstrate a significant rearrangement and destabilization of these highly conserved and functionally relevant structural elements in the DKC mutant CR7 sequence, suggesting a structural and functional explanation for the observed telomerase deficiency.
Conclusion
The solution methods used here provide a unique probe of multiple conformational states in equilibrium. The results of our structural and thermodynamic studies and the additional phylogenetic conservation found in this work, taken together with chemical probing (31) and in vitro and in vivo assays of telomerase activity (5, 8, 13, 14), point to the importance of a pseudoknot–hairpin equilibrium for telomerase function. Our results further indicate that the DKC mutation in the pseudoknot domain, which results in impaired telomerase function, does not significantly alter the structure of either the pseudoknot or stem 1 hairpin but rather changes their relative stabilities, resulting in an increased population of the hairpin compared with wild type. This structural equilibrium further implies that a change in the relative stabilities of the pseudoknot and hairpin conformations would have a critical effect on telomerase function. The catalytic protein subunit, hTERT, binds to the pseudoknot domain (10), and it is possible that its activity and/or ability to translocate depends on the interconversion between the two conformations. These studies provide strong support for a dynamic role for the RNA in telomerase function and provide direct structural and thermodynamic insight into how mutations in essential RNA domains may disrupt telomerase activity.
Supplementary Material
Acknowledgments
We thank Robert Peterson for help with the NMR spectroscopy and Evan Feinstein for figure preparation. This research was supported by the National Science Foundation and the National Institutes of Health (NIH) (to J.F.), a NIH National Research Service Award postdoctoral fellowship (to C.A.T.), and a European Molecular Biology Organization postdoctoral fellowship (to L.T.).
Abbreviations
- DKC
dyskeratosis congenita
- NOE
nuclear Overhauser effect
- HMQC
heteronuclear multiple quantum correlation
Footnotes
Data deposition: The chemical shift data and coordinates for the 18 lowest energy structures of HPWT have been deposited in the BioMagResBank and the Protein Data Bank, www.rcsb.org (PDB ID code 1NA2), respectively.
References
- 1.Dokal I. Br J Haematol. 2000;110:768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- 2.Montanaro L, Chilla A, Trere D, Pession A, Govoni M, Tazzari P L, Derenzini M. J Invest Dermatol. 2002;118:193–198. doi: 10.1046/j.0022-202x.2001.01634.x. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell J R, Wood E, Collins K. Nature. 1999;402:551–555. doi: 10.1038/990141. [DOI] [PubMed] [Google Scholar]
- 4.Vulliamy T J, Knight S W, Mason P J, Dokal I. Blood Cells Mol Dis. 2001;27:353–357. doi: 10.1006/bcmd.2001.0389. [DOI] [PubMed] [Google Scholar]
- 5.Ball S E, Gibson F M, Rizzo S, Tooze J A, Marsh J C W, Gordon-Smith E C. Blood. 1998;91:3582–3592. [PubMed] [Google Scholar]
- 6.Knight S W, Heiss N S, Vulliamy T J, Greschner S, Stavrides G, Pai G S, Lestringant G, Varma N, Mason P J, Dokal I, Poustka A. Am J Hum Genet. 1999;65:50–58. doi: 10.1086/302446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell J R, Cheng J, Collins K. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason P J, Dokal I. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 9.Chen J L, Blasco M A, Greider C W. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell J R, Collins K. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Rivera L, Blasco M A. J Biol Chem. 2001;276:5856–5865. doi: 10.1074/jbc.M008419200. [DOI] [PubMed] [Google Scholar]
- 12.Dragon F, Pogačić V, Filipowicz W. Mol Cell Biol. 2000;20:3037–3048. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Autexier C, Pruzan R, Funk W D, Greider C W. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 14.Tesmer V M, Ford L P, Holt S E, Frank B C, Yi X M, Aisner D L, Ouellette M, Shay J W, Wright W E. Mol Cell Biol. 1999;19:6207–6216. doi: 10.1128/mcb.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vulliamy T, Marrone A, Dokal I, Mason P J. Lancet. 2002;359:2168–2170. doi: 10.1016/S0140-6736(02)09087-6. [DOI] [PubMed] [Google Scholar]
- 16.Dieckmann T, Feigon J. J Biomol NMR. 1997;9:259–272. doi: 10.1023/a:1018622708674. [DOI] [PubMed] [Google Scholar]
- 17.Cromsigt J, van Buuren B, Schleucher J, Wijmenga S S. In: Methods in Enzymology. James T, Dotsch V, Schmitz U, editors. Vol. 338. San Diego: Academic; 2001. pp. 371–399. [DOI] [PubMed] [Google Scholar]
- 18.Wöhnert J, Ramachandran R, Gorlach M, Brown L R. J Magn Reson. 1999;139:430–433. doi: 10.1006/jmre.1999.1797. [DOI] [PubMed] [Google Scholar]
- 19.Sklenář V, Dieckmann T, Butcher S E, Feigon J. J Biomol NMR. 1996;7:83–87. doi: 10.1007/BF00190460. [DOI] [PubMed] [Google Scholar]
- 20.Luy B, Marino J P. J Am Chem Soc. 2000;122:8095–8096. [Google Scholar]
- 21.Dingley A J, Masse J E, Feigon J, Grzesiek S. J Biomol NMR. 2000;16:279–289. doi: 10.1023/a:1008307115641. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, Pok K, Butcher S E, Kang S, Chanfreau G, Feigon J. EMBO J. 2001;20:7240–7249. doi: 10.1093/emboj/20.24.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Alba E, Tjandra N. Prog Nucleic Magn Reson Spectrosc. 2002;40:175–197. [Google Scholar]
- 24.Losonczi J A, Andrec M, Fischer M W F, Prestegard J H. J Magn Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 25.Lavery R, Sklenar H. J Biomol Str Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 26.Lu X J, Shakked Z, Olson W K. J Mol Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 27.Theimer C A, Wang Y, Hoffman D W, Krisch H M, Giedroc D P. J Mol Biol. 1998;279:545–564. doi: 10.1006/jmbi.1998.1812. [DOI] [PubMed] [Google Scholar]
- 28.Lietzke S E, Barnes C L, Berglund J A, Kundrot C E. Structure (London) 1996;4:917–930. doi: 10.1016/s0969-2126(96)00099-8. [DOI] [PubMed] [Google Scholar]
- 29.Baeyens K J, Debondt H L, Holbrook S R. Nat Struct Biol. 1995;2:56–62. doi: 10.1038/nsb0195-56. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka Y, Kojima C, Yamazaki T, Kodama T S, Yasuno K, Miyashita S, Ono A, Kainosho M, Kyogoku Y. Biochemistry. 2000;39:7074–7080. doi: 10.1021/bi000018m. [DOI] [PubMed] [Google Scholar]
- 31.Antal M, Boros E, Solymosy F, Kiss T. Nucleic Acids Res. 2002;30:912–920. doi: 10.1093/nar/30.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilley D, Blackburn E H. Proc Natl Acad Sci USA. 1999;96:6621–6625. doi: 10.1073/pnas.96.12.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhattacharyya A, Blackburn E H. EMBO J. 1994;13:5721–5731. doi: 10.1002/j.1460-2075.1994.tb06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philippe C, Benard L, Portier C, Westhof E, Ehresmann B, Ehresmann C. Nucleic Acids Res. 1995;23:18–28. doi: 10.1093/nar/23.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia T B, SantaLucia J, Burkard M E, Kierzek R, Schroeder S J, Jiao X Q, Cox C, Turner D H. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 36.Gultyaev A P, Van Batenburg F H D, Pleij C W A. RNA. 1999;5:609–617. doi: 10.1017/s135583829998189x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews D H, Sabina J, Zuker M, Turner D H. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.