Abstract
One of the serious bottlenecks in structural genomics projects is overexpression of the target proteins in soluble form. We have applied the directed evolution technique and prepared soluble mutants of the Mycobacterium tuberculosis Rv2002 gene product, the wild type of which had been expressed as inclusion bodies in Escherichia coli. A triple mutant I6T/V47M/T69K (Rv2002-M3) was chosen for structural and functional characterizations. Enzymatic assays indicate that the Rv2002-M3 protein has a high catalytic activity as a NADH-dependent 3α, 20β-hydroxysteroid dehydrogenase. We have determined the crystal structures of a binary complex with NAD+ and a ternary complex with androsterone and NADH. The structure reveals that Asp-38 determines the cofactor specificity. The catalytic site includes the triad Ser-140/Tyr-153/Lys-157. Additionally, it has an unusual feature, Glu-142. Enzymatic assays of the E142A mutant of Rv2002-M3 indicate that Glu-142 reverses the effect of Lys-157 in influencing the pKa of Tyr-153. This study suggests that the Rv2002 gene product is a unique member of the SDR family and is likely to be involved in steroid metabolism in M. tuberculosis. Our work demonstrates the power of the directed evolution technique as a general way of overcoming the difficulties in overexpressing the target proteins in soluble form.
Large-scale genome sequencing projects have provided a huge amount of information on gene sequences. However, for a considerable fraction of the predicted gene products, we are far from being able to assign their functions. In some cases, there may be no sufficient sequence similarity to homologous proteins with known function. In other cases, functional assignment on the basis of sequence similarity alone is ambiguous, because proteins sharing conserved sequence motifs often serve a variety of molecular functions. As the three-dimensional structure of proteins is intimately coupled with the molecular function, the structure of a protein may provide clues for its molecular function. The validity of this approach has been demonstrated by several examples (1–3), and a number of large-scale structural genomics projects have been initiated (4, 5).
One of the most serious bottlenecks in structural genomics efforts lies in the expression of target proteins in soluble form (6, 7). This difficulty severely limits the overall success rate of current structural genomics projects. Many polypeptides fail to fold into their native state and accumulate as insoluble inclusion bodies when they are overexpressed heterologously in Escherichia coli, the most frequently used expression system at present. One of the most successful approaches for overcoming this difficulty is site-directed mutagenesis of one or a few amino acid residues. However, it generally requires extensive trial-and-errors to find out the proper amino acid substitutions, which will result in improved solubility of the expressed proteins, because it is difficult to predict the necessary changes. For example, a structural study on the catalytic domain of HIV integrase required a systematic replacement of hydrophobic residues (8, 9). As an efficient method of obtaining mutant proteins with improved solubility in E. coli expression systems, the directed evolution technique using GFP as a folding reporter was proposed (10). In this experiment, the gene encoding the target protein is subjected to random mutations and soluble mutants are selected from a mutant library of the target protein fused to the N terminus of GFP, because there is a good correlation between folding of the target protein expressed alone and the fluorescence of E. coli cells expressing GFP fusions (10). Here we report a successful application of the directed evolution approach to a target protein of the structural genomics project on Mycobacterium tuberculosis (11, 12).
The M. tuberculosis Rv2002 gene encodes a 260-residue protein, with a calculated molecular mass of 27,030 Da. It belongs to the short-chain dehydrogenase/reductase (SDR) family, because it contains the characteristic dinucleotide binding motif GXXXGXG (residues 14–20) and the YXXXK (residues 153–157) sequence motif. It has been annotated as fabG3, a homolog of β-ketoacyl ACP reductase (KAR) from M. tuberculosis (fabG1, Rv1483) (13), which is the second enzyme in fatty acid elongation cycle, on the basis of amino acid sequence similarity (identity of 31% in a 241-residue overlap). Among KARs, the highest sequence identity is observed with that from Bacillus halodurans (38% in a 244-residue overlap). It also shows significant sequence similarity toward l-3-hydroxyacyl-CoA dehydrogenase involved in fatty acid β-oxidation (35% identity in a 204-residue overlap with the one from rat brain; Swiss-Prot, O70351) and 3α, 20β-hydroxysteroid dehydrogenase (HSD) (49% identity in a 243-residue overlap with the one from Streptomyces hydrogenans; Swiss-Prot, P19992). Because it shows significant sequence similarity toward various SDR family enzymes with diverse functions, its molecular or biological function cannot be unambiguously inferred from its primary sequence data alone. Functional assignment of the Rv2002 gene product will be greatly facilitated by its structural and functional characterizations, for which a considerable amount of the protein is required. Because it was initially expressed as inclusion bodies in E. coli, soluble mutants were prepared by applying the GFP-based directed evolution technique and this enabled us to perform further studies on the triple mutant I6T/V47M/T69K, designated as Rv2002-M3. Crystallization of the triple mutant was reported previously (14). Here we report the results of our structural and functional characterizations. Our work suggests that the Rv2002 gene product is a unique member of the SDR family and may be involved in steroid metabolism in M. tuberculosis. This study also demonstrates that directed evolution is a powerful approach to overcoming the difficulties in protein overexpression.
Materials and Methods
GFP-Based Directed Evolution.
Each round of GFP-based directed evolution consisted of two stages (10). The first stage was preparation of a mutant library of GFP fusions by introducing random mutations into the Rv2002 gene through error-prone PCR and DNA shuffling. The next stage was selection of E. coli colonies from the mutant library, which showed brighter fluorescence compared with the wild type. GFP fused at the C terminus of the Rv2002 protein serves as a reporter for proper folding of the upstream protein. Mutant Rv2002 genes from the selected colonies, which showed enhanced fluorescence, were used for preparation of a mutant library in the next round. After three rounds of forward evolution without backcrossing with the wild type, we finally selected five mutants with the greatest fluorescence improvement and checked their solubility. Mutation sites were identified through DNA sequencing of both strands.
Preparation of mutant library.
The Rv2002 gene was amplified by PCR using the wild-type gene cloned into the C-terminal His-tagging vector of Waldo (10, 14) with Pfu (exo+) DNA polymerase (Stratagene), and the PCR product was randomly cleaved with DNase I (GIBCO/BRL) at 15°C for 3 min by using Mn2+ as the metal cofactor. DNA fragments were reassembled with Pfu (exo−) DNA polymerase (Stratagene) without primers of the Rv2002 gene. An additional PCR in the presence of the primers elongated the partially reassembled gene fragments to its full length. Reassembled genes were digested with NdeI and BamHI (New England Biolabs) and were ligated into the GFP-fusion vector (10) by using T4 DNA ligase (GIBCO/BRL) and transformed into DH10B cells (GIBCO/BRL) by electroporation. The plasmid library of mutants was recovered from the resuspension of mutant colonies on LB-agar plates.
Screening.
The mutant plasmid library was transformed into B834(DE3) cells (Novagen), and the cells were plated directly onto nitrocellulose membranes on a LB-agar plate. After incubation at 37°C for 10 h, the membrane was transferred onto a LB-agar plate containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated for 5–6 h for induction. The 40 brightest colonies were picked and transferred onto the master plate. The master plate was incubated at 37°C for 14–16 h, and its replica was made on a nitrocellulose membrane. The replica membrane was incubated on a LB-agar plate at 37°C for 8–10 h, transferred onto a LB-agar plate containing 1 mM IPTG, and incubated for an additional 4–6 h for induction. Colonies (10–20) showing significant fluorescence improvements over the wild type were selected, and the cell mass of selected colonies on the plate was recovered. A mixture of plasmids from them was used as the starting template for PCR in subsequent rounds of directed evolution.
Site-Directed Mutagenesis.
Three double mutants (I6T/V47M, I6T/T69K, V47M/T69K) and three single mutants (I6T, V47M, T69K) of Rv2002 were prepared by removing the mutations from Rv2002-M3 using the QuikChange Site-Directed Mutagenesis kit (Stratagene). S140A, E142A, and Y153F mutants of Rv2002-M3 were prepared with the same kit. The mutations were confirmed by sequencing.
Overexpression and Purification.
The soluble mutant Rv2002-M3 was overexpressed and purified as reported (14). The selenomethionine-substituted Rv2002-M3 protein was expressed in E. coli B834(DE3) cells, by using the M9 cell culture medium containing extra amino acids. DTT (10 mM) was added during purification. E142A, Y153F, and S140A mutants of the Rv2002-M3 protein were overexpressed and purified as Rv2002-M3.
Enzyme Assay.
Cofactor specificity.
Reductase activity was measured by using progesterone as a substrate in the presence NAD(P)H. Assays were performed at 30°C with the following components: 125 μM progesterone, 100 mM sodium cacodylate, pH 6.0, 150 μM NAD(P)H and 1.0 μM purified Rv2002-M3 protein. The conversion of NAD(P)H to NAD(P)+ was monitored spectrophotometrically at 340 nm.
Optimal pH.
The optimal pH for dehydrogenase activity was determined at 30°C by using androsterone as the substrate under conditions of 0.5 μM purified Rv2002-M3 protein, 0.5 mM NAD+, 50 μM androsterone, and 100 mM of an appropriate buffer. The optimal pH for reductase activity was determined with progesterone and NADH.
Specific activity toward putative substrates.
Reductase activity was measured at 30°C against acetoacetyl-CoA and progesterone, and dehydrogenase activity was measured against l-3-hydroxybutyric acid and five different steroid compounds (androsterone, epiandrosterone, 20α-hydroxyprogesterone, 20β-hydroxyprogesterone, and 17β-estradiol). The reaction mixture contained 100 mM sodium cacodylate (pH 6.0), 1 μM purified Rv2002-M3 protein, 125 μM of each putative substrate, and 0.15 mM NADH for reduction (or 1 mM NAD+ for oxidation).
Determination of kinetic parameters.
Km and kcat were determined on three steroidal substrates (androsterone, 20β-hydroxyprogesterone, and progesterone), for which relatively high activities were measured in the above assay. Changes in NADH concentration were monitored for the initial 5 min at 30°C for the substrate concentration ranging from 1 to 100 μM.
Crystallization, Data Collection, and Structure Determination.
Details of crystallization and crystallographic methods are published as supporting information on the PNAS web site, www.pnas.org. Table 2, summarizing the statistics for x-ray data collection, phasing, and model refinement, is also published as supporting information on the PNAS web site.
Results and Discussion
Preparation of Soluble Mutants by Directed Evolution.
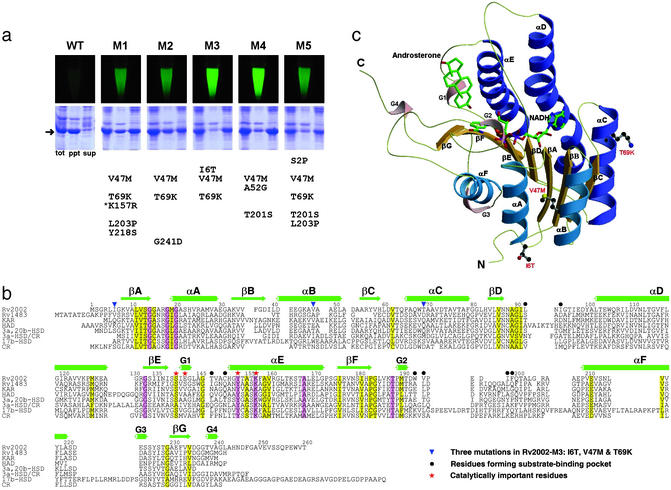
The wild-type Rv2002 with a C-terminal hexa-histidine tag was expressed as inclusion bodies in E. coli (Fig. 1a). Several approaches to overcoming this difficulty may be considered. The first approach is refolding. However, the yield of refolding of misfolded proteins is usually so low that refolding is not generally applicable for structural studies, which require a large amount of properly folded proteins. The second approach is introduction of point mutations by site-directed mutagenesis. This is an inefficient and limited way of exploring the sequence space for soluble expression, requiring extensive trial-and-errors, and is unlikely to succeed if multiple mutations are necessary for soluble expression. Another approach may be exhaustive trials of other cell-based or cell-free expression systems. However, it could be very time-consuming and costly to construct a number of different expression vectors, including eukaryotic expression systems, and to test them under different conditions. Gateway cloning technology (Invitrogen) offers convenience in construction of different expression vectors, but commercially available destination vectors for E. coli expression are currently very limited. Compared with the above approaches, directed evolution is generally applicable for many structural studies and offers an advantage that soluble mutants can be engineered conveniently in a short period without sacrificing the high yield, low cost, and speed of E. coli expression. Therefore, we applied the GFP-based directed evolution technique (10) to obtain soluble mutants of Rv2002, and several of them showed dramatically improved solubility on E. coli expression (Fig. 1a). Each of these mutants carried three to five point mutations, among which V47M and T69K were most common. All of the mutation sites fall outside of the conserved sequence motifs of the SDR family (Fig. 1b), except K157R in the mutant M1. We chose a triple mutant Rv2002-M3 for structural and functional studies, because it showed a maximum improvement in solubility and contained the smallest number of point mutations. Our subsequent structural analysis confirmed that the three point mutations (I6T/V47M/T69K) of Rv2002-M3 are all located far from the substrate-binding pocket, the cofactor-binding pocket, the catalytic site, and the subunit interface (Fig. 1c), as discussed in more detail below. Thus, it is reasonable to expect that these mutations would have only a minor effect on the function and structure.
Figure 1.
GFP-based directed evolution, sequence alignment, and overall subunit structure of the Rv2002-M3 protein. (a) Fluorescence of the resuspended cells harboring genes encoding the wild-type or mutant Rv2002 proteins in a GFP-fused form and expression test of the wild-type or mutant Rv2002 proteins in a nonfused form. WT, wild type; M1–M5, soluble mutants; tot, total cell; ppt, precipitant fraction; sup, supernatant fraction. The mutations of each mutant are listed. The arrow indicates the expressed Rv2002 proteins and the asterisk signifies the mutation at a conserved residue of the SDR family. (b) Sequence alignment of Rv2002 with other SDRs. Rv1483, β-ketoacyl ACP reductase (fabG1) from M. tuberculosis; KAR, β-ketoacyl ACP reductase from B. halodurans; HAD, l-3-hydroxyacyl-CoA dehydrogenase from rat brain; 3a,20b-HSD, 3α,20β-hydroxysteroid dehydrogenase from S. hydrogenans; 3a-HSD/CR, 3α-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni; 17b-HSD, 17β-hydroxysteroid dehydrogenase from human; CR, lung carbonyl reductase from mouse. This figure was produced with ALSCRIPT (32). (c) Ribbon diagram of the Rv2002-M3 monomer in complex with NADH and androsterone. Carbon, nitrogen, oxygen, and phosphorus atoms are in green (or black), blue, red, and purple, respectively. All figures of structures in this paper were produced with MOLSCRIPT (33), BOBSCRIPT (34), and RASTER3D (35).
Overall Tertiary and Quaternary Structures.
We have determined the crystal structures of the Rv2002-M3 protein as a binary complex with NAD+ at 1.8 Å resolution and as a ternary complex with androsterone and NADH at 2.4 Å. In both the binary and ternary complex structures, the protein model includes amino acid residues 2–245. The C-terminal 15 residues as well as the hexa-histidine tag have no electron density and are apparently disordered in the crystal. Each subunit comprises a single domain containing the characteristic dinucleotide-binding fold (Rossmann fold). Its central β-sheet consists of seven parallel β-strands βC–βB–βA–βD–βE–βF–βG and is flanked on each side by three parallel α-helices, (αA, αB, αF) or (αC, αD, αE) (Fig. 1c). Additionally, it contains four 310-helices. Conformations of the two (or four) monomers in the asymmetric unit of the binary (or ternary) complex crystal are essentially identical, with rms deviations of 0.17 Å (or 0.06–0.08 Å) in the binary (or ternary) complex structure for 244 Cα atom pairs. Two loop regions (residues 52–54 and 131–132) and both N- and C-terminal regions show the largest deviations. Structural changes on binding androsterone are mainly localized to the substrate-binding loops. Corresponding subunits in the binary complex with NAD+ and in the ternary complex with androsterone and NADH show rms deviations of 0.21–0.22 Å for 244 Cα atom pairs, with the largest Cα deviations of 0.74–1.17 Å at Ala-52, Asp-53, Glu-98, Asp-99, and Trp-193. The latter three residues are close to the substrate binding loops, whereas the former two residues belong to a loop with high B-factors. In the crystal, four chemically identical subunits form a tetramer of the 222 molecular symmetry with approximate dimensions of 65 Å × 65 Å × 75 Å. The buried solvent-accessible surface area in the interface between subunits related by the P/Q/R axis is 1,430/1,500/770 Å2 per monomer. The P axis interface, formed by the residues 202–240, encompass the helix αF, strand βG, and 310-helix G3. Hydrophobic residues of helices αD and αE that are exposed on the subunit surface contribute mainly to the Q axis interface by forming a four-helix bundle about the Q axis. The two subunits related by the R axis swap their C-terminal loop regions (residues 241–245). The R axis interface also includes the 310-helix G4 and two loop regions (residues 145–147 and 197–200).
Cofactor and Substrate Specificities.
The reductase activity of the Rv2002-M3 protein was measured by using progesterone as the substrate in the presence of either NADH or NADPH. The optimum pH for the reductase activity was found to be ≈6.0 (data not shown), similarly to other SDRs (15). The reductase activity measurements showed a definite preference of NADH as the cofactor (Table 1). This cofactor specificity is consistent with the structural observation. In the crystal structures of both the binary and ternary complexes (Fig. 2a), the side chain oxygen atoms of Asp-38 form hydrogen bonds with two oxygen atoms of the adenosine ribose of NAD(H), thus restricting the binding of NADP(H). A similar mode of NAD(H) binding was observed in other NAD(H)-dependent enzymes (16, 17). In comparison, two basic residues make strong electrostatic interactions with the 2′-phosphate group of NADPH in the NADPH-dependent enzymes (18).
Table 1.
Steady-state kinetic analysis of the Rv2002-M3 protein
| Substrate | kcat, min−1 | Km, M | kcat/Km, min−1 M−1 | Activity*, % |
|---|---|---|---|---|
| Oxidation | ||||
| Androsterone | 7.6 | 2.4 × 10−5 | 3.1 × 105 | 100 |
| Epiandrosterone | — | — | — | 3 |
| 20α-hydroxyprogesterone | — | — | — | 2 |
| 20β-hydroxyprogesterone | 4.3 | 1.7 × 10−5 | 2.6 × 105 | 69 |
| 17β-estradiol | — | — | — | 4 |
| l-3-hydroxybutyric acid | — | — | — | ND† |
| Reduction | ||||
| Progesterone | 1.2 | 3.3 × 10−6 | 3.6 × 105 | 22 |
| Progesterone, with NADPH‡ | — | — | — | ND† |
| Acetoacetyl-CoA | — | — | — | 1 |
Relative to the specific activity on androsterone, which is set to 100%.
No detectable activity was measured with 1.0 μM purified Rv2002-M3 protein.
All other putative substrates were assayed with NAD+ (for oxidation) or NADH (for reduction).
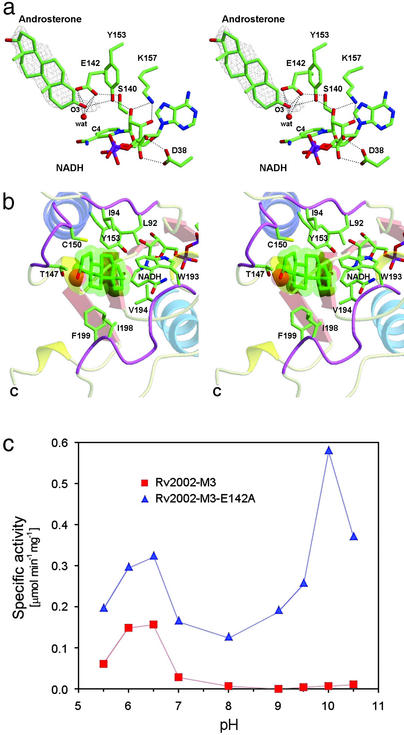
Figure 2.
Catalytic site, substrate-binding pocket, and the effect of Glu-142 on catalysis. (a) Stereo view of the catalytic site of Rv2002-M3 in complex with androsterone and NADH. Glu-142 is present near the Ser-140/Tyr-153/Lys-157 catalytic triad. The final (2Fo − Fc) electron density map calculated by using 20–2.4 Å data are contoured at 1 σ for the androsterone molecule. Possible hydrogen bonds are shown as dashed lines. (b) Binding of androsterone. Three loop regions, which interact with androsterone, are shown in purple. For NADH, only the nicotinamide part is shown. (c) The effect of Glu-142 on dehydrogenase activity. The Rv2002-M3-E142A mutant recovers the dehydrogenase activity at basic pH, which is characteristic of other SDRs.
To investigate the substrate specificity, we checked the catalytic activity of Rv2002-M3 against various putative substrates, including acetoacetyl-CoA, l-3-hydroxybutyric acid, and several steroidal compounds. Rv2002-M3 showed no detectable activity for oxidation of l-3-hydroxybutyric acid and only an insignificant activity for reduction of acetoacetyl-CoA. On the other hand, we could measure significant activities for oxidation of androsterone (3α-hydroxy-5α-androstan-17-one) and 20β-hydroxyprogesterone (4-pregnen-20β-ol-3-one), and for reduction of progesterone (4-pregnen-3,20-dione). The oxidation activities against 17β-estradiol (1,3,5-estratriene-3,17β-diol), epiandrosterone (3β-hydroxy-5α-androstan-17-one), and 20α-hydroxyprogesterone (4-pregnen-20α-ol-3-one) were very low (Table 1). To summarize, Rv2002-M3 showed the highest activity as NAD+-dependent 3α, 20β-HSD among the enzymatic activities tested. Our structural and functional characterizations of the Rv2002-M3 protein indicate that the Rv2002 gene product is less likely to play a catalytic role as either KAR (generally NADPH-dependent) in the fatty acid synthetic pathway or l-3-hydroxyacyl-CoA dehydrogenase (generally NAD+-dependent) in the fatty acid β-oxidation pathway. It is more likely to play an uncharacterized role in steroid metabolism in M. tuberculosis. Interestingly, a possible link between steroid and M. tuberculosis infection and intracellular survival was proposed (19, 20). Further studies on the role of steroid and steroid metabolism in M. tuberculosis could provide new insights into its pathogenesis.
Cofactor and Substrate Binding at the Active Site.
The modes of cofactor binding in both the binary complex with NAD+ and the ternary complex with androsterone and NADH are similar. The NE and NH1 atoms of Arg-17 form hydrogen bonds with two oxygen atoms of the phosphate group in the adenine side of NAD(H). Asp-38 forms hydrogen bonds with two oxygen atoms of the adenosine ribose and plays a key role in determining the cofactor specificity, as mentioned above. We attempted cocrystallization of the Rv2002-M3 protein with acetoacetyl-CoA, 17β-estradiol, progesterone, and androsterone in the presence of either NAD+ or NADH. However, only the androsterone complex gave an interpretable electron density for the bound substrate in the substrate-binding pocket (Fig. 2a), whereas a poorly defined electron density was observed for progesterone and no electron density was observed for acetoacetyl-CoA and 17β-estradiol. These crystallographic observations are in good accordance with the results of our enzymatic assays toward these putative substrates (Table 1).
The steroidal ring of androsterone is bound in a pocket formed by the three loop regions, 92–94, 147–150, and 193–199, making contacts with the side chains of Leu-92, Ile-94, Thr-147, Cys-150, Tyr-153, Val-194, Ile-198, and Phe-199 (Fig. 2b). The reactive O3 atom of androsterone is directed toward the nicotinamide ring of NADH. The 147–150 loop is adjacent to the conserved sequence motif YXXXK (residues 153–157), whereas the 92–94 and 193–199 loops are highly variable in sequence and length among SDR family members (Fig. 1b). A structure-based sequence alignment (Fig. 1b) indicates that the 193–199 loop is shorter by five residues compared with that of 3α, 20β-HSD from S. hydrogenans (16, 21).
Catalytic Triad and Glu-142 in the Active Site.
A possible catalytic mechanism proposed for the SDR family involves a catalytic triad consisting of conserved Ser, Tyr, and Lys residues (16, 18, 22, 23). The active site of Rv2002-M3 has a corresponding catalytic triad Ser-140/Tyr-153/Lys-157. The tyrosine residue was proposed to play a key role as a catalytic acid/base in reduction/oxidation reaction. The lysine residue was proposed to have dual roles. One is to contribute to positioning and orientation of NADH through hydrogen bonding to two oxygen atoms of ribose in the nicotinamide side of NADH (Fig. 2a). The other is to facilitate the formation of the phenolate ion by lowering the pKa value of the tyrosine hydroxyl group. The conserved serine residue was proposed to form hydrogen bonds with the substrate, the reaction intermediate, and the product and/or with the hydroxyl group of the conserved tyrosine residue. When we prepared Y153F and S140A mutants of Rv2002-M3, the enzymatic activities for both oxidation of androsterone and reduction of progesterone were completely lost, suggesting a similar mechanism for Rv2002-M3 as other SDRs (24–26).
However, the structure reveals a unique feature of the active site of Rv2002-M3, i.e., the presence of Glu-142 near the catalytic residues, Tyr-153 and Ser-140, and the substrate (Fig. 2a). In other SDR enzymes (15, 16, 18, 27–30), a glycine, alanine, serine, or valine residue is frequently present at the corresponding position of Glu-142 (Fig. 1b). Interestingly, the two carboxylic oxygen atoms of Glu-142 are within the distance of possible hydrogen bonds with the O3 atom of androsterone (3.45–3.48 Å). The O3 atom of androsterone also forms a hydrogen bond with a nearby water molecule (2.85 Å). This water molecule is 3.39 Å away from the hydroxyl oxygen atom of Tyr-153 and 3.43 Å away from the C4 atom of nicotinamide ring, the site of hydride transfer in NADH (Fig. 2a). In the structure of the binary complex with NAD+, there are three additional water molecules, which are excluded from the active site on binding androsterone.
In both structures of the binary and ternary complexes of Rv2002-M3, the hydroxyl group of Ser-140 points away from androsterone or Tyr-153, and forms a hydrogen bond with one of the two carboxylic oxygen atoms of Glu-142 (Fig. 2a). This is different from other SDRs, in which the hydroxyl group of the conserved serine residue is oriented toward the reactive oxygen atom of the substrate or the hydroxyl group of the catalytic tyrosine residue. As mentioned above, the side chain of Glu-142 is located in the proximity of androsterone in the Rv2002-M3 structure, with its side chain interacting with the O3 atom of the substrate and forming a strong hydrogen bond with Ser-140 (2.75 Å between OE2 of Glu-142 and OG of Ser-140). Because this unusual Glu occupies a key position in the active site of Rv2002-M3, we explored its possible role by mutagenesis.
Glu-142 Reverses the Effect of Lys-157.
Rv2002-M3 shows a pH optimum at 6.0–6.5 for its dehydrogenase activity (Fig. 2c), whereas other SDRs were reported to have the optimum pH between 8 and 10 for the oxidation reaction (15). To check whether this difference originates from the presence of Glu-142 in the active site, we prepared the E142A mutant of Rv2002-M3. Its optimum pH for the reduction of progesterone shifted slightly from 6.0 to 6.5 (data not shown) but its dehydrogenase activity (for oxidation of androsterone) showed dual pH optima, one at pH 6.0–6.5 and the other at pH 10 (Fig. 2c). We interpret this result as follows. Glu-142 of Rv2002-M3 is not directly involved in catalysis but its negative charge counteracts against the positive charge of Lys-157, thus restoring the normal pKa of Tyr-153. This pushes the second pH optimum of Rv2002-M3 from ≈10 to ≈12, at which pH the protein is unstable and loses the catalytic activity. As a consequence, Rv2002-M3 with Glu-142 at the active site does not show a pH optimum at ≈10 for the dehydrogenase activity, unlike other classical SDRs, which lack an equivalent Glu. Tyr-153 must be involved in the oxidation reaction at acidic pH through some unknown mechanism, because the Y153F mutant of Rv2002-M3 completely lost activities at both acidic and basic pHs. To summarize, Glu-142 reverses the effect of Lys-157 in influencing the pKa of Tyr-153 and its presence in the active site makes Rv2002 a unique member of the SDR family.
Roles of Mutations in Solubility Improvement.
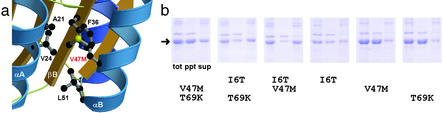
How do I6T/V47M/T69K mutations contribute to the improved solubility of the E. coli-expressed Rv2002 protein? The I6T mutation site is in the N-terminal loop, on the molecular surface (Fig. 1c), with the OG atom of Thr-6 interacting with the carbonyl oxygen atoms of Gly-3 and Thr-6 itself through hydrogen bonding. The mutation V47M on helix αB (Fig. 1c) increases the hydrophobic contact with Ala-21 and Val-24 in helix αA, Phe-36 in the adjacent strand βB, and Leu-51 in helix αB (Fig. 3a). It seems to contribute to a tighter packing of the hydrophobic core, which is composed of three secondary structure elements (strands βA, βB, and helix αA), and consequently to the overall stability of the subunit. T69K on helix αC (Fig. 1c) and I6T certainly should increase the intrinsic solubility of the folded protein, because these substitutions occur on the molecular surface and enhance the polar characteristics of the molecule. Other mechanisms may also contribute to soluble expression, as evidenced by higher solubility of the double mutant I6T/V47M compared with that of V47M/T69K (see below). A major role of V47M mutation may be lowering the kinetic barriers in folding pathway of Rv2002 by enhancing the stability of the above-mentioned hydrophobic core, which is formed by five residues (Ala-21, Val-24, Phe-36, Val-47, and Leu-51). All of these residues are in the N-terminal side of the polypeptide chain and the formation of this hydrophobic core in the early stage of folding pathway may be facilitated by the V47M substitution. An attractive suggestion would be that the substitutions I6T and T69K primarily change the intrinsic solubility and the V47M substitution affects the folding kinetics. This suggestion is consistent with the idea that the solubility of a recombinant protein is determined not only by the intrinsic solubility of the folded protein but also by the folding pathway in vivo (31).
Figure 3.
V47/M mutation in Rv2002-M3 and expression test. (a) V47M seems to contribute to a tighter packing of the hydrophobic core. (b) Expression test of single or double mutants, which have one or two of the three point mutations I6T/V47M/T69K. tot, total cell; ppt, precipitant fraction; sup, supernatant fraction.
Are all three point mutations I6T/V47M/T69K required for soluble expression? To address this question, we prepared six mutants carrying one or two of the above mutations. All three single mutants (I6T, V47M, or T69K) were expressed as mainly inclusion bodies. On the other hand, two double mutants, I6T/V47M and I6T/T69K, were highly soluble on E. coli expression and the other double mutant, V47M/T69K, showed ≈30% soluble expression (Fig. 3b). For Rv2002, it would have been difficult, if not impossible, to discover the soluble mutants by a more rational approach of designing or predicting the point mutations. In comparison, the GFP-based directed evolution approach searches the mutation space in an efficient way and thus allows one to obtain the desired soluble mutants more readily. It is expected that the directed evolution approach to overcoming the difficulties in protein overexpression will play an important role in the future structural genomics research.
Supplementary Material
Acknowledgments
We thank Professor N. Sakabe and his staff at beamline BL-18B of the Photon Factory, Japan, and the staff of beamline 6B at Pohang Light Source for assistance during data collection. We also thank Professor Kyeong Kyu Kim at Sungkyunkwan University for allowing data collection on the R-Axis IV++ system. This work was supported by the Korea Ministry of Science and Technology (NRL-2001, Grant M1-0104-00-0132). J.K.Y. is a recipient of the BK21 Fellowship.
Abbreviations
- SDR
short-chain dehydrogenase/reductase
- NAD
nicotinamide adenine dinucleotide
- HSD
20β-hydroxysteroid dehydrogenase
- KAR
β-ketoacyl ACP reductase
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1NFR for the NAD+ complex of the selenomethionine crystal, 1NFF for the NAD+ complex of the native crystal, and 1NFQ for the androsterone/NADH complex of the native crystal).
References
- 1.Lima C D, Klein M G, Hendrickson W A. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 2.Hwang K Y, Chung J H, Kim S-H, Han Y S, Cho Y. Nat Struct Biol. 1999;6:691–696. doi: 10.1038/10745. [DOI] [PubMed] [Google Scholar]
- 3.Lee J Y, Kwak J E, Moon J, Eom S H, Liong E C, Pedelacq J-D, Berendzen J, Suh S W. Nat Struct Biol. 1999;8:789–794. doi: 10.1038/nsb0901-789. [DOI] [PubMed] [Google Scholar]
- 4.Burley S K. Nat Struct Biol. 2000;7:932–934. doi: 10.1038/80697. [DOI] [PubMed] [Google Scholar]
- 5.Blundell T L, Mizuguchi K. Prog Biophys Mol Biol. 2000;73:289–295. doi: 10.1016/s0079-6107(00)00008-0. [DOI] [PubMed] [Google Scholar]
- 6.Christendat D, Yee A, Dharamsi A, Kluger Y, Savchenko A, Cort J R, Booth V, Mackereth C D, Saridakis V, Ekiel I, et al. Nat Struct Biol. 2000;7:903–909. doi: 10.1038/82823. [DOI] [PubMed] [Google Scholar]
- 7.Christendat D, Yee A, Dharamsi A, Kluger Y, Gerstein M, Arrowsmith C H, Edwards A M. Prog Biophys Mol Biol. 2000;73:339–345. doi: 10.1016/s0079-6107(00)00010-9. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins T M, Engelman A, Ghirlando R, Craigie R. Proc Natl Acad Sci USA. 1995;92:6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 10.Waldo G S, Standish B M, Berendzen J, Terwilliger T C. Nat Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 11.Terwilliger T C. Nat Struct Biol. 2000;7:935–939. doi: 10.1038/80700. [DOI] [PubMed] [Google Scholar]
- 12.Goulding C W, Apostol M, Anderson D H, Gill H S, Smith C V, Kuo M R, Yang J K, Waldo G S, Suh S W, Chauhan R, et al. Curr Drug Targets Infect Dis. 2002;2:121–141. doi: 10.2174/1568005023342551. [DOI] [PubMed] [Google Scholar]
- 13.Banerjee A, Sugantino M, Sacchettini J C, Jacobs W R., Jr Microbiology. 1998;144:2697–2707. doi: 10.1099/00221287-144-10-2697. [DOI] [PubMed] [Google Scholar]
- 14.Yang J K, Yoon H-J, Ahn H J, Lee B I, Cho S, Waldo G S, Park M S, Suh S W. Acta Crystallogr D. 2002;58:303–305. doi: 10.1107/s0907444901018789. [DOI] [PubMed] [Google Scholar]
- 15.Breton R, Housset D, Mazza C, Fontecilla-Camps J C. Structure (London) 1996;4:905–915. doi: 10.1016/s0969-2126(96)00098-6. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh D, Wawrzak Z, Weeks C M, Duax W L, Erman M. Structure (London) 1994;2:629–640. doi: 10.1016/s0969-2126(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 17.Varughese K I, Skinner M M, Whiteley J M, Matthews D A, Xuong N H. Proc Natl Acad Sci USA. 1992;89:6080–6084. doi: 10.1073/pnas.89.13.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka N, Nonaka T, Nakanishi M, Deyashiki Y, Hara A, Mitsui Y. Structure (London) 1996;4:33–45. doi: 10.1016/s0969-2126(96)00007-x. [DOI] [PubMed] [Google Scholar]
- 19.Av-Gay Y, Sobouti R. Can J Microbiol. 2000;46:826–831. [PubMed] [Google Scholar]
- 20.Gatfield J, Pieters J. Science. 2000;288:1647–1650. doi: 10.1126/science.288.5471.1647. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh D, Erman M, Wawrzak Z, Duax W L, Pangborn W. Structure (London) 1994;2:973–980. doi: 10.1016/s0969-2126(94)00099-9. [DOI] [PubMed] [Google Scholar]
- 22.Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffery J, Ghosh D. Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 23.Oppermann U C T, Filling C, Jörnvall H. Chem Biol Interact. 2001;130–132:699–705. doi: 10.1016/s0009-2797(00)00301-x. [DOI] [PubMed] [Google Scholar]
- 24.Obeid J, White P C. Biochem Biophys Res Commun. 1992;188:222–227. doi: 10.1016/0006-291x(92)92373-6. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, Jiang J C, Lin Z-G, Lee W R, Baker M E, Chang S H. Biochemistry. 1993;32:3342–3346. doi: 10.1021/bi00064a017. [DOI] [PubMed] [Google Scholar]
- 26.Oppermann U C T, Filling C, Berndt K D, Persson B, Benach J, Ladenstein R, Jörnvall H. Biochemistry. 1997;36:34–40. doi: 10.1021/bi961803v. [DOI] [PubMed] [Google Scholar]
- 27.Fisher M, Kroon J T M, Martindale W, Stuitje A R, Slabas A R, Rafferty J B. Structure (London) 2000;8:339–347. doi: 10.1016/s0969-2126(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 28.Powell A J, Read J A, Banfield M J, Gunn-Moore F, Yan S D, Lustbader J, Stern A R, Stern D M, Brady R L. J Mol Biol. 2000;303:311–327. doi: 10.1006/jmbi.2000.4139. [DOI] [PubMed] [Google Scholar]
- 29.Grimm C, Maser E, Mobus E, Klebe G, Reuter K, Ficner R. J Biol Chem. 2000;275:41333–41339. doi: 10.1074/jbc.M007559200. [DOI] [PubMed] [Google Scholar]
- 30.Mazza C, Breton R, Housset D, Fontecilla-Camps J C. J Biol Chem. 1998;273:8145–8152. doi: 10.1074/jbc.273.14.8145. [DOI] [PubMed] [Google Scholar]
- 31.Georgiou G, Valax P. Curr Opin Biotechnol. 1996;7:190–197. doi: 10.1016/s0958-1669(96)80012-7. [DOI] [PubMed] [Google Scholar]
- 32.Barton G J. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 33.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 34.Esnouf R M. J Mol Graphics. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 35.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.