Abstract
The inhibitory interaction of phospholamban (PLN) with the sarco(endo)plasmic reticulum Ca2+ ATPase isoform 1 (SERCA1a) was modeled on the basis of several constraints which included (i) spontaneous formation of SS-bridges between mutants L321C in transmembrane helix 4 (M4) of SERCA1a and N27C in PLN and between V89C (M4) and V49C (PLN); (ii) definition of the face of the PLN transmembrane helix that interacts with SERCA; (iii) cross-linking between Lys-3 of PLN and Lys-397 and Lys-400 of SERCA2a. The crystal structure of SERCA1a in the absence of Ca2+, which binds PLN, was used as the structure into which an atomic model of PLN was built. PLN can fit into a transmembrane groove formed by the juxtaposition of M2, the upper part of M4, M6, and M9. In the SERCA1a structure with bound Ca2+, this groove is closed, accounting for the ability of Ca2+ to disrupt PLN–SERCA interactions. Near the cytoplasmic surface of the bilayer, the PLN helix is disrupted to prevent its collision with M4. The model can be extended into the cytoplasmic domain so that Lys-3 in PLN can be cross-linked with Lys-397 and Lys-400 in SERCA1a with little unwinding of the N-terminal helix of PLN.
Sarco(endo)plasmic reticulum Ca2+ ATPases (SERCAs) have proven to be exceptionally useful models for the investigation of structure-function relationships and the mechanism of action of P-type cation pumps (1–4). SERCA1a is also the first P-type ATPase for which high-resolution crystal structures have been presented in both E1 (5) and E2 (6) conformations. The high-resolution structures have not only defined the gross movements of the major actuator (A), nucleotide binding (N), phosphorylation (P), and Ca2+ binding domains, but have provided precise information on the relationship among individual amino acids and helices.
The activity of SERCA2a is regulated by phospholamban (PLN) (7, 8), a member of a family of low-molecular-mass transmembrane proteins that includes sarcolipin (SLN) (9–11). Because PLN has been demonstrated to be a major regulator of the kinetics of cardiac contractility through its effects on SERCA2a in the heart (12, 13), it is a potential drug target for management of cardiac function. Accordingly, it has been of great interest to determine the sites of interaction between PLN and SERCA2a or other SERCA isoforms.
James et al. (14) cross-linked PLN to SERCA2a by using a cross-linking agent (the Denny–Jaffe reagent) to show that Lys-3 of PLN lies within 15 Å of both Lys-397 and Lys-400 in the sequence Lys–Asp–Asp–Lys-400 in what is now recognized as the cytosolic N domain of SERCA2a. Studies using chimera formation and mutagenesis confirmed that the sequence, Lys–Asp–Asp–Lys–Pro–Val–402 is involved in PLN–SERCA2a interactions (15). Further studies then showed that a series of charged and hydrophobic amino acids between Glu-2 and Ile-18 in PLN were also mutation-sensitive in the assay used for measurement of Ca2+ affinity of SERCA2a (16).
The C-terminal region of PLN, involving either Leu-28–Leu-52 or Asn-30–Leu-52 fused to a variety of epitopes, was shown to retain inhibitory function (17). Mutagenesis of the transmembrane helix of PLN established that mutations of amino acids located on one face of the helix lost the ability to interact with SERCA2a (18). These amino acids were proposed to be the ones that normally interact with SERCA2a. Mutation of amino acids on the other face frequently disrupted formation of the PLN pentamer (18–20). Because the monomeric mutants were superinhibitory, it was deduced that the PLN monomer is the inhibitory species and that an increase in PLN monomer content could lead to superinhibition through mass action and, potentially, an increased affinity of mutant PLN for SERCA (18, 21). Indeed, it has become clear that residues forming a “leucine zipper” in the PLN pentameter may also interact, in the monomeric state, with SERCA residues (22). In an attempt to discover the sites in SERCA1a that would interact with the inhibitory face of PLN mutants, L321A in M4 and V795A, L802A(V), T805A, and F809A in M6 were shown to have diminished ability to interact functionally with PLN (23).
Further investigation of the PLN sequence, Met-20–Asn-30 (24), revealed that PLN mutants N27A, Q29A, and N30A were also superinhibitory, presumably because their dissociation from the inhibited PLN–SERCA2a complex was slowed. A further attempt to discover the site in SERCA1a that might interact with Asn-27 or Asn-30 led to mutagenesis of the loop between M6 and M7 (L67) (25). Mutagenesis of Asp-813 in SERCA1a led to a spectrum of results that were consistent with a site of interaction between Asp-813 and Asn-27 or Asn-30 in PLN. However, recent structural results (5) show that interaction at this site is unlikely to occur. The Asp813A mutation disrupts hydrogen bonding to Asn-755 in M5 and to Ser-917 in L89. The extensive rearrangements in the stalk region that result are likely to be an indirect, rather than a direct cause of loss of the ability of PLN to interact with SERCA1a. In recent experiments, cross-linking between the PLN mutant N30C and the natural Cys-318 in SERCA2a has been shown (26). In these experiments, the cross-linker, bis-maleimidohexane, was sufficiently long to bridge about 10 Å between Cys side chains.
In the present study, we have evaluated all of these observations in relation to the recent structure of SERCA1a in the E2 conformation (6), which binds PLN, and in the E1Ca2+ conformation (5), which does not bind PLN. We have noted that the movement of M2 in the conformational transition from E1 to E2 leads to the opening of a groove between M2 and M9 into which PLN can fit (6). In this groove, PLN lies along the exposed faces of M4 and M6 in SERCA1a. New cross links between Asn-27 in PLN and Leu-321 in SERCA1a, at the cytosolic boundary, and between Val-49 in PLN and Val-89 in SERCA1a, near the luminal boundary, define two well separated interaction sites. With these fixed sites, it was possible to deduce the remaining transmembrane interaction sites on the basis of the known structures of SERCA1a and PLN and the results of mutagenesis. The model shows that PLN interacts with M2, M4, M6, and M9 as it traverses the membrane and extends through the cytosol toward another well defined PLN binding site (Lys–Asp–Asp–Lys–Pro–Val-402) in the N domain of SERCA1a.
Materials and Methods
Materials.
Enzymes for DNA manipulation were obtained from New England Biolabs and Amersham Pharmacia. G-Sepharose and a chemiluminescence kit for measurement of coimmunoprecipitation were purchased from Pierce.
Oligonucleotide-Directed Mutagenesis and Expression.
Site-directed mutagenesis of SERCA1a and PLN was carried out as described (15, 16, 18, 23). The coexpression of various combinations of wild-type or mutant PLN and wild-type or mutant SERCA1a in HEK-293 cells and the preparation of microsome fractions were described (15, 16, 18, 23).
Site-Directed Cross-Linking of Amino Acids in PLN with Amino Acids in SERCA1a.
The introduction of Cys residues at specific sites in PLN and SERCA1a by PCR-based mutagenesis and disulfide cross-linking between Cys residues was carried out as described by Rice et al. (27). Microsomal fractions were isolated from HEK-293 cells coexpressing mutant PLN and mutant SERCA1a. After incubation with 1 mM glutathione for 5 min, 20 μl of the reduced samples in 0.15 M KCl/0.25 M sucrose/10 mM Tris⋅HCl, pH 7.5/20 μM CaCl2/5 mM MgCl2 were mixed with 20 μl of 140 mM KCl/5 mM MgCl2/5 mM EGTA/400 μM Na vanadate/25 mM Pipes, pH 7, 150 mM sucrose to induce the E2 conformation. Cross-linking was compared in the presence and in the absence of the oxidant copper phenanthroline (0.3 mM CuSO4 and 0.9 mM 1,10-phenanthroline). After a 10-min incubation at room temperature, an equal volume of 2× loading buffer containing 25 mM N-ethylmaleimide was added, and the samples were subjected to SDS/PAGE.
To determine the effect of elevated Ca2+ on the formation of cross-links, 10 mM CaCl2 replaced 5 mM EGTA and 400 μM Na vanadate in the reaction mixture, thereby inducing the E1Ca2+ conformation. Oxidant was included in these experiments.
To determine the effect of phosphorylation of PLN by protein kinase A (PKA) on the formation of cross-links, 16 μg of microsomal protein in 16 μl of 0.15 M KCl/0.25 M sucrose/10 mM Tris⋅HCl, pH 7.5/20 μM CaCl2/5 mM MgCl2 were phosphorylated by the addition of 2 μl of cAMP-dependent PKA catalytic subunit (Sigma) (3,000 units/ml) and 2 μl of 0.5 mM ATP to yield a final ATP concentration of 50 μM. After 5 min at 30°C, 20 μl of 0.15 M KCl/5 mM MgCl2/10 mM EGTA/800 μM Na vanadate/10 mM Tris⋅HCl, pH 7.5/0.25 M sucrose was added to the phosphorylated samples to drive the enzyme into the E2 conformation and allow cross-links to form in the absence of oxidant. After a 10-min incubation at room temperature, an equal volume of 2× loading buffer containing 25 mM N-ethylmaleimide was added, and the samples were subjected to SDS/PAGE.
Ca2+ Transport Activity.
Measurements of Ca2+ transport activity in microsomal fractions were carried out as described (15, 16, 18, 23).
Modeling.
Modeling was initiated with the solution structures of PLN, determined by NMR (28–30). For the initial model, 1FJK was used for the transmembrane segment, and 1PLP was used for the cytoplasmic segment. 1IWO was used for the SERCA1a model. Because thapsigargin (TG) in the absence of Ca2+ does not diminish physical interactions between PLN and SERCA (31), the use of 1IWO for modeling is justified. The model was constructed manually by using turbo-frodo (http://afmb.cnrs-mrs.fr) and refined by energy minimization and molecular dynamics using presto (32).
Results
Cross-Linking of PLN to SERCA1a.
Intramolecular cross-linking between Cys-substituted amino acids in M4 and M6 has been reported (27), but later, extensive attempts to create cross-links between Cys-substituted amino acids inside the transmembrane sectors of PLN and SERCA1a have been unsuccessful. Nevertheless, a cross-link between the PLN mutant N30C and endogenous Cys-318 in SERCA2a, which was disrupted by PLN phosphorylation and elevated Ca2+, has been reported (26).
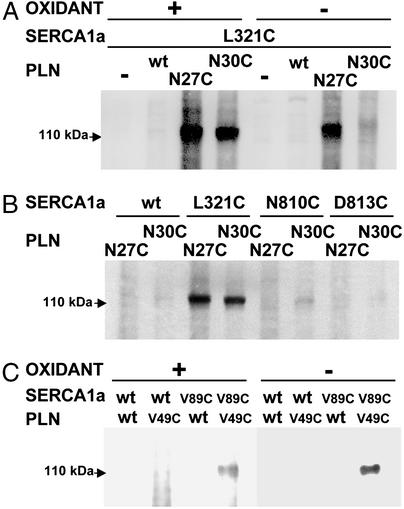
Mutation of Leu-321 to Ala diminishes PLN–SERCA interaction (23). Because this residue lies near the cytosolic boundary of M4, attempts were made to cross-link L321C to PLN mutants N27C and N30C. Cross-links were formed between L321C and N27C and between L321C and N30C in the presence of the oxidant, Cu-phenanthroline (Fig. 1A). The very strong cross-link between L321C and N27C also formed without oxidants, but the cross-link between L321C and N30C required oxidants (Fig. 1A). These results indicate that these residues are very close to each other, but that Asn-27 is closer than Asn-30 to Leu-321.
Figure 1.
Detection of disulfide cross-linked products. Microsomes prepared from transfected HEK-293 cells were reduced with 1 mM glutathione, then incubated for 10 min in the presence of 0.3 mM Cu(II) (1,10-phenanthroline)3, as described in Materials and Methods. Between 2 and 5 μg of total protein were loaded in each lane, and samples were separated by SDS/PAGE, transferred to nitrocellulose, and stained with antibody 1D11 against PLN to detect a band that represents a cross-link between PLN and SERCA1a with a mass of ≈115 kDa. (A) A strong cross-link forms between SERCA1a mutant L321C and PLN mutant N27C in the presence and in the absence of the oxidant, Cu-phenanthroline. A strong cross-link also forms between L321C and N30C, but requires oxidant. (B) A weak cross-link forms between SERCA1a mutant N810C and PLN mutant N30C in the presence of oxidant, but not between N810C and N27C. No cross-links are formed between SERCA1a mutant D813C and PLN mutant N27C or N30C. (C) A strong cross-link forms between SERCA1a mutant V89C and PLN mutant V49C in the presence and in the absence of the oxidant, Cu-phenanthroline.
Earlier studies indicated the possibility that amino acids in the L67 loop might interact with Asn-27 or Asn-30 in PLN (25). Data presented in Fig. 1B provide a hint of a very weak interaction of SERCA1a mutant N810C with PLN mutant N30C, but not with N27C. The weak interaction between the former two residues depended on the presence of oxidant (not shown). No cross-linking was observed between the SERCA1a mutant D813C and the PLN mutants N27C or N30C (Fig. 1B). On the basis of early modeling in this study, a cross-link between SERCA1a mutant V89C and PLN mutant V49C was predicted to occur near the luminal surface of these two interacting molecules. Data presented in Fig. 1C show that a strong cross link was formed between V49C in PLN and V89C in SERCA1a in the presence and in the absence of Cu-phenanthroline.
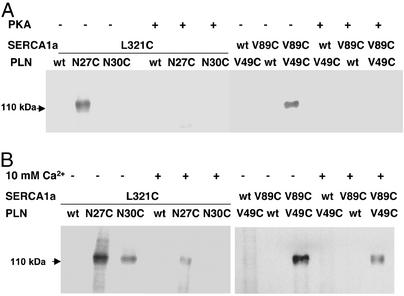
The cross-links that formed between SERCA1a mutant L321C and PLN mutants N27C or N30C and those that formed between SERCA1a mutant V89C and PLN mutant V49C were diminished by either phosphorylation of PLN with PKA (Fig. 2A) or by elevation of Ca2+ to 10 mM (Fig. 2B). All of the cross-links that were observed were susceptible to reduction by DTT (not shown).
Figure 2.
Prevention of the formation of disulfide cross-links by phosphorylation of PLN or the addition of 10 mM Ca2+. (A) Cross-linking was carried out in the absence of the oxidant, as described in the legend to Fig. 1, except that PLN in the microsomal fractions was phosphorylated by PKA before cross-linking was initiated. (B) Cross-linking was carried out in the presence of the oxidant, as described above, except that 10 mM CaCl2 was present in the reaction mix.
Constraints in the Construction of the Model for PLN-SERCA1a Interactions.
Modeling of the interaction between PLN and SERCA1a is subject to a number of constraints, as follows.
Constraint 1.
The formation of SS-bridges between N27C (without oxidants) or N30C (requiring Cu-phenanthroline) in PLN and L321C in SERCA1a fixes the position of the cytoplasmic end of the transmembrane segment of PLN with respect to M4.
Constraint 2.
The formation of SS-bridges between V49C near the C-terminal end of PLN and V89C near the luminal end of M2 in SERCA1a (without oxidants), fixes the position of PLN with respect to M2.
Constraint 3.
Alanine-scanning mutagenesis of PLN has defined the face of the transmembrane helix that interacts between PLN monomers to form a PLN pentamer (18–20) and the face that interacts with SERCA1a (18). Because mutation of amino acids Leu-31, Asn-34, Phe-35, Ile-38, and Leu-42 results in marked loss of PLN inhibitory function, these residues are expected to lie on a narrow face of the PLN transmembrane α-helix.
Constraint 4.
Lys-3 of PLN can be cross-linked to Lys-397 and Lys-400 of SERCA2a over a distance of ≈15 Å by using the Denny–Jaffe reagent (14). Alanine-scanning mutagenesis of domain Ia of PLN (Fig. 3) showed loss of functional interaction with SERCA2a for PLN mutants E2A, V4A, L7A, R9A, I12A and R14A (16). Among this group, only Glu-2 is a likely candidate for interaction with Lys-397 or Lys-400, whereas Lys-3 might interact electrostatically with Asp-398 or Asp-399. Because PLN domain Ia (Fig. 3) containing these amino acids has been shown to be helical in solution (28–30), Val-4, Leu-7, and Ile-12, are likely to lie on the same face of the domain Ia α-helix, where they might interact with SERCA.
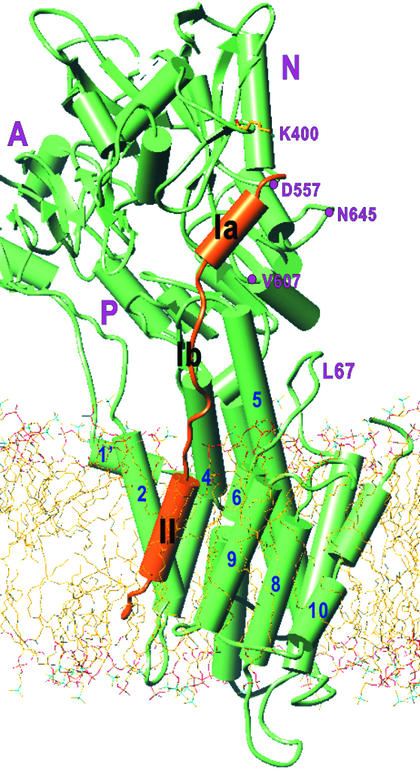
Figure 3.
Overall view of an atomic model of PLN bound to SERCA1a in a lipid bilayer. α-Helices are represented by cylinders and β-sheets by arrows. Orange, PLN; green, SERCA. The atomic model of PLN was constructed from two NMR structures (1FJK and 1PLP) and refined with molecular dynamics and energy minimization. The model of SERCA was derived from that for the E2 form of rabbit SERCA1a (1IWO); three cytoplasmic domains (A, N, and P) and 10 transmembrane helices (1–10) are marked. PLN domain Ia is α -helical and lies in a groove in the N domain (K400, D557, V607, and N645 are shown) of SERCA1a. Domain Ib is not helical. Domain II is the transmembrane α-helix, running roughly parallel to the M9 helix of SERCA in the groove formed by M2, M4, M6, and M9. The PLN–SERCA complex was embedded in a lipid bilayer generated by molecular dynamics simulation of dioleoyl phosphatidyl choline.
Constraint 5.
PLN can be phosphorylated by PKA at Ser-16 (33, 34). From the crystal structure of this kinase with an inhibitor peptide, the segment from about Arg-9 to Glu-19 would have to be unwound to be able to interact with PKA (35). Although this constraint implies the potential for unwinding of PLN domain Ia, it does not mandate the unwinding of this segment when PLN is bound to SERCA.
Modeling of the Transmembrane Domain.
Because the tightest constraints were those that define the helical segment between PLN Asn-27 and Val-49 and its interaction with M2, M4, M6, and M9 of SERCA in the E2 conformation, modeling was initiated for the transmembrane interaction (Figs. 4 and 5). Constraint 1 brings Asn-27 of PLN slightly above Leu-321 of SERCA1a. This is only possible if the C-terminal helix of the PLN solution structure is disrupted around Asn-30, such that the polypeptide is unwound in the N-terminal direction, because a continuous helix would collide with Arg-324 and Arg-325 on M4. Cys-318 lies within 10 Å of Asn-30 in PLN, as reported (26) but these residues are too far distant to form a significant interaction site directly; besides, Cys-318 points in the wrong direction (Fig. 5).
Figure 4.
Interactions between the transmembrane segment of PLN and SERCA1a. (A) Water-accessible surface of PLN and SERCA1a in the E2 form. Color represents lipophilicity (brown, hyrdophobic; sky blue, hydrophilic; green, neutral) as calculated with SYBYL6.8 (Tripos Associates, St. Louis). White dots outline PLN. (B and C) PLN in a ball-and-stick model in atom color. SERCA appears in green in E2 (B, PDB ID code 1IWO) and E1Ca2+ (C, PDB ID code 1EUL). Transmembrane helices of SERCA are numbered. Some of the residues on the M9 and PLN helices that make van der Waals contacts are also numbered, as well as T17, one of the two residues in PLN that are phosphorylated. Orange circles in A and B indicate the loop connecting M9 and M10 that pushes the C-terminal part of PLN toward M2. The red circle in C shows that the PLN helix collides with the M2 helix of SERCA in the E1Ca2+ form due to the large movement of M2 toward M9. As a result, PLN will be dissociated from SERCA by Ca2+ binding. Horizontal bars indicate the boundaries of the hydrophobic core of the lipid bilayer.
Figure 5.
Details of the cytoplasmic half of the transmembrane region in a view looking down the transmembrane helix of PLN. Only M4, M6, and M9 helices of SERCA (green) and PLN (yellow) are shown. Note that the transmembrane helices of SERCA do not run parallel to the PLN helix and that Leu-321 in SERCA appears to guide the path of PLN just above the membrane surface. A potential hydrogen bond between Asn-34 in PLN and Thr-805 in SERCA is indicated by a dashed line.
Unwinding of the PLN helix at Asn-30 would allow the face of the remaining C-terminal helix formed by Leu-31, Phe-35, Ile-38 and Leu-42 to be oriented toward the M9 helix of SERCA1a (Constraint 3; Fig. 5). These bulky side chains form a rough hydrophobic surface that is complementary to that formed by M9 (Figs. 4 A and B; Table 1). Asn-34 is of particular interest, because it is a polar residue that resides within a hydrophobic environment in the PLN transmembrane helix. In our model, Thr-805 on M6 is directly apposed to Asn-34 on the SERCA1a side (Fig. 5).
Table 1.
Interactions between PLN and SERCA1a in the proposed model
| PLN | SERCA1a
|
|||
|---|---|---|---|---|
| M2 | M4 | M6 | M9 | |
| M20 | K328 | |||
| Q23 | K328, K329 | |||
| A24 | R325 | |||
| Q26 | R324 | |||
| N27 | L321, R324, R325 | |||
| L28 | W932 | |||
| Q29 | R324, R324 | |||
| N30 | T317, A320, L321, R324 | |||
| L31 | T805, F809 | |||
| N34 | T317 | L802, T805 | ||
| F35 | L802 | L939 | ||
| L37 | L797 | |||
| I38 | V798, L802 | L943 | ||
| F41 | I97 | L797 | ||
| L42 | L943, L953 | |||
| L44 | L96 | |||
| I45 | V93 | W794 | I956, F957 | |
| C46 | P952 | |||
| I48 | L96 | |||
| V49 | V89 | |||
| L51 | I85 | |||
SERCA1a residues in the transmembrane sequences indicated are proposed to form hydrophobic (van der Waals) interactions with the PLN residue in the same row. SERCA1a residues in bold letters are proposed to form polar interactions with the PLN residue in the same row. SERCA1a residues in italics are those for which van der Waals contacts are likely to cause repulsion.
From this point, we propose that the PLN transmembrane helix runs parallel to the M9 helix (Fig. 3), bringing the C-terminal segment close to the luminal surface where M2 and M9 meet. The C-terminal segment is helical in the model derived from PLN in chloroform-methanol (1FJK), but, after energy minimization and molecular dynamics calculation, it is predicted to be unwound on interaction with SERCA1a. This is because the loop (orange circles in Fig. 4) connecting M9 and M10 is kinked toward M2, near the luminal surface around M9, and would collide with an extended helix. Unwinding of the C-terminal segment is also indicated through the cross-linking of the Cys mutants of Val-49 in PLN and Val-89 in M2 of SERCA1a (Fig. 1C and Constraint 2), as these residues can come close only in the unwound state. The carbonyl groups of the three residues at the C terminus, Met-50, Leu-51, and Leu-52, then emerge from the hydrophobic core of the membrane (Fig. 4).
Modeling of the Cytosolic Domains.
In extending the model from the C terminus of PLN domain Ib to the N terminus, another NMR structure, 1PLP, was used for guidance. The results of mutation indicate that the interaction between PLN domain Ia and SERCA1a is hydrophobic, suggesting that domain Ia lies in a groove. Because of the position of Lys-400, only one such groove can be identified (Fig. 3). This groove is formed by a β-sheet which is constricted by the loop containing KDDKDV402 on one side and Pro-603 and Asp-557 on the other side. The distance between the two Lys residues cross-linked by the Denny–Jaffe reagent contains an ambiguity of ≈15 Å, depending on the conformations of the two residues. However, the fact that mutation of Ile-12 to Ala led to loss of inhibitory function (Constraint 4) determined the position of the lower end of the domain Ia helix. Connecting the two α-helical segments, forming domains Ia and II, to domain Ib was straightforward, without much unwinding of the helices.
In the final model, the distance between the amine of Lys-3 in PLN and those of Lys-397 and Lys-400 in SERCA1a is 8–15 Å, depending on the side chain conformations, compatible with cross-linking through the Denny–Jaffe reagent. Lys-3 is able to form a salt bridge with Asp-399, satisfying Constraint 4. The phosphorylated residues, Ser-16 and Thr-17, in a segment of PLN that would not be associated with SERCA (Fig. 4), are certainly accessible to PKA. The model presented was energy minimized and optimized by molecular dynamics calculations, which unwound the helices further. These calculations must be considered rather preliminary, however, particularly for the cytosolic part, because the calculations are done with proteins in vacuo.
Discussion
Structural Aspects of PLN–SERCA1a Interactions in PLN Domain II.
The transmembrane segment of PLN is predicted to lie in a groove formed by transmembrane helices M2, M4, M6, and M9 in SERCA1a in the E2 conformation (Figs. 4 and 5). The dimensions of this groove are reduced during the transition of E2 (Fig. 4B) to the E1Ca2+ (Fig. 4C) conformation by the movement of M2 toward M9. In fact, the position of PLN in E2 is similar to that of M2 in E1Ca2+; in both cases, the PLN helix stabilizes surrounding transmembrane helices. The positions are not identical because the M2 helix does not unwind higher up and, therefore, “pushes” against M4, causing it to remain further away from M9 than does PLN. The C-terminal helix of PLN ends at Leu-31 (Fig. 4), at the interface between the cytosol and the lipid bilayer, consistent with our cross-linking results. The structure deviates here from the starting structure, 1FJK, which has a longer helix. The complete match of the lipophilicity profile of PLN and SERCA in these regions is demonstrated in Fig. 4A.
Fig. 5 shows a view looking down the transmembrane helix of PLN from the cytoplasmic side. The face formed by Phe-35, Leu-42, and Ile-45 orients toward M9. Leu-31, Asn-34, Ile-38, Cys-41, and Ile-45 form a face oriented toward M6. Interaction with M4 seems insignificant in the transmembrane region (Table 1). Residues located on the face of the PLN helix opposite to M4 (Phe-32, Ile-33, Leu-37, Ile-40, Leu-43, Leu-44, Ile-47, Met-50, Leu-51) are involved in pentamer formation and would, potentially, be free to interact with another molecule of PLN. However, M4 would interfere with pentamer formation by separating the face formed by Leu-37, Leu-44, and Leu-51 from the other residues that are involved in pentamer formation (Fig. 5). The residues involved in interaction with M2 are Ile-48, Val-49, and Leu-52 (Table 1); all of these residues impair PLN function when mutated to Ala (18), again in complete agreement with the model.
In our model, Leu-37 is predicted to interact with Leu-797 in M6 (Table 1). In mutagenic studies of “leucine zipper” residues (22), the inhibitory function of PLN was highly enhanced in the L371 mutant, even though this mutation does not induce monomer formation. This effect is readily explained by additional hydrophobic interactions with Ala-313 (M4) in our model. Mutation to Ala, which induced monomer formation, also enhanced inhibitory function, largely through mass action. Mutation to Phe, which also induced monomer formation, enhanced inhibitory function weakly, perhaps because of mass action combined with steric hindrance. Further interactions, consistent with mutagenic data (22), are predicted between Leu-44 and Leu-96 in M2 and between Leu-51 and Ile-85 in M2. However, no interactions with SERCA are predicted for Ile-40 or Ile-47 in our model.
Both positive and negative results obtained through mutagenesis of M6 are largely consistent with our model. The mutant L802A, which results in the complete loss of PLN function, interacts with Asn-34 Phe-35 and Ile-38 (Table 1). On the PLN side, mutations N34A, F35A, and I38A largely abolish PLN inhibitory function. Asn-34 also forms a hydrogen bond with Thr-805 (Fig. 5), consistent with the loss of PLN function that is associated with mutation of Thr-805. This hydrogen bond is unique in the transmembrane region and possibly determines the orientation of the PLN transmembrane helix. Leu-31 interacts with both Thr-805 and Phe-809, and individual mutations to all three of these residues disturb inhibitory function. It is of interest that Leu-802 and Thr-805 in M6, both interacting with Asn-34 in PLN, interact with Val-104 and Gln-108 in M2, respectively, in the PLN-free E1Ca2+ conformation, again suggesting a similarity between M2 and PLN.
Attempts to provide biochemical evidence for interactions of PLN with M9 have shown that the mutants L939A and S940A retained normal PLN function. The mutant L943A was not expressed. The P952C/wt, L953C/wt, M955C/wt, and I956C/wt mutants did not form cross-links. These negative data are consistent with our model.
Structural Aspects of Interactions in PLN Domain Ib.
The superinhibitory nature of the N27A and N30A mutants can be explained by their excessively tight contacts with residues on M4 (Table 1); this is particularly true for Asn-27. According to the initial energy-minimized model, the side chain of Asn-27 collides with the main chain of Arg-324 and Arg-325; this collision will be largely removed by mutation to Ala. This finding suggests that the affinity of PLN is delicately adjusted by nature. In humans, Asn-27 is replaced by Lys, but this substitution may enhance interaction, because the lysyl side chain is less bulky around Cγ.
Structural Aspects of Interactions in PLN Domain Ia.
The similarity between the sequences of PLN domain Ia and the inhibitory peptide of PKA (35) suggests that PLN bound to SERCA might take a similar conformation of the inhibitor bound to PKA. The sequence of the inhibitor is TTYADFIASGRTGRRNAIHD and the corresponding PLN N-terminal sequence is MEKVQYLTRSAIRRASTIE. From Met-1 to Glu-19, PLN would be 1 aa shorter than this inhibitor at the N terminus, with the C-terminal consensus sequence IRRASTIE being well conserved. Arg-11, Arg-14, and Arg-15 in the inhibitor, corresponding to Ser-10, Arg-13, and Arg-14 in PLN, appear to play important roles in defining the conformation of the inhibitor by forming salt bridges with Glu residues in PKA. When PLN binds to PKA, similar salt bridges would form. In our model of PLN bound to SERCA, these three residues in PLN do not form strong interactions with SERCA; instead they are exposed to solvent and are available for PKA binding. This lack of strong electrostatic interactions makes a similar conformation for PLN bound to SERCA and PLN bound to PKA unlikely. The PLN domain Ia helix deviates from the canonical α-helix conformation around Ser-10 and Ala-11 in 1FJK, suggesting that it might unwind easily.
Comparative Aspects of Thapsigargin and PLN Inhibition of SERCA.
TG is a very potent inhibitor of SERCA. With a dissociation constant in the picomolar range, TG can virtually titrate SERCA molecules (36). The dissociation constant of the PLN–SERCA interaction has not yet been measured. Even though inhibitory interaction is extensive, the concentration of the two molecules in the confined space of the sarcoplasmic reticulum membrane is very high and their affinity might be rather low (submillimolar). For both TG and PLN, the transmembrane interaction is almost completely hydrophobic with only one hydrogen bond being formed in either case. The shape of TG fits very well into a cavity formed by M3, M5, and M7 (6). For PLN, the binding cavity is shallow and the PLN molecule does not follow the shape of the cavity with high fidelity (Fig. 5). Large empty spaces are observed between PLN and M4/M2 because M2, M4, M6, and M9 helices do not run parallel to each other (Figs. 3 and 4). Moreover, the superinhibitory effect of PLN and SLN together (37), suggests that the cavity might be large enough to form sites for both PLN and SLN. Modeling of this potential tripartite structure supports this idea (M.A., K. Kurzydlowski, S. De Leon, M. Tada, C.T., and D.H.M., unpublished data). It appears that transmembrane contacts between PLN and SERCA are adjusted very precisely so that contacts are only mildly attractive; extensive tight contacts that might provide an excessively high affinity between PLN and SERCA are not formed. Mutation can increase PLN–SERCA affinity, as exemplified by the superinhibitory action of the domain Ib mutants, N27A and N30A (24).
Acknowledgments
This work was supported by grants from the Ministry of Education, Science, Sports, Culture and Technology of Japan (to C.T.) and the Heart and Stroke Foundation of Ontario (to D.H.M.), and by Canadian Institutes for Health Research Grant MT-12545 (to D.H.M.).
Abbreviations
- SERCA
sarco(endo)plasmic reticulum Ca2+-ATPase
- PLN
phospholamban
- SLN
sarcolipin
- PKA
protein kinase A
- TG
thapsigargin
Footnotes
This contribution is part of the special series of Inaugural articles by members of the National Academy of Sciences elected on May 1, 2001.
References
- 1.Moller J V, Juul B, le Maire M. Biochim Biophys Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 2.Mintz E, Guillain F. Biochim Biophys Acta. 1997;1318:52–70. doi: 10.1016/s0005-2728(96)00132-6. [DOI] [PubMed] [Google Scholar]
- 3.MacLennan D H, Rice W J, Green N M. J Biol Chem. 1997;272:28815–28818. doi: 10.1074/jbc.272.46.28815. [DOI] [PubMed] [Google Scholar]
- 4.McIntosh D B. Nat Struct Biol. 2000;7:532–535. doi: 10.1038/76735. [DOI] [PubMed] [Google Scholar]
- 5.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 6.Toyoshima C, Nomura H. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 7.Tada M, Kadoma M. BioEssays. 1989;10:157–163. doi: 10.1002/bies.950100505. [DOI] [PubMed] [Google Scholar]
- 8.Simmerman H K, Jones L R. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 9.Odermatt A, Taschner P E, Scherer S W, Beatty B, Khanna V K, Cornblath D R, Chaudhry V, Yee W C, Schrank B, Karpati G, et al. Genomics. 1997;45:541–553. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- 10.Odermatt A, Becker S, Khanna V K, Kurzydlowski K, Leisner E, Pette D, MacLennan D H. J Biol Chem. 1998;273:12360–12369. doi: 10.1074/jbc.273.20.12360. [DOI] [PubMed] [Google Scholar]
- 11.Hellstern S, Pegoraro S, Karim C B, Lustig A, Thomas D D, Moroder L, Engel J. J Biol Chem. 2001;276:30845–30852. doi: 10.1074/jbc.M102495200. [DOI] [PubMed] [Google Scholar]
- 12.Luo W, Grupp I L, Harrer J, Ponniah S, Grupp G, Duffy J J, Doetschman T, Kranias E G. Circ Res. 1994;75:401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- 13.Koss K L, Kranias E G. Circ Res. 1996;79:1059–1063. doi: 10.1161/01.res.79.6.1059. [DOI] [PubMed] [Google Scholar]
- 14.James P, Inui M, Tada M, Chiesi M, Carafoli E. Nature. 1989;342:90–92. doi: 10.1038/342090a0. [DOI] [PubMed] [Google Scholar]
- 15.Toyofuku T, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1994;269:22929–22932. [PubMed] [Google Scholar]
- 16.Toyofuku T, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1994;269:3088–3094. [PubMed] [Google Scholar]
- 17.Kimura Y, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1996;271:21726–21731. doi: 10.1074/jbc.271.36.21726. [DOI] [PubMed] [Google Scholar]
- 18.Kimura Y, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1997;272:15061–15064. doi: 10.1074/jbc.272.24.15061. [DOI] [PubMed] [Google Scholar]
- 19.Adams P D, Arkin I T, Engelman D M, Brunger A T. Nat Struct Biol. 1995;2:154–162. doi: 10.1038/nsb0295-154. [DOI] [PubMed] [Google Scholar]
- 20.Simmerman H K, Kobayashi Y M, Autry J M, Jones L R. J Biol Chem. 1996;271:5941–5946. doi: 10.1074/jbc.271.10.5941. [DOI] [PubMed] [Google Scholar]
- 21.Autry J M, Jones L R. J Biol Chem. 1997;272:15872–15880. doi: 10.1074/jbc.272.25.15872. [DOI] [PubMed] [Google Scholar]
- 22.Cornea R L, Autry J M, Chen Z, Jones L R. J Biol Chem. 2000;275:41487–41494. doi: 10.1074/jbc.M008195200. [DOI] [PubMed] [Google Scholar]
- 23.Asahi M, Kimura Y, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1999;274:32855–32862. doi: 10.1074/jbc.274.46.32855. [DOI] [PubMed] [Google Scholar]
- 24.Kimura Y, Asahi M, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 1998;273:14238–14241. doi: 10.1074/jbc.273.23.14238. [DOI] [PubMed] [Google Scholar]
- 25.Asahi M, Green N M, Kurzydlowski K, Tada M, MacLennan D H. Proc Natl Acad Sci USA. 2001;98:10061–10066. doi: 10.1073/pnas.181348298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones L R, Cornea R L, Chen Z. J Biol Chem. 2002;277:28319–28329. doi: 10.1074/jbc.M204085200. [DOI] [PubMed] [Google Scholar]
- 27.Rice W J, Green N M, MacLennan D H. J Biol Chem. 1997;272:31412–31419. doi: 10.1074/jbc.272.50.31412. [DOI] [PubMed] [Google Scholar]
- 28.Mortishire-Smith R J, Pitzenberger S M, Burke C J, Middaugh C R, Garsky V M, Johnson R G. Biochemistry. 1995;34:7603–7613. doi: 10.1021/bi00023a006. [DOI] [PubMed] [Google Scholar]
- 29.Pollesello P, Annila A, Ovaska M. Biophys J. 1999;76:1784–1795. doi: 10.1016/S0006-3495(99)77339-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamberth S, Schmid H, Muenchbach M, Vorherr T, Krebs J, Carafoli E, Griesinger C. Helv Chim Acta. 2000;83:2141–2152. [Google Scholar]
- 31.Asahi M, McKenna E, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 2000;275:15034–15038. doi: 10.1074/jbc.275.20.15034. [DOI] [PubMed] [Google Scholar]
- 32.Morikami K, Nakai T, Kidera A, Saito M, Nakamura H. Comput Chem. 1992;16:243–248. [Google Scholar]
- 33.Simmerman H K, Collins J H, Theibert J L, Wegener A D, Jones L R. J Biol Chem. 1986;261:13333–13341. [PubMed] [Google Scholar]
- 34.Fujii J, Ueno A, Kitano K, Tanaka S, Kadoma M, Tada M. J Clin Invest. 1987;79:301–304. doi: 10.1172/JCI112799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knighton D R, Zheng J H, Ten Eyck L F, Xuong N H, Taylor S S, Sowadski J M. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 36.Lytton J, Westlin M, Hanley M R. J Biol Chem. 1991;266:17067–17071. [PubMed] [Google Scholar]
- 37.Asahi M, Kurzydlowski K, Tada M, MacLennan D H. J Biol Chem. 2002;277:26725–26728. doi: 10.1074/jbc.C200269200. [DOI] [PubMed] [Google Scholar]