Abstract
Relaxation values reflecting residue-specific line broadening revealed amino acids in the DNA-binding domain of PU.1 on a surface potentially involved in protein–protein interactions. Mutation of these amino acids did not cause protein unfolding but destabilized PU.1–DNA binding. Addition of IFN response factor 4 to form the ternary complex recovered binding stability. Fluorescence quenching experiments proved that this surface of PU.1 interacts with IFN response factor 4 during binding. Our results provide evidence that residues that display increased conformational exchange can be used to predict areas of protein–protein interactions.
Transcription factors function at the most important regulatory level of gene expression. Most factors belong to families with conserved DNA-binding domains (DBD), consensus DNA-binding sequences, and similar structural motifs. Many of these protein–DNA interactions are well defined by crystallographic and NMR structures. The role of structural flexibility in protein–protein interactions between transcription factors and associated proteins is of great significance to understanding events in transcriptional regulation. Dynamic properties of proteins are basic features that control specificity of action of highly homologous family members with selected DNA-binding sequences. These properties also distinguish between protein partners as components of signal transduction pathways in cells.
PU.1 is a hematopoiesis-specific ets family transcription factor expressed in myeloid and B cells (1, 2). PU.1 binds to DNA and recruits a transactivation partner, IFN regulatory factor 4 (IRF4) (3), to an adjacent DNA sequence (4). Phosphorylation of S148 in the PEST (proline, glutamine, serine, and threonine rich) domain of PU.1 (2) is essential for strong binding of IRF4 in vitro and, ostensibly, an in vivo requirement for B cell maturation. However, regions of the PU.1 DBD may also interact with IRF4. A weak interaction was detected between PU.1 and IRF4 in the absence of S148 phosphorylation, but did not result in IRF4 binding to DNA (5). A two-step binding process was proposed for these interactions (6). Using fluorescence anisotropy (7) and quantitative hydroxyl radical footprinting (QHRF) (8), the presence of the PU.1 DBD bound to DNA resulted in optimal binding of IRF4 to the complex. Furthermore, weak binding of IRF4 DBD to DNA, in the absence of PU.1, was reported when the first nineteen N-terminal amino acids were deleted. This suggests a mechanism for recruitment of IRF4 by PU.1 whereby the inhibitory effect of the IRF4 DBD N-terminal domain is relieved by an interaction with the PU.1 DBD. The cooperative interaction of IRF4 DBD with PU.1 DBD/λB 3′ enhancer DNA binary complex also shows that these regions of the proteins interact in the absence of the PU.1 PEST domain and the IRF4 interaction domain (ID) (7). These results strongly suggest that interactions between PU.1 and IRF4 DBD have physiological consequences for transcription regulation.
The motional properties of proteins determine important aspects of biological specificity and can be extracted efficiently from NMR relaxation data (9–12). NMR relaxation studies of free PU.1 DBD at high and low concentrations provided residue-specific backbone dynamic information (13). These studies indicated a high degree of flexibility in the loop between helices α2 and α3 in the free state. Although the crystal structure of DNA-bound PU.1 DBD has been reported (14) and secondary protein structure elements in NMR studies of the free PU.1 DBD suggested a similar tertiary structure (13), the dynamics study provided new functional insights on this transcription factor. The importance of specific amino acids in the minimal IRF4 ID (residues 245–450) were also shown to be necessary for binding to PU.1 (15). Here, we report mutational and biophysical data confirming that conformational exchange processes identified from analysis of NMR relaxation data for the PU.1 DBD (13) reveal a previously unreported surface of the protein involved in stabilization of the PU.1/λB DNA binary complex and in its interaction with IRF4.
Materials and Methods
Relaxation Data.
The conditions and procedures for NMR spectroscopy and data analysis have been reported (13). The typical relaxation parameters, R1, the longitudinal relaxation rate constant, and R2, the transverse relaxation rate constant, were obtained in that study. Here, we calculate (R2/R1)high concentration:(R2/R1)low concentration ratios [(R2/R1)H:(R2/R1)L] (9, 16, 17) and R2R1 values (19) for PU.1 DBD by using relaxation constants obtained at 2.5- and 0.3-mM concentrations.
Protein Expression and Purification.
Gene constructs for IRF4 (Pip), ΔIRF4 (ΔPip), and IRF4-K94E (Pip-K94E) in pcDNA3 (Invitrogen) have been reported (15). PU(129–272) in pcDNA3 was prepared by standard PCR and cloning techniques, and constructs in pET (Novagen) and pGEX (Amersham Pharmacia) vectors were prepared by PCR using the pcDNA3 constructs as templates. Sequencing demonstrated fidelity. Amino acid mutations in PU.1 constructs were made using oligonucleotides (Genset, San Diego) and the QuikChange kit (Stratagene). The TNT Quick Coupled Transcription/Translation System (Promega) was used for in vitro protein expression with the pcDNA3 constructs, according to the manufacturer's protocol. The concentration of synthesized protein was estimated using parallel reactions incorporating [35S]methionine (ICN) measured by trichloroacetic acid precipitation and scintillation counting. BL21(DE3)pLysS competent bacteria (Active Motif, Carlsbad, CA) were transformed with PU(129–272) and mutant genes in pGEX-6P-1 for expression as GST fusion proteins. IRF4, ΔIRF4, ΔIRF4-K94E, and IRF4 DBD genes in pET28b were expressed as His-tag fusion proteins. Proteins were isolated by affinity chromatography on Glutathione-Sepharose 4B (Amersham Pharmacia) and Ni-NTA Agarose (Qiagen, Valencia, CA). GST fusion proteins were digested with PreScission protease and purified by FPLC on a Mono S column (Amersham Pharmacia).
Electrophoretic Mobility Shift Assay (EMSA).
EMSA was performed as reported (15). DNA oligonucleotides (Genset, San Diego) containing the λB PU.1- and IRF4-binding sites were as described (15) and labeled with [γ-32P]ATP (ICN) by using polynucleotide kinase (Roche Molecular Biochemicals). Dried gels were exposed to a phosphor screen and scanned on a PhosphorImager, and bands were quantitated using imagequant software (Molecular Dynamics).
Spectrofluorometry.
Monobromotrimethyl-ammoniobimane bromide (M-1380, Molecular Probes) was dissolved in water, quantitated spectrophotometrically, and reacted with PU(129–272)-M187C protein in 20 mM Hepes (pH 7.5). The conjugated protein was purified by FPLC and phosphorylated with Casein Kinase II (New England Biolabs). We calculated the percentage conjugation from molar concentrations of both dye and protein, corrected for absorption overlap at 280 nm. Protein concentrations were confirmed by triplicate Bio-Rad protein assays using a standard curve prepared with purified PU(129–272) protein assayed spectrophotometrically by using its extinction coefficient. Fluorescence measurements were made on a Shimadzu RF-1501 (Kyoto) spectrofluorometer in binding buffer [20 mM Hepes (pH 7.9)/75, 150, or 300 mM NaCl/0.5 mM EDTA/0.1% Tween 20]. Samples were scanned from 390–550 nm. The conjugated protein showed an emission (Em) maximum at 464 nm (published 472 ± 4 nm), when excited at the published absorbance maximum of 378 nm. Readings at the Em maximum were corrected for baseline differences between samples and for dilution effects (always <3%).
Circular Dichroism (CD) and Calorimetry.
Proteins in 10 mM phosphate buffer (pH 7.5) at 5 μM were scanned five times from 260–180 nm and a buffer blank was scanned three times by using an Aviv 62A DS spectrometer (Aviv Associates, Lakewood, NJ). The protein CD spectra were corrected for the buffer contribution. CD thermal denaturation was done from 25 to 90°C at 222 nm with proteins at 40 μM. Differential scanning calorimetry was performed on an N-DSC II calorimeter (Calorimetry Sciences, Provo, UT) at a scanning rate of 1 K/min under 3.0 atm (1 atm = 101.3 kPa) of pressure in 10 mM phosphate (pH 7.5) at 0.5 mg/ml.
Surface Plasmon Resonance (SPR).
Protein interaction kinetics were investigated by SPR as reported (18), except that the buffer contained 150 mM NaCl at a flow rate of 100 μl/min. A BIACORE 3000 (Biacore AB, Uppsala) was used with sensor chips SA for attachment of biotinylated λB DNA oligos. Data were evaluated using the BIAEVALUATION 3.0 program.
Results
Identification of Residues with Increased Conformational Exchange.
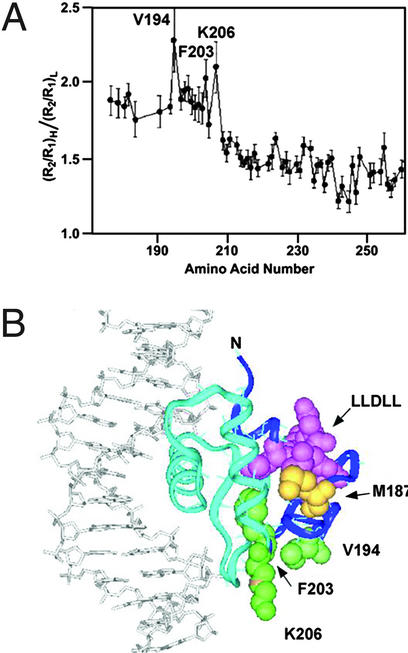
Protein interactions between transcription factors are likely to be determinants of specificity. It has been suggested that increased NMR relaxation values may predict amino acid residues involved in protein interactions (17). A concentration-dependent increase in the value of R2, the transverse relaxation rate constant, has been reported (13). In the present study, the PU.1 DBD residues most affected by high conformational-exchange contributions were extracted from plots of the (R2/R1)H:(R2/R1)L ratios for each amino acid (Fig. 1A) by using existing data collected at two concentrations (13). We observed notably higher (R2/R1)H:(R2/R1)L ratios (>1.7) for 29 residues clustered at the N terminus. The highest ratios (>2.0) existed for residues V194, F203, and K206, suggesting that they may be specific protein contact residues. We also calculated R2R1 values (data not shown) from those data to identify residues with faster chemical exchange contributions that are independent of rotational anisotropy effects (19). This data treatment also yielded higher R2R1 values for the 29 N-terminal amino acids.
Figure 1.
(A) Plot of residue-specific NMR relaxation constants ratios for PU.1 DBD suggests amino acids involved in protein–protein interactions. Data were obtained on 0.3- and 2.5-mM samples of PU.1 protein. (R2/R1)high concentration:(R2/R1)low concentration ratios were calculated for each amino acid and plotted. Residues with the largest ratios (>2.0) are labeled. (B) Ribbon representation of PU.1 DBD bound to DNA with residues of interest rendered in CPK format: dark blue, N-terminal 29 residues with highest Rex values; green, V194, F203, and K206; yellow, M187; pink, LLDLL hydrophobic motif.
We mapped the positions of the three amino acids with the highest (R2/R1)H:(R2/R1)L ratios on the PU.1 DBD–DNA binary complex crystal structure (14). The first 29 residues, including V194, F203, K206, and a hydrophobic LLDLL motif, are on the surface consisting of helix α1 and sheets β1 and β2. M187 is in the loop between α1 and β1. As shown (Fig. 1B), all of these elements project away from the DNA-binding region, and the V194 and K206 residues are solvent exposed. Although these amino acids have no direct role in the binding of the transcription factor to DNA (14), they may participate in protein interactions.
Mutational Studies.
PU.1 does not homodimerize to bind DNA (14). We hypothesized that, in vivo, the three amino acids in Fig. 1 with the highest (R2/R1)H:(R2/R1)L values interact with other transcription factors, including IRF4. To test the involvement of these three amino acids in ternary complex formation, conservative substitutions were made in full-length PU.1, changing V194 to alanine, glycine, or phenylalanine, F203 to valine or isoleucine, and K206 to serine, and combining of two or three of these mutations. Many of the mutations were also made in PU(129–272), a ternary complex-forming truncated protein with only the DBD and PEST domains that restricts the interaction possibilities with IRF4 (Fig. 2). Also, M187 was mutated to cysteine to attach a fluorescent probe to test for protein–protein interaction on this surface.
Figure 2.
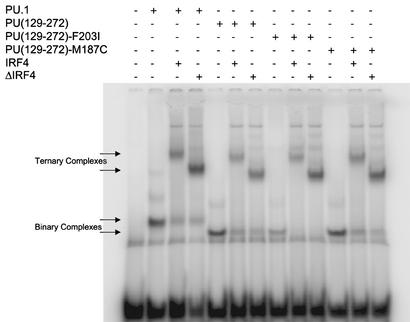
EMSA shows destabilization and restabilization of PU.1–DNA binding. Proteins were synthesized in vitro and mixed as indicated with λB enhancer sequence oligonucleotides containing the PU.1 and IRF4 binding sites labeled with 32P as probe. Binary and ternary complexes are indicated by arrows.
Proteins were produced in vitro, and the formation of a binary complex with λB DNA probe was shown by slower-migrating bands on native acrylamide gels (Fig. 2). For ternary complex formation, full-length IRF4(1–450), IRF4 DBD(1–149), or the minimal construct for ternary complex formation, IRF4(1–149/245–450) [ΔIRF4, previously called ΔPip (15)] was used. The supershifted bands arose from IRF4 binding to form ternary complex and were not due to dimerization of PU.1 bound to DNA, because the ternary complexes were supershifted with anti-hemagglutinin (HA) antibody binding to HA-tagged IRF4 proteins (ref. 15; see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org).
Fig. 2 shows the gel from a representative EMSA experiment of in vitro synthesized protein. Full-length PU.1 shifts the DNA probe (lane 2), and IRF4 and ΔIRF4 supershift according to their sizes (lanes 3 and 4). Likewise, PU(129–272) shifts the DNA probe (lane 5), the band moving faster than for the larger PU.1, and is supershifted by both IRF4 proteins (lanes 6 and 7). The F203I mutation of PU.1 resulted in significantly reduced binding to λB probe (lane 8), showing that binding stability was impaired, although F203 is not a contact residue in the crystal structure of PU.1 (ref. 14; Fig. 1B). Addition of IRF4 or ΔIRF4 protein stabilized the mutant binary complex and supershifted (lanes 9 and 10) nearly as much probe as wild type (lanes 6 and 7). Primary EMSA data for all constructs containing the F203I or F203V mutations showed the same effect of weakening the binary complex and the subsequent restabilization of the ternary complex on addition of IRF4 (see Figs. 5 and 6, which are published as supporting information on the PNAS web site). This stabilization was a specific protein effect, because addition of BSA did not produce the same result.
In agreement with published data (7), more DNA probe was shifted by the ternary complex than for the binary complex with wild-type PU.1 when IRF4 was not limiting. The destabilization of binary complex due to the F203I mutation was quantitated by calculating the stabilization on addition of IRF4 as ratios of probe shifted by ternary complex:binary complex for mutant versus wild-type protein. This calculation produced a value showing the fold increase in DNA probe shifted by the mutant ternary complex, relative to its binary complex, over the DNA probe shifted by the wild-type ternary complex relative to its binary complex. All data were from paired bands for wild-type and mutant PU.1 proteins with the same amount of IRF4 or ΔIRF4 added, with none of the pixels in the bands at the maximum value, and with not all of the probe shifted or all of the binary complex supershifted. These conditions guarantee that neither the probe nor the binary complex are limiting, that the band pixel values directly correlate with actual probe disintegrations per minute, and that the quantity of supershifting IRF protein is the same for both reactions. Using ratios minimizes the effect of minor differences in PU.1 protein concentrations between wild-type and mutant samples, and allows data from different experiments to be combined. These ratios (Table 1) show that on addition of ΔIRF4, the mutations K206S and V194A synergize with the F203I mutation to nearly double the binary complex destabilization effect from 6.7 for F203I alone to 13.3 for the triple mutant, although neither V197 nor S206 mutations alone or in combination with each other have as much influence on binary complex stability (see Fig. 6). The M187C mutation shows only a minor effect (Fig. 2, lanes 11–13, Table 1). Also, both IRF4 and ΔIRF4 are equivalent in their abilities to make ternary complex (Table 1; ref. 15). ΔIRF4 does not bind λDNA by itself (see Fig. 7, which is published as supporting information on the PNAS web site). To investigate the interactions between PU.1 and IRF4, we transferred gene constructs of PU(129–272) wild type, PU(129–272) F203I, and ΔIRF4 into bacterial expression vectors for production of larger quantities of proteins.
Table 1.
Mutation of residues with high Rex values causes destabilization of binary complex that is stabilized with the formation of ternary complex
| PU(129–272) mutation | IRF protein | n | Fold stabilization ±SD |
|---|---|---|---|
| F203I/K206S/V197A | IRF4 | 2 | 12.1 ± 3.7 |
| F203I/K206S/V197A | ΔIRF4 | 4 | 13.3 ± 1.9 |
| F203I | ΔIRF4 | 2 | 6.7 ± 2.3 |
| M187C | ΔIRF4 | 2 | 1.6 ± 0.3 |
EMSA band volumes were quantitated and the ratio between that of the binary and ternary complex was calculated. The ratio for mutant protein was divided by the ratio for wild-type protein to produce a value for the stabilization effect. n, number of replicates.
Conformational Integrity of Mutant Proteins.
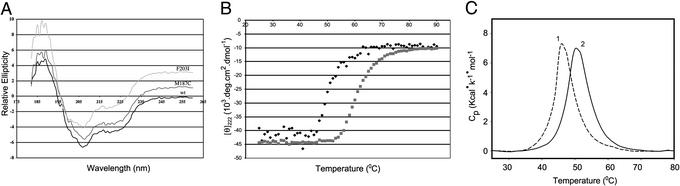
Mutations of the most prominent residues shown in Fig. 1A, V194, F203, and K206, were designed to minimize conformational changes in the PU.1 protein. Of the single mutations, F203I had the greatest destabilization effect on binary complex (Table 1; see Fig. 6). Yet by three different criteria, F203I is folded (Fig. 3). CD spectra show the wild-type and mutant proteins to be virtually identical, whereas repeated thermal denaturation and calorimetry experiments show F203I to be more stable than wild-type PU.1. The M187C mutation was critical for the fluorescence quenching experiments and showed a pattern of ellipticity very similar to the wild-type and F203I proteins (Fig. 3A).
Figure 3.
Stability of PU.1 proteins. (A) CD relative ellipticity. PU(129–272) wild type and mutants F203I and M187C proteins were scanned from 260 to 180 nm. Spectra corrected for buffer contribution are plotted at the same scale. The wild type is plotted on the true axes; the mutants are offset on the y axis for clarity. (B) CD thermal denaturation. Wild type (⧫) and F203I mutant ( ) at 40 μM have Tm = 51 and 61°C, respectively, monitored at 222 nm between 25 and 90°C. (C) Differential scanning calorimetry. Scans obtained for wild type (1) and F203I mutant (2). The assay solutions contained 0.5 mg/ml protein and 10 mM phosphate (pH 7.5). Analysis of the DSC data yielded Tm and ΔHcal values of 46.9 ± 0.9°C and 51.3 ± 9.5 kcal/mol for wild type, and 51.1 ± 0.8°C and 51.1 ± 5.9 kcal/mol for F203I mutant.
) at 40 μM have Tm = 51 and 61°C, respectively, monitored at 222 nm between 25 and 90°C. (C) Differential scanning calorimetry. Scans obtained for wild type (1) and F203I mutant (2). The assay solutions contained 0.5 mg/ml protein and 10 mM phosphate (pH 7.5). Analysis of the DSC data yielded Tm and ΔHcal values of 46.9 ± 0.9°C and 51.3 ± 9.5 kcal/mol for wild type, and 51.1 ± 0.8°C and 51.1 ± 5.9 kcal/mol for F203I mutant.
Determination of Dissociation Constants.
To ensure that fluorescence quenching experiments would be done at concentrations above the dissociation constant (Kd), binding was measured for binary and ternary complexes of λB double-stranded DNA, PU(129–272)-M187C, and ΔIRF4. EMSA (Fig. 2, lanes 5–7 and 11–13, Table 1) and CD (Fig. 3) comparisons reveal no significant differences between PU(129–272) wild type and the M187C mutant. These studies were carried out on a Biacore instrument using SPR. Oligos were biotinylated on the 5′ end of the coding strand and attached to streptavidin-coated flow cells. For binary complexes, triplicate experiments were run at concentrations of 15.6–500 nM protein. For ternary complexes, the concentration of PU(129–272)-M187C protein was kept constant at 100 nM and ΔIRF4 was injected at concentrations of 15.6–500 nM, also in triplicate. Sensogram curves did not describe simple kinetic interactions, so dissociation constants were obtained by plotting the equilibrium binding values to determine the half-maximal concentrations. The binary complex showed a Kd of 45 ± 2, representing the dissociation of PU(129–272) from the DNA, whereas the ternary complex had a Kd of 119 ± 11, representing the dissociation of ΔIRF4 from the binary complex (Table 2).
Table 2.
SPR determination of binary and ternary complex dissociation constants
| Protein | DNA | Method | KD × 109, M |
|---|---|---|---|
| PU(129–272) | λB 32-mer | SPR | 45 ± 2 |
| PU(106–272)-His | λB 100 bp | QHRF | 58* |
| PU(106–272)-His | λB 20-mer | FA | 160† |
| PU(161–272) | λB 100 bp | QHRF | 15* |
| PU(161–272) | λB 20-mer | FA | 692* |
| PU(160–272) | 16-mer | SPR | 170‡ |
| ΔIRF4 + PU(129–272) | λB 32-mer | SPR | 119 ± 11 |
| IRF4(20–137) − PU | λB 100 bp | QHRF | 559* |
| IRF4(20–137) + PU(161–272) | λB 100 bp | QHRF | 117* |
| IRF4(20–137) + PU(106–272) | λB 100 bp | QHRF | 190* |
| IRF4(20–137) − PU | λB 20-mer | FA | 910† |
Fluorescence Quenching.
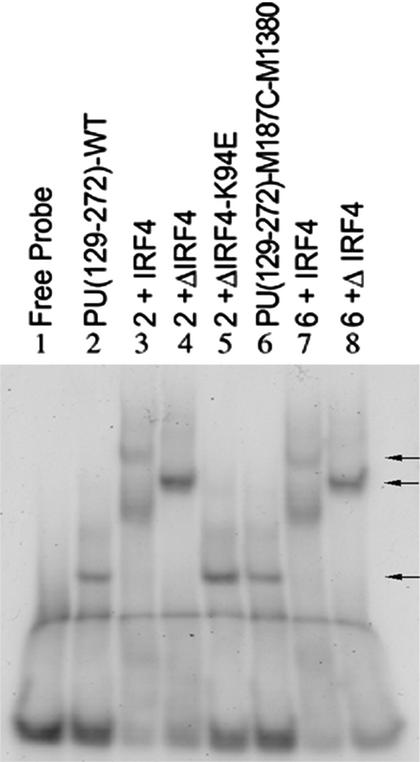
M187 (Fig. 1B, yellow) is located spatially close to V194, F203, and K206, and away from the DNA-binding elements, so this residue was mutated to cysteine to allow introduction of a unique fluorescent probe (M-1380). The probe was attached to PU(129–272)-M187C via a maleimide moiety. The conjugated protein still formed binary and ternary complexes (Fig. 4, lanes 6–8). Double-stranded λB oligos were brought to 1.0 μM in binding buffer and phosphorylated PU(129–272)-M187C-M1380 was added to 500 nM. Titrations were then carried out using concentrated solutions of ΔIRF4 or ΔIRF4-K94E. The mutant protein does not bind DNA (ref. 15; Fig. 4, lane 5), but has the same extinction coefficient as ΔIRF4, which allowed us to use titrations of the mutant protein for baseline values in the experiments. The ionic strength was varied to discourage nonspecific aggregation of PU.1. BSA and ovalbumin were similar to ΔIRF4-K94E in their effects (data not shown), proving that nonspecific protein interactions were unable to stabilize the ternary complexes. The same experiment using IRF4 DBD wild type and IRF4 DBD-K94E proteins produced no quenching under our conditions, showing that this effect is not due to interactions between the DBDs of PU.1 and IRF4 but must involve the IRF4 ID (data not shown). Table 3 shows that addition of ΔIRF4 quenched the fluorescence of the M-1380 molecule by 80% at 150 mM NaCl, relative to the baseline fluorescence of the binary complex, when compared with the fluorescence quenching of the control having the same concentration of ΔIRF4-K94E.
Figure 4.
Fluorophore-conjugated PU.1 forms binary and ternary complexes (indicated by arrows). Bacterially synthesized proteins were combined as shown with λB enhancer DNA probe.
Table 3.
Fluorescence quenching of the M-1380 fluor attached to the PU(129–272)-M187C residue at different concentrations of IRF4 proteins and salt
| [NaCl], nM | ΔIRF4 | K94E | Quench, % |
|---|---|---|---|
| 75 | 292 | 422 | 64.5 |
| 150 | 288 | 457 | 84.3 |
| 300 | 263 | 396 | 60.4 |
Values are arbitrary units of fluorescence at the emission maximum of 466 nm. Binary complex is present in all experiments at 0.5 μM and percent quench is based on its fluorescence. K94E, ΔIRF4-K94E.
Discussion
We previously reported (13) NMR relaxation parameters (R1, R2, and nuclear Overhauser effects) for 2.5- and 0.3-mM samples of PU.1 DBD. The main variation in the relaxation parameters obtained at high versus low concentrations was an increase in the decay of the transverse coherence rate, R2, at the higher concentration (13). R2 is highly sensitive to processes that involve conformational exchange. Therefore, the derived quantity, designated Rex, represents the excess contribution to the decay of transverse coherence when conformational exchange processes are present (9). In the N-terminal region of PU.1 DBD, 29 amino acids, including V194, F203, and K206, exhibit residue-specific chemical exchange contributions assumed to arise from transient oligomerization (13, 17). The data suggested that at 0.3 mM, PU.1 is primarily a monomer, whereas at 2.5 mM, PU.1 is likely multimeric (13). Homomultimerization was the sole logical explanation because internal conformational exchange within the monomer (11) would give equivalent Rex terms at high and low concentrations.
Calculation of (R2/R1)H:(R2/R1)L ratios (Fig. 1A) allows extraction of Rex contributions that arise specifically from conformational exchange as the concentration is increased. To distinguish residues subject to exchange broadening from effects due to rotational anisotropy, the diffusion tensors were calculated for each residue in the protein (9, 13, 20). R2R1 values for the 0.3-mM PU.1 DBD sample (data not shown) were also higher in the N-terminal region, confirming that the effects we observed were due to true Rex contributions, not effects due to anisotropy (19). Why does the N-terminal region of the DBD exhibit conformational exchange? Because PU.1 functions in a ternary complex with IRF4, not as a homodimer (14), we hypothesized that this flexible region of PU.1 DBD, prone to oligomerization, corresponded to the IRF4 binding site. We believe that conformationally flexible regions of proteins are likely to serve as binding sites; correspondingly, NMR relaxation is an ideal technique to identify such sites.
Helix α3 of PU.1 lies within the major groove of the DNA while the loop between sheets β3 and β4 and the loop between α2 and α3 interact with the phosphate backbone. V194 is in β1, F203 in β2, and K206 at the end of the short loop between β2 and α2; none of them are in structural elements known to be involved in DNA binding (14). In the published alignment of all known ets domain proteins, F203 is an insertion that is unique to PU.1 and SpiB, whereas K206 and V194 are only weakly conserved within the ets family (21). Fig. 1B shows that the amino acids with the highest Rex values project away from the DNA contact region. We have shown that mutations in these amino acids modulate ternary complex formation by PU.1, DNA, and IRF4. Helix α1 contains the sequence LLDLL (Fig. 1B), a hydrophobic motif essential for the binding of many transcriptional coactivators (22) and absolutely conserved in all ets domains (21). The presence of this motif on the PU.1 DBD surface exhibiting high Rex values suggests that it participates in PU.1 interactions with partner transcription factors, such as IRF4. In fact, the IRF4 ID has a conserved domain that contains a complementary hydrophobic motif between two helices with charged residues believed to participate in specific PU.1 recognition (15).
EMSA experiments with conservative mutations of V194, F203, and K206 show that all are important for stability of the binary complex of PU.1 with the λB enhancer sequence (Table 1, Fig. 2; see Fig. 6). Although F203I was the single mutation that most significantly destabilized the binary complex, combination with mutations in V194 and K206 caused a more severe effect (Table 1; see Fig. 6). These results are consistent with the location of the residues in the highly conserved minimal DBD of PU.1 (2), although the residues are unique or not highly conserved.
We observed that the addition of IRF4 or ΔIRF4 in EMSA experiments increased the amount of ternary complex formed with all PU.1 constructs to levels nearly equivalent to complexes formed with wild-type PU.1. The most dramatic effects were found for the mutant combinations (Table 1; and see Fig. 6), indicating that the interaction of IRF4 stabilizes the weakened DNA binding of the PU.1 mutant proteins. BSA added to EMSA binding reactions in place of IRF4 proteins had no effect (data not shown), confirming that nonspecific protein interactions cannot stabilize binary complexes with mutant PU.1 proteins. Also, CD spectral scans and thermal denaturation and calorimetry show that the F203I protein is folded (Fig. 3), indicating that the mutation did not cause denaturation that could alter binding to DNA. Although the exact causes of destabilization of the binary complex are not presently understood, in all of the cases, IRF4 addition resulted in stabilization of ternary complex, indicating that the surface of PU.1 containing the mutated residues interacts with IRF4.
To probe directly for the interaction between the PU.1 surface that contains F203, we placed a fluorescent molecule on this surface to test for quenching during binding. The conservative mutation of PU(129–272)-M187 to cysteine provided a unique fluorophore attachment site (Fig. 1B) in the middle of the putative ID. This mutation to cysteine does not alter the protein structure (Fig. 3) or the ability to form binary complex (Figs. 2 and 4, Table 1). These experiments required the use of DNA and protein concentrations well above the Kd values to ensure that the binding kinetics were not limiting. SPR was used to obtain dissociation constants for the binary and ternary complexes bound to λB double-stranded DNA (Table 2). The binding kinetics were complex, so dissociation constants were extracted by plotting equilibrium distributions for replicate determinations over a range of protein concentrations. To assess any contribution of protein–protein interactions not dependent on DNA binding, SPR experiments were run with ΔIRF4-GST fusion protein attached to flow cells by means of an anti-GST antibody and phosphorylated PU(129–272)-M187C. No interaction was detected in the absence of DNA (data not shown).
A survey of the literature indicates that experimentally determined Kd values for binary and ternary complexes vary greatly, depending on the method of measurement, the exact PU.1 and IRF4 constructs, and, perhaps, the length of the λB-containing DNA (Table 2 and ref. 7). Although we have not determined specific reasons for differences between our values and those of others, our 45 nM value agrees well with the 58 nM value determined by QHRF. Fluorescence anisotropy suggested weaker binding in general, which may be due to the shorter length of the λB oligos used in that study (7). Likewise, the use of a short, optimized 16-mer DNA fragment in our previous study (18) yielded a Kd of 170 nM for PU(160–272), higher than the value determined in the present study for PU(129–272). Table 2 shows that our ΔIRF4 binds to the PU(129–272)–DNA binary complex with the same affinity (117 nM vs. 119 nM) as the minimal DBD of IRF4 (residues 20–137) with the shorter PU(161–272) determined by QHRF (8), even though PU(161–272) lacks the PEST domain containing S148 needed for IRF4 recruitment.
Although comparisons of the specific Kd values in the various experiments are difficult, all of these data indicate that there is considerable interaction between the DBDs of PU.1 and IRF4, but that stabilization of the full-length protein ternary complex requires further interaction between other domains of the two proteins. This is consistent with the two-step interaction process between PU.1 and IRF4 proposed by others (6). It is also in agreement with the effect of specific mutations we made in the ID of IRF4 (15) and the binding differences we now report from EMSA experiments (Fig. 2). For the present study, these data indicated that we could run the fluorescence quenching experiments with PU.1 at a concentration of 500 nM, ≈10-fold higher than the Kd, resulting in nearly all of the PU.1 protein being complexed with DNA.
PU(129–272) derivatized with the fluorophore M-1380 formed ternary complexes in EMSA experiments (Fig. 2). Increasing salt concentrations were used to discourage nonspecific PU.1/PU.1 interactions that might mask the fluorescent probe and sequester it from interactions with IRF4. The quenching of the fluorescent signal of M-1380 attached to M187C provided conclusive proof that IRF4 interacts with that region of PU.1 on the same surface containing the amino acids with the highest Rex values. We interpret the finding that, at equivalent concentrations of ΔIRF4, the maximal quenching effect occurred at 150 mM NaCl to mean that PU.1/PU.1 interactions interfered with fluorescence at 75 mM NaCl, whereas at 300 mM NaCl binary and ternary complex formation was being adversely affected by the high ionic strength. The lack of fluorescence quenching with only IRF4 DBD shows that the IRF4 ID is needed for the quenching effect seen.
In conclusion, the data in the present study show that the measurement of relaxation values can be a useful probe to suggest regions of interaction between proteins, although the general applicability of this method remains to be determined. Biophysical and biochemical methods demonstrated that three amino acids with the highest Rex values (13) are located in a region of PU.1 that interacts with IRF4. These results are consistent with recent reports proposing Rex as a measurement of conformational changes in biologically functional time scales (μs–ms; reviewed in refs. 9, 10, and 23). Pfuhl et al. (17) demonstrated that only residues in the known dimer interface of the CD2 crystals showed increased Rex values as a function of concentration in the solution NMR studies. Volkman et al. (11) postulated that the coexistence of two conformations of nitrogen regulatory protein C (NtrC) led to increased Rex values, even before phosphorylation of the aspartate residue needed to generate the active conformation. Our present work shows that Rex values measured in PU.1 DBD identified a new region of interaction with its partner transcription factor, IRF4, further extending the scope of this emerging use of Rex as a biophysical diagnostic probe of molecular interactions.
Supplementary Material
Acknowledgments
We thank Art Palmer and Cathy Royer for helpful discussions on biophysical approaches, George Fortes for initial fluorescence experimental design and implementation, and Dan Sem (Triad Therapeutics) for access to instrumentation. This work was supported by National Institutes of Health Grant 5 R01 GM 56354-03 (to N.A.-M.).
Abbreviations
- CD
circular dichroism
- DBD
DNA-binding domain
- EMSA
electrophoretic mobility-shift assay
- ID
interaction domain
- IRF
IFN regulatory factor
- PEST
proline, glutamine, serine, and threonine rich
- QHRF
quantitative hydroxyl radical footprinting
- SPR
surface plasmon resonance
References
- 1.Karim F D, Urness L D, Thummel C S, Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A, Gunther C V, Nye J A, et al. Genes Dev. 1990;4:1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- 2.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 3.Mittrucker H W, Matsuyama T, Grossman A, Kundig T M, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi P S, Mak T W. Science. 1997;275:540–543. [PubMed] [Google Scholar]
- 4.Pongubala J M, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pongubala J M, Van Beveren C, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 6.Perkel J M, Atchison M L. J Immunol. 1998;160:241–252. [PubMed] [Google Scholar]
- 7.Yee A A, Yin P, Siderovski D P, Mak T W, Litchfield D W, Arrowsmith C H. J Mol Biol. 1998;279:1075–1083. doi: 10.1006/jmbi.1998.1838. [DOI] [PubMed] [Google Scholar]
- 8.Gross P, Yee A A, Arrowsmith C H, Macgregor R B., Jr Biochemistry. 1998;37:9802–9811. doi: 10.1021/bi9731448. [DOI] [PubMed] [Google Scholar]
- 9.Palmer A G, III, Kroenke C D, Loria J P. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 10.Wand A J. Nat Struct Biol. 2001;8:926–931. doi: 10.1038/nsb1101-926. [DOI] [PubMed] [Google Scholar]
- 11.Volkman B F, Lipson D, Wemmer D E, Kern D. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 12.Evenas J, Malmendal A, Akke M. Structure (Cambridge, UK) 2001;9:185–195. doi: 10.1016/s0969-2126(01)00575-5. [DOI] [PubMed] [Google Scholar]
- 13.Jia X, Lee L K, Light J, Palmer A G, III, Assa-Munt N. J Mol Biol. 1999;292:1083–1093. doi: 10.1006/jmbi.1999.3123. [DOI] [PubMed] [Google Scholar]
- 14.Kodandapani R, Pio F, Ni C Z, Piccialli G, Klemsz M, McKercher S, Maki R A, Ely K R. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 15.Ortiz M A, Light J, Maki R A, Assa-Munt N. Proc Natl Acad Sci USA. 1999;96:2740–2745. doi: 10.1073/pnas.96.6.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kay L E, Torchia D A, Bax A. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 17.Pfuhl M, Chen H A, Kristensen S M, Driscoll P C. J Biomol NMR. 1999;14:307–320. doi: 10.1023/a:1008319917267. [DOI] [PubMed] [Google Scholar]
- 18.Pio F, Assa-Munt N, Yguerabide J, Maki R A. Protein Sci. 1999;8:2098–2109. doi: 10.1110/ps.8.10.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kneller J M, Lu M, Bracken C. J Am Chem Soc. 2002;124:1852–1853. doi: 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- 20.Lee L K, Rance M, Chazin W J, Palmer A G., III J Biomol NMR. 1997;9:287–298. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]
- 21.Laudet V, Hanni C, Stehelin D, Duterque-Coquillaud M. Oncogene. 1999;18:1351–1359. doi: 10.1038/sj.onc.1202444. [DOI] [PubMed] [Google Scholar]
- 22.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, et al. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishima R, Torchia D A. Nat Struct Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.