Abstract
We report experiments with Xenopus laevis, using both intact embryos and ectodermal explants, showing that the transcription factor AP2α is positively regulated by bone morphogenetic protein (BMP) and Wnt signaling, and that this activation is an essential step in the induction of neural crest (NC). Ectopic expression of AP2α is sufficient to activate high-level expression of NC-specific genes such as Slug and Sox9, which can occur as isolated domains within the neural plate as well as by expansion of endogenous NC territories. AP2α also has the property of inducing NC in isolated ectoderm in which Wnt signaling is provided but BMP signaling is minimized by overexpression of chordin. Like other NC regulatory factors, activation of AP2α requires some attenuation of endogenous BMP signaling; however, this process occurs at a lower threshold for AP2α. Furthermore, AP2α expression domains are larger than for other NC factors. Loss-of-function experiments with antisense AP2α morpholino oligonucleotides result in severe reduction in the NC territory. These results support a central role for AP2α in NC induction. We propose a model in which AP2α expression, along with inactivation of NC inhibitory factors such as Dlx3, establish a feedback loop comprising AP2α, Sox9, and Slug, leading to and maintaining NC specification.
Keywords: bone morphogenetic protein‖Wnt‖Sox9‖Slug‖morpholino antisense oligonucleotides
Neural crest (NC) cells are a distinguishing feature of vertebrate embryos. They arise from the boundary between the dorsal/neural plate and ventral/epidermal territories as the result of inductive interactions, both between these tissues and with other embryonic cell types (1, 2). Evidence obtained primarily from Xenopus strongly suggests that two independent signals are necessary for NC induction: a bone morphogenetic protein (BMP) signal, which must be partially modulated (3–5), and a separate input that can be either a canonical Wnt signal (6–9), retinoic acid (10), or fibroblast growth factor signal (11). During neurulation, NC cells undergo a classical epithelial-mesenchymal transition and commence an elaborate migration throughout the body, following prescribed routes, and subsequently differentiate into a host of terminal cell types including craniofacial bones and cartilage, elements of the peripheral nervous system, and melanocytes (12, 13).
The molecular basis for NC specification is still incompletely understood. A diverse assortment of regulatory factors has been implicated in the early steps in this process. Among these are zinc finger factors Snail (14), Slug (15, 16), Zic3 and Zic5 (17, 18), the helix–loop–helix factor Twist (19), the winged helix factor FoxD3 (20, 21), the paired class homeodomain Pax3 (22), the high mobility group factor Sox9 (23), and the translational initiation factor eIF4AII (24). To varying degrees these factors are either capable of inducing NC marker gene expression when expressed at ectopic locations or are required for NC as shown by gene targeting or other loss-of-function studies.
The transcription factor AP2 comprises a family with four members in the human genome, with functional and developmental expression data available on the AP2α (25, 26), AP2β (27), and AP2γ (28) genes. The fourth member, AP2δ, has only recently been described (29). Homologs of AP2α have been reported to be expressed in the early stages of NC development of mouse, fish, and frog, as well as in other tissues including epidermis (26, 30–33). In this article we have adopted the name AP2α to denote the single AP2 isoform that has been described to date in Xenopus, which is most closely homologous to mammalian AP2α and thus probably the homolog of this gene (34). In the mouse, gene targeting of AP2α resulted in a perinatal lethal phenotype characterized by absence or severe reduction of cranial bones of NC origin, delayed neural fold elevation, extensive failure of neural tube closure, failure of ventral body wall closure, lateral displacement of facial primordia, as well as limb and other defects (35, 36). These results implicated AP2α in NC development, at least in the rostral compartment. However, some NC-derived cranial structures persisted, including the hyoid bone, Meckel's cartilage, and mandibular tissue. In addition, the NC marker genes Twist and Pax3 continued to be expressed in homozygous knockout mice (35), making it difficult to assign a specific role for AP2α.
To further elucidate this potential role for AP2α, we have turned to Xenopus laevis embryos, which are ideally suited for gene regulatory and cell signaling studies at early gastrula and neurula stages. We present the results of both gain-of-function and loss-of-function experiments in X. laevis, supporting an essential role for AP2α gene function at the earliest stage of NC development. Furthermore, key differences between AP2α and other NC regulators reveal interesting features of the response to BMP and Wnt signaling in this process.
Materials and Methods
Embryo Manipulation.
Embryos were obtained from adult X. laevis by hormone-induced egg laying and artificial fertilization with standard methods and staged according to Nieuwkoop and Faber (37). For ectodermal explants, full-length capped transcripts encoding AP2α (31), chordin (Chd) (38), or Xwnt-3a (39) were injected into two sites in the animal hemisphere of the one-cell embryos. After injection, the embryos were cultured in 4% Ficoll/1× modified Ringer's solution (MR) for 90 min and then transferred to 0.3× MR. Ectodermal explants were removed at stages 7 and 8 and cultured until stages 14 and 15 then processed for Northern blot analysis. For whole-embryo experiments, embryos were injected into one cell at the two-cell stage or into a single animal hemisphere cell at the 8- to 16-cell stage with either plasmid DNA (to avoid axis duplication) encoding Xwnt-1 or Xwnt-3a, or RNA encoding GSKβ, AP2α or morpholino oligonucleotides (MOs) (GeneTools, Philomath, OR), mixed with β-galactosidase RNA as a lineage tracer. The AP2α MO sequence was 5′-GGC GAT CCT GCC ATT CCC CCA TTT T-3′ (translational start codon underlined), and the control, which was mismatched at four positions (lowercase), was 5′-GGC cAT CgT GCC ATT CCC gCA TAT T-3′. Embryos were fixed for in situ hybridization at mid neural plate stage in 4% paraformaldehyde and stained for β-galactosidase activity by using 5-bromo-4-chloro-3-indolyl-β-d-galactoside (GIBCO/BRL) or 6-chloro-3-indolyl-β-d-galactoside (Research Organics). Coupled in vitro transcription/translation reactions with AP2α plasmid DNA were carried out according to the manufacturer's instructions (Promega).
Northern Blot Hybridization.
RNAs were isolated and analyzed by using denaturing methylmercury hydroxide RNA gels as described (40). Probes for expression of AP2α (31), Slug (16), Sox9 (23), Xtwi (19), Sox2 (41), Otx2 (42), Dlx3 (43), and Dlx5 (44) were labeled with 32P-dCTP by primer extension (Amersham Pharmacia). Staining with 18S ethidium bromide was used to monitor equal loading of embryonic RNA samples.
In Situ Hybridization.
Whole-mount in situ hybridization was carried out according to Harland (45), with some modifications (44). Antisense probes labeled with digoxigenin or fluorescein were synthesized by using a Roche Molecular Biochemicals RNA labeling kit with cDNA templates encoding AP2α, Slug, Sox9, and Sox2. For histology, stained embryos were embedded into hydroxyethyl methacrylate, and 12-μm sections were cut on a rotary microtome and briefly counterstained with eosin.
Results
Regulation of AP2α Expression in NC by Wnt Signaling.
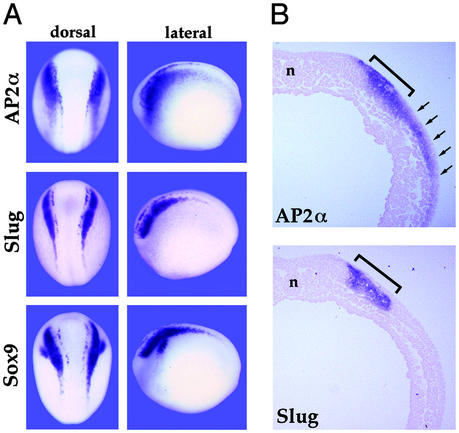
At open neural plate stages, Xenopus AP2α transcripts are highly abundant in premigratory NC, particularly in the cranial region, but are also evident in prospective trunk NC cells. Interestingly, the rostral expression domain extends over a larger area than that of other NC regulatory factors, i.e., Slug and Sox9 (Fig. 1A). This pattern is evident in cross sections comparing AP2α and Slug expression (Fig. 1B). AP2α RNA is present at lower but significant levels in the epidermis, expression that also depends on BMP signaling (34). AP2α RNA has not been detected in any mesodermal or endodermal tissues through stage 20 (data not shown).
Figure 1.
Comparison of neurula stage expression of AP2α and other NC marker genes. (A) Whole-mount in situ hybridization with probes for AP2α, Slug, and Sox9 are shown in dorsal and lateral views. AP2α transcripts appear to extend more laterally than for the other two genes. This is shown more clearly in cross section in B, where the bracket indicates the Slug domain and the arrows point to the more extensive lateral AP2α. Note that under the conditions used here the epidermal expression of AP2α, which is considerably lower than for NC, is not apparent. The notochord is indicated by n. (Magnifications: A, ×7; B, ×42.)
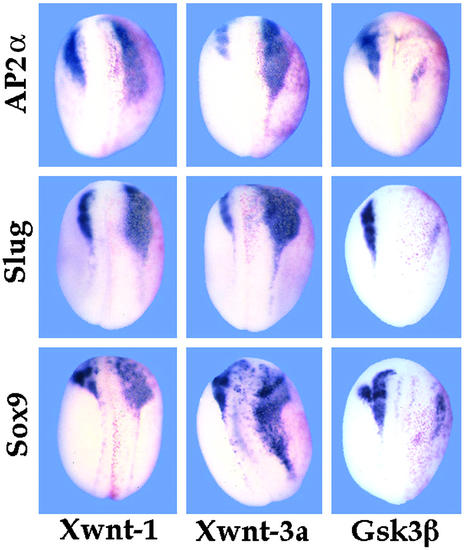
Several lines of evidence suggest that induction of NC in vertebrate development depends on multiple signals, including BMPs and Wnt-class factors (5–9, 46–48). To test whether the NC-specific expression of AP2α conformed to this pattern with respect to Wnt signaling, embryos were injected into one blastomere at the two-cell stage with plasmid DNA encoding Xwnt-1 (49) or Xwnt-3A (39). DNA was used instead of RNA to circumvent axis duplication (7). As shown in Fig. 2, these treatments resulted in significant lateral expansion of the cranial NC territory as indicated by hybridization with NC markers such as Sox9 (23) or Slug (16). Wnt signaling can be inhibited at the intracellular level by overexpression of GSK3β (50), which results in reduction or elimination of NC marker gene expression (Fig. 2). In both types of experiment, AP2α behaved in a manner similar to that of Sox9 and Slug, indicating that like these other genes, AP2α expression in NC is under the positive control of Wnt signaling.
Figure 2.
NC expression of AP2α depends on Wnt signaling. Whole-mount in situ hybridizations with NC markers Sox9, Slug, and AP2α to embryos injected into one cell at the two-cell stage with 100 pg of plasmid DNA encoding Xenopus Xwnt-1 (Left) and Xwnt-3A (Center), or 1 ng RNA encoding GSK3β (Right), along with a β-galactosidase lineage tracer (red staining). In all cases the injected side is oriented to the right (red staining). Both Wnt treatments resulted in lateral and posterior expansion of NC territory, including expression of AP2α as well as the other NC markers. The reciprocal treatment with GSK3β, which interferes with downstream Wnt signaling (50), had the opposite effect: AP2α, Sox9 and Slug all were repressed. (Magnification: ×15.)
AP2α Is Required for NC Induction.
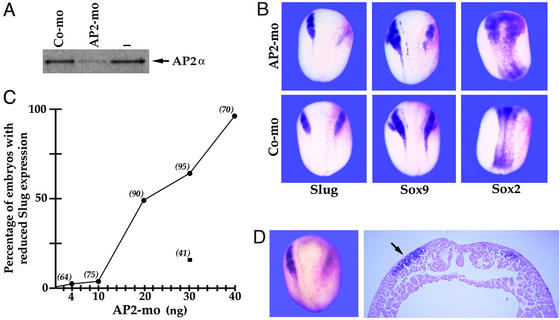
Transgenic mice lacking AP2α gene function have major craniofacial deficiencies, suggesting a function for this factor in the formation and/or subsequent development of the cranial NC cells that give rise to facial bones and cartilage (35, 36). To evaluate the importance of AP2α in the initial induction of NC, we used antisense MOs (51) to interfere with AP2α translation in Xenopus embryos. As shown in Fig. 3A, the AP2α MO strongly repressed in vitro translation of synthetic AP2α mRNA, whereas a control MO incorporating four mismatches (of 25; see Materials and Methods) had much less effect. When injected into one of the two blastomeres of a two-cell embryo, the AP2α MO, but not the control MO, strongly inhibited expression of NC markers, such as Slug and Sox9 (Fig. 3B). This effect may be accompanied by some lateral expansion of neural plate, as indicated by Sox2 expression (Fig. 3B Upper Right), although this is difficult to evaluate because of morphological effects on neural fold elevation noted below. The inhibitory effect of the AP2α MO was dose dependent, as shown in Fig. 3C, reaching nearly 100% at 40 ng injected. The control MO was not tested at all concentrations, but at 30 ng had little effect. The reduction in NC gene expression was also accompanied by a suppression of neural fold elevation on the injected side (Fig. 3D), an effect that has also been observed with loss-of-function experiments targeting Sox9 (23). We have obtained essentially identical results by using overexpression of dominant negative AP2α derivatives lacking an activation domain or fused to the repressor domain of the Drosophila engrailed factor (data not shown).
Figure 3.
Loss of function by MO antisense injections. (A) In vitro-coupled transcription/translation reactions with plasmid encoding the AP2α ORF in the presence of either control (Co-mo) or perfect-match AP2α MO (AP2-mo; 50 ng; see Materials and Methods for sequences). (B) Neurula stage embryos injected into one cell at the two-cell stage with 30 ng of AP2α MO or the control MO along with β-galactosidase mRNA as a lineage tracer and probed for expression of NC markers Slug and Sox9 and the neural plate marker Sox2. The injected sides are on the right in these dorsal views (reddish β-galactosidase staining). The AP2α MO reduces NC and slightly expands neural plate laterally. (C) AP2α-MO inhibits Slug expression in a dose-dependent manner (●). Values in parentheses indicate the number of embryos analyzed for each concentration of injected MO. Injection of 30 ng of a control MO had a minor effect on Slug expression (■). (D) Cross section of AP2α MO-injected embryo showing reduction in Slug. The injected side is to the right and the uninjected side is to the left (arrow). This also shows the reduced elevation of neural fold after AP2α MO injection. (Magnifications: B, ×7; D, ×42.)
Ectopic AP2α Transforms Neural Plate to NC.
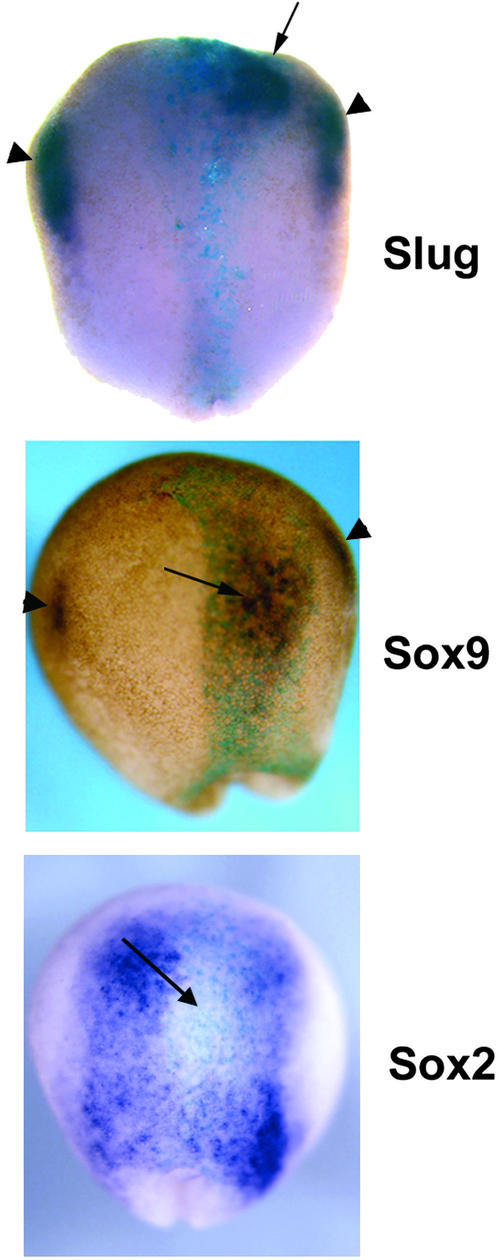
The preceding data support the conclusion that AP2α is required early in NC development. To determine whether this factor is sufficient for NC specification, we carried out ectopic expression experiments. RNA encoding AP2α was injected at the 8- to 16-cell stage into blastomeres predicted from the Xenopus embryonic fate map to include neural plate tissue among their progeny (52). The result of this ectopic expression, shown in Fig. 4, was that strong activation of NC-specific gene expression (e.g., Slug and Sox9) was elicited. Importantly, in many cases this conversion to NC comprised isolated domains within the neural plate, which were physically separated from endogenous NC territories, as opposed to contiguous, lateral expansions of NC into the neural plate and epidermis (which was also observed). Ectopic expression of AP2α was also found to repress neural plate gene activity, as exemplified by Sox2 (Fig. 4 Bottom). In these experiments, induction of NC was not observed in the most posterior region of the neural plate.
Figure 4.
Ectopic expression of AP2α converts neural plate into NC. Whole-mount in situ hybridization of probes for Slug, Sox9 (NC), and Sox2 (neural plate) to embryos injected at the 16-cell stage with 100 pg of RNA encoding AP2α, along with β-galactosidase lineage tracer RNA (5-bromo-4-chloro-3-indolyl-β-d-galactoside, turquoise staining). Discrete domains of intense ectopic expression of both NC marker genes was observed (arrows) within the neural plate. Endogenous Slug and Sox9 expression is indicated by the arrowheads. Concomitant repression of neural plate identity also occurred, as indicated by the reduction in Sox2 expression (arrow). (Magnification: ×22.)
Differential Response of NC Gene Expression to a BMP Signaling Gradient.
In Xenopus (5, 44, 53) and zebrafish (32), the degree of BMP signal attenuation plays an important role in determining the type of ectodermal tissue induced during gastrulation. To examine this in the context of NC induction, increasing doses of RNA encoding Chd were injected, to simulate a gradient of BMP activity, along with a constant dose of Xwnt-3a RNA. Expression of AP2α and other regulatory factors was then monitored by Northern blot. The results, shown in Fig. 5A, indicate that AP2α differs from Sox9, Slug, and Xtwi with respect to the lower threshold for activation; AP2α (which is expressed at low level in uninjected ectoderm, presumably as part of the epidermal program) is significantly up-regulated by the lowest (10 pg) dose of Chd, whereas the other three genes were not similarly activated until 10 times more Chd RNA was injected. Expression of Dlx3, which we have reported to act as an antagonist of NC induction (54) was reduced to an essentially constant baseline by the 10-pg Chd dose, whereas Dlx5 expression was reduced at the lower Chd doses, then partially recovered at higher doses, similar to what has been observed in the absence of Wnt overexpression (44). Neural plate marker genes Sox2 and Otx2 were repressed in animal caps under all conditions. When BMP signaling was inhibited to an even higher extent, by injection of more Chd RNA (Fig. 5B), the NC marker genes were down-regulated compared with lower Chd doses, whereas the neural plate marker Sox2 was activated. This finding supports the idea that some level of BMP signaling is essential for NC induction, but not, as revealed by the continued repression of the anterior neural marker Otx2, for posterior transformation of ectoderm by Wnt signaling. The effective elimination of BMP signaling in this experiment is reflected in the virtually complete repression of Dlx5, which shows a lower sensitivity to BMP signal interference than Dlx3 (44, 54). The loss of NC under very high Chd/very low BMP signal conditions can be reversed by coinjection of AP2α mRNA, as shown in Fig. 5C. This restored expression of Sox9, Slug, and Xtwi (endogenous levels of AP2α were not measured because of interference from injected RNA) and repressed Sox2, indicating that under these conditions AP2α was able to transform neural plate tissue into NC, analogous to what was observed in the intact embryo (Fig. 4).
Figure 5.
Differential control of NC factors by BMP signal attenuation. Shown is Northern blot analysis of RNA isolated from animal caps derived from embryos injected at the one-cell stage with a mixture of RNAs encoding Chd, Xwnt-3A, and AP2α. Uninjected (UI) embryo animal cap RNA and whole embryo (W) RNA are shown for comparison. Ethidium bromide staining of 18S ribosomal RNA is shown to confirm equal lane loading. All samples were cultured to stages 14 and 15. (A) Shallow gradient of Chd dosage ranging from 10 pg to 2.5 ng. AP2α up-regulation was elicited by as little as 10 pg of Chd RNA, whereas other NC markers (Sox9, Slug, and Xtwi) required more complete inhibition of BMP signaling (100–300 pg of Chd RNA). In all injected samples, the BMP-dependent homeobox gene Dlx3, and, to a lesser extent, Dlx5, was repressed, as were Sox2 and Otx2. (B) Steeper gradient of Chd dosage resulting in complete inhibition of BMP signaling. The results are similar to A, except the highest Chd dose (3 ng), which suppresses NC gene expression, restores the general neural plate marker Sox2 expression but not the anterior neural marker Otx2. The completeness of BMP signal inhibition can be judged by the reduction of Dlx5 transcripts to background levels. (C) Induction of NC gene expression in the absence of BMP signaling by AP2α. A very high Chd dose (4 ng) prevents BMP signaling, induces Sox2, and blocks NC induction as in B, whereas coinjection of 100 pg of AP2α RNA results in reactivation of NC markers and suppression of Sox2. The anterior neural plate marker Otx2 continues to be repressed under these conditions, presumably because of Wnt signaling.
Discussion
The results presented here strongly support the conclusion that the transcription factor AP2α is essential for the specification of NC in Xenopus. Therefore the deficiencies in NC derivatives found in AP2α knockout mice are most likely the result of an initial failure to specify NC progenitors as opposed to subsequent abnormalities in migration or differentiation. Comparing the properties and functions of AP2α to that of other factors furthermore suggests that AP2α might occupy a distinct level in the hierarchy of NC regulation. For instance, overexpression of ectopic AP2α regularly induces NC within the neural plate, whereas Slug (9) and to a lesser extent Sox9 (23) induce NC via expansion of existing domains and do not induce NC within the prospective CNS. During development, AP2α is initially activated throughout the ectoderm at the beginning of gastrulation (34) and is subsequently up-regulated in NC, whereas ectodermal expression of Sox9 and Slug initiates as part of the NC pathway, and thus follows that of AP2α. Another difference in expression is spatial, with a more lateral boundary for AP2α, particularly in comparison to Slug (Fig. 1). This finding may be at least due to differential regulation by a morphogenetic gradient of BMP signaling. In the artificially generated gradient of BMP signaling in animal caps expressing Chd, AP2α was activated by Wnt signaling at higher BMP levels than Slug, Xtwi, or Sox9 (Fig. 5). AP2α was also able to induce Slug and Sox9 and repress neural plate in the absence of BMP signaling (Figs. 4 and 5C). One interpretation of this finding is that AP2α expression is the primary essential target of BMP signaling in NC specification.
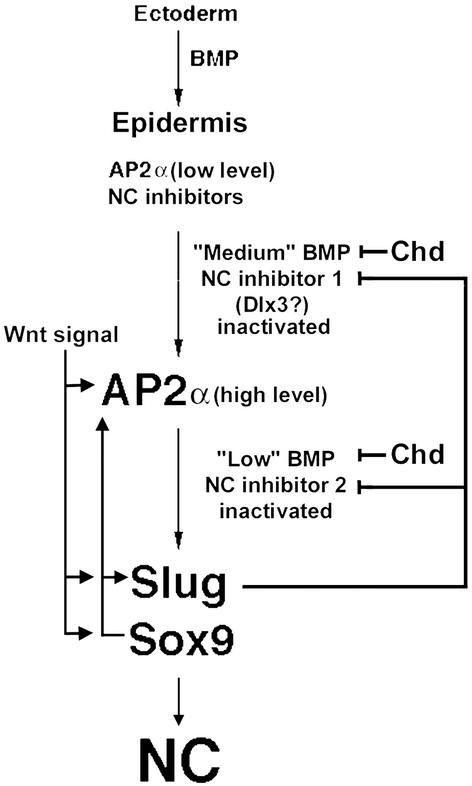
An attempt to systematically rationalize these observations is summarized in Fig. 6. This model proposes that naive ectoderm requires BMP signaling to activate relatively low-level AP2α expression throughout the ectoderm at the beginning of gastrulation, as part of the epidermal specification pathway (34) and that this AP2α expression is a prerequisite for subsequent NC induction. This requirement accounts for the observation that AP2α can induce NC in the midline of the neural plate, but overexpression of Wnt factors or Sox9 and Slug typically does not, because the prerequisite AP2α levels are absent in this region (and bypassed by ectopic AP2α introduced into the neural plate). For AP2α up-regulation and NC initiation to take place, in addition to a Wnt signal, BMP signaling must be partially attenuated, symbolized by Chd in Fig. 6. In this model Wnt is meant as a shorthand for factors, including retinoids and fibroblast growth factor signaling, in addition to the Wnt/β-catenin pathway, and Chd for the various BMP antagonists emanating from the organizer region. As shown in Fig. 5, injection of 10 pg Chd RNA, along with 300 pg of Xwnt-3a RNA, results in essentially complete up-regulation of AP2α. This treatment is insufficient, however, to activate the other NC markers Sox9, Slug, or Twist, which require a 10-fold higher Chd RNA dose. In view of the different thresholds for up-regulating AP2α compared with activation of other NC factors, we propose that this BMP signal attenuation involves two steps: an initial BMP reduction that results in the silencing of the first of two NC inhibitory factors and, in conjunction with Wnt signaling, in up-regulation of AP2α from the basal, epidermal expression to that of NC. This step is followed by a second phase of BMP attenuation (see below). A candidate for the first inhibitor is the homeodomain gene Dlx3. As shown in Fig. 5, this factor is repressed to a baseline level by the same dose of Chd (10 pg), in conjunction with Xwnt-3a that triggers up-regulation of AP2α. In addition, Dlx3 can antagonize Wnt/β-catenin signaling in early development and prevent NC induction in vivo (54, 55). Transcripts of the Dlx3 gene are excluded from the NC, at least in cranial region, and the Dlx3 gene has an intermediate dependence on BMP signaling, becoming silenced under BMP attenuation conditions that do not inactivate AP2α expression (ref. 43; T.L. and T.D.S., unpublished data).
Figure 6.
A model for NC induction. In the ectoderm, BMP signaling initiates and supports the expression of epidermal AP2α, at a basal level, and NC inhibitors. Because BMP signaling is attenuated by antagonists (Chd), this sequentially results in down-regulation of two NC inhibitors, the first of which may be the homeodomain gene Dlx3. With the first inhibitor removed, Wnt signaling results in up-regulation of AP2α, and, after silencing of the second inhibitor, induction of other factors such as Slug and Sox9. Slug reinforces the repression of the NC inhibitors, whereas Sox9 and AP2α positively regulate each other and Slug. All three factors are required for full induction and/or maintenance of NC. See Discussion for details.
After the increase and decrease, respectively, of AP2α and Dlx3, a secondary, more severe reduction in BMP signaling (also in conjunction with Wnt signaling) could then lead to activation of the remaining NC program. We have no candidate to mediate this secondary effect, but we predict the control will be negative in nature based on the paradigm of Dlx3 and suggest that another as yet unidentified NC antagonist exists, exhibiting less sensitivity to BMP attenuation compared with Dlx3. In this model, the critical function of Slug, which functions as a transcriptional repressor (56), is to stabilize the silencing of Dlx3 and the other NC inhibitors, which were extinguished as the result of BMP signal attenuation. This finding might account for the capacity of Slug to induce NC in explants when Wnt signaling is provided (9). Sox9, like AP2α, is a transcriptional activator, so these factors could establish an obligatory feedback mechanism that is necessary for continued expression of all three regulatory genes. This process would then lead to activation of additional, NC-specific target genes and specification of this tissue. Identification of these downstream targets will be an important goal of future research.
Finally, it is important to recognize that there are other regulatory factors that can induce or influence NC when experimentally expressed. For example, the RNA helicase eIF4AII has been reported to autonomously induce NC in ectodermal explants (24). We would predict that such a function resides downstream from the Chd and Wnt signaling events, but could either be upstream or downstream from the obligatory feedback/maintenance mechanism that we have portrayed as having three components but almost certainly is more complicated, including other factors that can induce NC in vivo. It should be possible to answer these kinds of questions with suitable coexpression experiments in Xenopus, which will continue to be a productive approach to this complex and important problem.
Acknowledgments
We thank Natasha Saint-Germain for technical help. J.-P.S.-J. is supported by grants from the National Institutes of Health (DE014212) and the March of Dimes.
Abbreviations
- NC
neural crest
- BMP
bone morphogenetic protein
- MO
morpholino oligonucleotide
- Chd
chordin
References
- 1.Selleck M A, Bronner-Fraser M. Development (Cambridge, UK) 1995;121:525–538. doi: 10.1242/dev.121.2.525. [DOI] [PubMed] [Google Scholar]
- 2.Mayor R, Young R M, Vargas A. Curr Top Dev Biol. 1999;43:85–113. doi: 10.1016/s0070-2153(08)60379-8. [DOI] [PubMed] [Google Scholar]
- 3.Liem K F, Tremml G, Roelink H, Jessell T M. Cell. 1995;82:969–979. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 4.Liem K F, Tremml G, Jessell T M. Cell. 1997;91:127–138. doi: 10.1016/s0092-8674(01)80015-5. [DOI] [PubMed] [Google Scholar]
- 5.Marchant L, Linker C, Ruiz P, Guerrero N, Mayor R. Dev Biol. 1998;198:319–329. [PubMed] [Google Scholar]
- 6.Ikeya M, Lee S M, Johnson J E, McMahon A P, Takada S. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 7.Saint-Jeannet J P, He X, Varmus H E, Dawid I B. Proc Natl Acad Sci USA. 1997;94:13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorsky R I, Moon R T, Raible D W. Nature. 1998;396:370–373. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 9.LaBonne C, Bronner-Fraser M. Development (Cambridge, UK) 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- 10.Simeone A, Acamoira D, Arcioni L, Bancinelli E, Mavilio F. Nature. 1990;34:763–766. doi: 10.1038/346763a0. [DOI] [PubMed] [Google Scholar]
- 11.Holowacz T, Sokol S. Dev Biol. 1999;205:296–308. doi: 10.1006/dbio.1998.9108. [DOI] [PubMed] [Google Scholar]
- 12.LaBonne C, Bronner-Fraser M. Annu Rev Cell Dev Biol. 1999;15:81–112. doi: 10.1146/annurev.cellbio.15.1.81. [DOI] [PubMed] [Google Scholar]
- 13.LeDouarin N M, Kalcheim C. The Neural Crest. 2nd ed. Cambridge, U.K.: Cambridge Univ. Press; 1999. [Google Scholar]
- 14.Essex L J, Mayor R M, Sargent M G. Dev Dyn. 1993;198:108–122. doi: 10.1002/aja.1001980205. [DOI] [PubMed] [Google Scholar]
- 15.Nieto M A, Sargent M G, Wilkinson D G, Cooke J. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 16.Mayor R, Morgan R, Sargent M G. Development (Cambridge, UK) 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- 17.Nakata K, Nagai T, Aruga J, Mikoshiba K. Proc Natl Acad Sci USA. 1997;94:11980–11985. doi: 10.1073/pnas.94.22.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakata K, Koyabu Y, Aruga J, Mikoshiba K. Mech Dev. 2000;99:83–91. doi: 10.1016/s0925-4773(00)00480-9. [DOI] [PubMed] [Google Scholar]
- 19.Hopwood N D, Pluck A, Gurdon J B. Cell. 1989;59:893–903. doi: 10.1016/0092-8674(89)90612-0. [DOI] [PubMed] [Google Scholar]
- 20.Dirksen M L, Jamrich M. Dev Dyn. 1995;17:107–116. doi: 10.1002/dvg.1020170203. [DOI] [PubMed] [Google Scholar]
- 21.Sasai N, Mizuseki K, Sasai Y. Development (Cambridge, UK) 2001;128:2525–2536. doi: 10.1242/dev.128.13.2525. [DOI] [PubMed] [Google Scholar]
- 22.Bang A G, Papalopulu N, Kintner C, Goulding M D. Development (Cambridge, UK) 1997;124:2075–2085. doi: 10.1242/dev.124.10.2075. [DOI] [PubMed] [Google Scholar]
- 23.Spokony R F, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet J P. Development (Cambridge, UK) 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- 24.Morgan R, Sargent M G. Development (Cambridge, UK) 1997;124:2751–2760. doi: 10.1242/dev.124.14.2751. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell P J, Wang C, Tjian R. Cell. 1987;50:847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- 26.Williams T, Admon A, Luscher B, Tjian R. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- 27.Moser M, Imhof A, Pscherer A, Bauer R, Arselgruber W, Sinowatz F, Hofstadter F, Schule R, Buettner R. Development (Cambridge, UK) 1995;121:2779–2788. doi: 10.1242/dev.121.9.2779. [DOI] [PubMed] [Google Scholar]
- 28.Chazaud C, Oulad-Abdelghani M, Bouillet P, Decimo D, Chambon P, Dolle P. Mech Dev. 1996;54:83–94. doi: 10.1016/0925-4773(95)00463-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhao F, Satoda M, Licht J D, Hayashizaki Y, Gelb B D. J Biol Chem. 2001;276:40755–40760. doi: 10.1074/jbc.M106284200. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell P J, Timmons P M, Hebert J M, Rigby P W, Tjian R. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 31.Snape A M, Winning R S, Sargent T D. Development (Cambridge, UK) 1991;113:283–293. doi: 10.1242/dev.113.1.283. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen V H, Schmid B, Trout J, Connors S A, Ekker M, Mullins M C. Dev Biol. 1998;199:93–110. doi: 10.1006/dbio.1998.8927. [DOI] [PubMed] [Google Scholar]
- 33.Epperlein H, Meulemans D, Bronner-Fraser M, Steinbeisser H, Selleck M A. Development (Cambridge, UK) 2000;127:2751–2761. doi: 10.1242/dev.127.12.2751. [DOI] [PubMed] [Google Scholar]
- 34.Luo T, Matsuo-Takasaki M, Thomas M L, Weeks D L, Sargent T D. Dev Biol. 2002;245:136–144. doi: 10.1006/dbio.2002.0621. [DOI] [PubMed] [Google Scholar]
- 35.Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell P J. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon A P, Flavell R A, Williams T. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- 37.Nieuwkoop P D, Faber J. Normal Table of Xenopus laevis (Daudin) Amsterdam: North–Holland; 1967. [Google Scholar]
- 38.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont L K, DeRobertis E M. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolda S L, Moody C J, Moon R T. Dev Biol. 1993;155:46–57. doi: 10.1006/dbio.1993.1005. [DOI] [PubMed] [Google Scholar]
- 40.Sargent T D, Jamrich M, Dawid I B. Dev Biol. 1986;114:238–246. doi: 10.1016/0012-1606(86)90399-4. [DOI] [PubMed] [Google Scholar]
- 41.Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Development (Cambridge, UK) 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- 42.Pannese M, Polo C, Andreazzoli M, Vignali R, Kablar B, Barsacchi G, Boncinelli E. Development (Cambridge, UK) 1995;1221:707–720. doi: 10.1242/dev.121.3.707. [DOI] [PubMed] [Google Scholar]
- 43.Feledy J A, Beanan M J, Sandoval J J, Goodrich J S, Lim J H, Matsuo-Takasaki M, Sato S, Sargent T D. Dev Biol. 1999;212:455–464. doi: 10.1006/dbio.1999.9374. [DOI] [PubMed] [Google Scholar]
- 44.Luo T, Matsuo-Takasaki M, Lim J H, Sargent T D. Int J Dev Biol. 2001;45:681–684. [PubMed] [Google Scholar]
- 45.Harland R M. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 46.Chang C, Hemmati-Brivanlou A. Dev Biol. 1998;194:129–134. doi: 10.1006/dbio.1997.8820. [DOI] [PubMed] [Google Scholar]
- 47.Deardorff M A, Tan C, Saint-Jeannet J-P, Klein P S. Development (Cambridge, UK) 2001;128:3655–3663. doi: 10.1242/dev.128.19.3655. [DOI] [PubMed] [Google Scholar]
- 48.Garcia-Castro M I, Marcelle C, Bronner-Fraser M. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- 49.McMahon A P, Moon R T. Cell. 1989;58:1075–1084. doi: 10.1016/0092-8674(89)90506-0. [DOI] [PubMed] [Google Scholar]
- 50.He X, Saint-Jeannet J-P, Woodgett J, Varmus H E, Dawid I B. Nature. 1995;372:677–679. [Google Scholar]
- 51.Summerton J, Weller D. Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 52.Moody S. Dev Biol. 1987;119:560–578. doi: 10.1016/0012-1606(87)90059-5. [DOI] [PubMed] [Google Scholar]
- 53.Wilson P A, Lagna G, Suzuki A, Hemmati-Brivanlou A. Development (Cambridge, UK) 1997;124:3177–3184. doi: 10.1242/dev.124.16.3177. [DOI] [PubMed] [Google Scholar]
- 54.Luo T, Matsuo-Takasaki M, Sargent T D. Mol Rep Dev. 2001;60:331–337. doi: 10.1002/mrd.1095. [DOI] [PubMed] [Google Scholar]
- 55.Beanan M J, Feledy J A, Sargent T D. Mech Dev. 2000;91:227–235. doi: 10.1016/s0925-4773(99)00303-2. [DOI] [PubMed] [Google Scholar]
- 56.LaBonne C, Bronner-Fraser M. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]