Abstract
Early in development, a part of the embryo is set aside to become the germ cell lineage that will ultimately differentiate to form sperm and eggs and transmit genetic information to the next generation. Men with deletions encompassing the Y-chromosome DAZ genes have few or no germ cells but are otherwise healthy, indicating they harbor specific defects in formation or maintenance of germ cells. A DAZ homolog, DAZL (DAZ-Like), is found in diverse organisms, including humans and is required for germ cell development in males and/or females. We identified proteins that interact with DAZ proteins to better understand their function in human germ cells. Here, we show that PUM2, a human homolog of Pumilio, a protein required to maintain germ line stem cells in Drosophila and Caenorhabditis elegans, forms a stable complex with DAZ through the same functional domain required for RNA binding, protein–protein interactions and rescue of Pumilio mutations in flies. We also show that PUM2 is expressed predominantly in human embryonic stem cells and germ cells and colocalizes with DAZ and DAZL in germ cells. These data implicate PUM2 as a component of conserved cellular machinery that may be required for germ cell development.
All stem cells have potential to differentiate or proliferate mitotically. These potentials must be balanced for the stem cell population to be maintained. If differentiation exceeds proliferation, the stem cell population is not maintained. Evidence in humans suggests that the DAZ genes function early in the germ line stem cells. Men with deletions encompassing the Y-chromosome DAZ gene cluster have defects in spermatogenesis that are detected initially in the stem cell population. These men frequently lack all germ cells, including the spermatogonial stem cells, and only somatic cells are present in testicular tissue (1–3). In addition, expression of the DAZ gene and its ancestral, autosomal homolog, DAZL, only occurs in germ cells; DAZ is expressed in males, and DAZL is expressed in males and females (4, 5). Evidence from model organisms also suggests the DAZ genes function in germ cell maintenance. The Xenopus homolog of DAZ, Xdazl, is expressed in a region of the early oocyte that contains the germ plasm that is required for formation and maintenance of the germ cell lineage (6, 7). Inhibition of Xdazl leads to loss of the primordial germ cells (6, 7). Finally, the DAZ homolog in mice, Dazl, is most abundantly expressed in premeiotic germ cells; disruption of this gene causes depletion of germ cells beginning prenatally (8–10). DAZ and DAZL homologs may function interchangeably as suggested by the observation that a human DAZ transgene can partially rescue a mouse Dazl mutation (11). To shed light on how DAZ genes might function in human germ cells, we sought to identify proteins that interact with DAZ proteins.
Materials and Methods
Two-Hybrid Screening of DAZ-Interacting Proteins.
The yeast two-hybrid system was used to identify proteins that interact with a DAZ:GAL4 DNA-binding domain fusion protein (CLONTECH). This construct was derived from a cDNA that encodes complete RNA-binding and DAZ repeat domains (pRR102). A library of testis cDNAs fused to the activation domain of GAL4 was screened three times according to the manufacturer's instructions. Twenty-three clones were obtained. Those that encoded proteins that interacted with the DNA-binding domain, interacted with unrelated laminin protein, or activated transcription independent of DAZ were not pursued.
Deletion Analysis.
Deletions of the PUM2 ORF were constructed by PCR amplification of overlapping fragments of the PUM2 cDNA and insertion into the pACT2 vector. Constructs were designed to encode fusion proteins in the correct reading frame, were transformed into yeast with the DAZ:GAL4 DNA-binding domain construct and screened for interaction by 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) filter assays. For DAZ deletions, primers were used to clone fragments of DAZ cDNA adjacent to the GAL4 DNA-binding domain within the pAS2 vector to produce fusion proteins. Constructs were transformed into yeast with PUM2:GAL4 activation domain construct and screened for interaction as above.
Coimmunoprecipitation of Human PUM2 and DAZ.
Yeast cells that expressed fusion proteins were pelleted, lysed and resuspended in lysis buffer (50 mM Tris⋅HCl, pH 7.5/1 mM EDTA/150 mM NaCl/10% glycerol/1% Nonidet P-40) with a mixture of protease inhibitors (CLONTECH). The PUM2 cDNA was cloned in frame with the GAL4 activation domain and a hemagglutinin (HA) tag and used to express protein for immunoprecipitation from yeast. HA antibodies covalently linked to inert beads (Babco, Richmond, CA) were incubated with yeast supernatants, washed with lysis buffer three times, resuspended in 1 mM glycine (pH 2.8), and incubated at 30°C to elute proteins. Coimmunoprecipitation from COS-7 cells was accomplished by transfection with a pBud vector (Invitrogen) containing the PUM2 cDNA fused to an HA tag. Cells were cultured for ≈48 h, lysed with lysis buffer as above, and incubated with DAZ proteins that were translated in vitro from a pET-29a(+) construct (Novagen) that encoded DAZ in frame with an S tag. Cell supernatant with S-DAZ, HA-PUM2, and both proteins were incubated with S-tag beads for 1–2 h, beads were washed, and proteins were eluted by denaturing at 95°C. Coimmunoprecipitation in mouse testis extract was accomplished by transforming BL21-Gold(DE3) pLys bacteria cells (Stratagene) with a pET-29a(+) construct (Novagen) that encoded DAZ in frame with an S tag. Bacteria cells were sonicated in lysis buffer as above and supernatants containing S-DAZ and containing the S Tag alone but no DAZ-protein fusion were precleared with protein A beads for 30 min, and incubated with S-tag beads for 1 h at room temperature. S-beads were washed, and incubated with testis supernatant for 2 h at 4°C. S-beads were washed, and eluted by denaturing at 95°C. Protein electrophoresis and Western blots were described (4).
Expression Analysis.
Northern hybridization and RT-PCR were performed as described (1). Tissues were homogenized in Trizol according to instructions (GIBCO/BRL, Bethesda). One microgram of total RNA was reverse-transcribed by using oligo(dT) primers (Roche Applied Sciences). cDNA (100 ng) was subjected to RT-PCR. Primers were (5′ to 3′): PUM1, AAAAACCTGAGAAGTTTGAATTGT and GCAAGACCAAAAGCAGAGTTG; PUM2, ACCAACATTCCTTGGTGAG and ATCAGGACCCCAAGAAGAGG.
Immunohistochemistry.
PUM2 antibodies were produced in rabbits with the peptide PNPTANKPLVEEFSNPETQN (Research Genetics, Huntsville, AL). This peptide is only present in PUM2; it is not in PUM1. PUM2 antisera were collected after 8 weeks. Immunohistochemistry was as described (4). Human testis sections were from a 20-week-old fetus and a 38-year-old man with a seminoma; sections were not in contact with the seminoma. Human ovary sections were from a 19-week-old fetus and 37- and 44-year-old women. Mouse sections were from 60-day-old mice. PUM2 and preimmune antisera were used at dilutions of 1:200 for histology and 1:700 for Western blots. Cell types were assigned by standard criteria (12).
Three-Hybrid Assay.
Yeast strain L40c, which contains the gene that encodes the LexA-MS2 fusion protein integrated into the chromosome, was transformed with RNA hybrid vector pIII/MS2-2/NRE (13). pIII/MS2-2/NRE contains the binding sites for MS2 coat protein linked to the NRE sequence. To assay for PUM2 interaction with the NRE, PUM2 cDNA was cloned into the pACT2 vector, which encodes the GAL4 activation domain. In addition, to assay for PUM2/DAZL/NRE complex formation, the PUM2 cDNA was cloned into the p412ADH plasmid and transformed with a DAZL fusion construct, containing the GAL4 activation domain in the pACT2 vector. Negative controls are as shown (Fig. 6). 5-Bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) filter assays were as described (13).
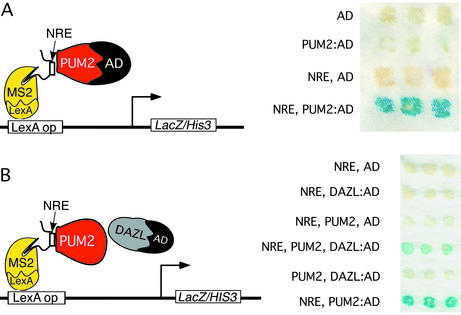
Figure 6.
Interaction of PUM2, DAZL, and NRE RNA. (A) Schematic of components used to detect PUM2/NRE interaction. Positive RNA–protein interaction was detected when the NRE and PUM2:AD were present as assayed by 5bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) assay (Right). No interaction was detected with GAL4 activation domain (AD) or PUM2:AD without the NRE or with NRE and AD alone. (B) Schematic of components used to detect NRE/PUM2/DAZL complex. A ternary complex was formed with NRE, PUM2, and DAZL:AD present (Left). No interaction was detected with the NRE and the AD or DAZL:AD alone or with the NRE with PUM2 and the AD alone. No interaction was detected in the absence of the NRE, or with PUM2 and DAZL:AD present. Positive control interaction of the NRE with PUM2:AD is shown.
Results
Identification of DAZ-Interacting Proteins.
We sought to identify proteins that interact with human DAZ proteins via a yeast two-hybrid screen. We identified seven proteins that potentially interact with DAZ (Table 1). These proteins are notable in several ways. (i) Five of the candidate interacting proteins contain domains predicted to bind RNA (PUM2, BOL, DZIP1, DZIP2, and DAZL). DAZ homologs have also been shown to bind RNA and associate with ribosomes (14–16). This finding suggests that DAZ proteins may function with interacting proteins to regulate translation. (ii) Four of the proteins are encoded by loci shown genetically to be required for fertility in other organisms (PUM2, BOL, hQK3, and DAZL) (1, 8, 17–19). (iii) Five of the proteins have been shown biochemically to interact with DAZ [BOL, DZIP1, DZIP2, hQK3 (data not shown)] and DAZL (14, 20, 21). These observations bolstered our confidence that we had identified legitimate partners.
Table 1.
Genes that encode proteins that potentially interact with DAZ
| Gene | Expression | Motif(s) |
|---|---|---|
| PUM2 (2) | Male/female germ cells and ES cells | PUF repeat |
| hQK3 (1) | Upregulated in ovary and testis* | None |
| BOL (1) | Male germ cells (25) | RRM, DAZ repeat |
| DZIP1 (6) | Testis and specific tissues | Zinc finger |
| DZIP2 (1) | Male germ cells* | Zinc finger |
| DZIP3 (2) | Unknown | None |
| DAZL (1) | Male/female germ cells (4, 5, 8) | RRM, DAZ repeat |
Parentheses indicate the number of clones obtained. The expression given was demonstrated in this report, unless indicated otherwise.
Expression determined by F.L.M. and R.A.R.P. (data not shown).
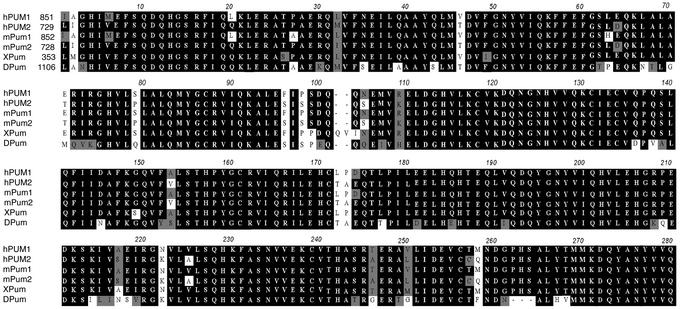
We further characterized interaction of DAZ with PUM2, a homolog of Drosophila Pumilio. Analysis of the PUM2 protein sequence we obtained (GenBank accession no. AF272350) indicated that the protein was nearly identical to that described in GenBank accession no. XM_015812. The exception was that the PUM2 protein encoded by the clone we isolated had an insertion of 79 aa at the N terminus. Overall, human PUM2 protein shares 75% identity with a second human protein, PUM1. PUM2 protein is also 96% identical to a mouse Pum2; 85% identical to the single known member of the family in frogs, XPum; and 35% identical to Drosophila Pumilio. The similarity between PUM2 and other Pumilio proteins was especially striking when we compared the RNA-binding domains of proteins from several organisms, including flies (Fig. 1). Remarkably, the human PUM2 RNA-binding domain is 80% identical to that of Drosophila Pumilio, the essential functional domain in fly embryogenesis (22).
Figure 1.
Alignment of RNA-binding domains of Pumilio. Sequences are from human (hPUM1 and hPUM2), mouse (mPum1 and mPum2), frog (XPum), and flies (DPum).
Interaction of DAZ and PUM2.
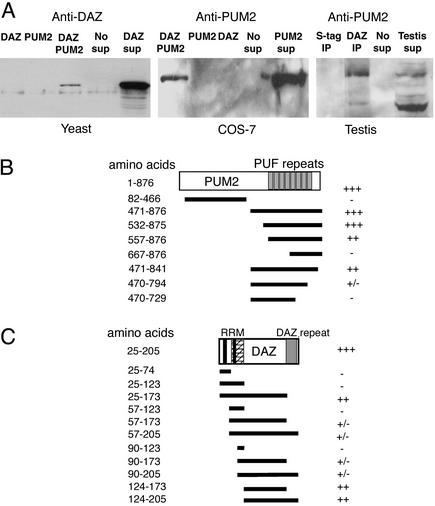
To verify interaction of the PUM2 and DAZ proteins that we observed in the two-hybrid system, we pursued biochemical isolation of PUM2 and DAZ complexes. We began with coimmunoprecipitation from yeast strains that expressed DAZ alone, PUM2 alone, and DAZ and PUM2 proteins together (Fig. 2A Left). We used antibodies that bind a tag on PUM2 to isolate it from yeast supernatants, and assayed for the presence of interacting DAZ protein by Western analysis. As shown in Fig. 2A Left, DAZ protein was coimmunoprecipitated only from supernatant that expressed both DAZ and PUM2. We extended these results to primate COS-7 cells (Fig. 2A Center). Antibodies that bind a tag on DAZ were incubated in supernatant from COS-7 cells containing DAZ and PUM2 or each protein alone. Coimmunoprecitation of PUM2 protein was detected by Western analysis only from cell supernatants where both DAZ and PUM2 proteins were expressed (Fig. 2A Center). In a similar manner, we also determined whether DAZ and PUM2 proteins interact in the testis (Fig. 2A Right). To accomplish this, we incubated mouse testis with human DAZ protein coimmunoprecipitated from bacterial extracts expressing DAZ. Association of DAZ and PUM2 occurred only when testis extracts were incubated with DAZ and was not observed in control extracts prepared from bacterial cells that lacked DAZ expression (Fig. 2A Right). These data establish that DAZ and PUM2 can form a stable complex.
Figure 2.
Interaction of PUM2 and DAZ proteins. (A) The DAZ protein was coimmunoprecipitated with PUM2 when HA beads were incubated with supernatant from yeast that expressed both DAZ and PUM2 fused to an HA tag; shown is DAZ protein detected by Western analysis (Left). DAZ protein was not coimmunoprecipitated with HA beads from yeast that expressed DAZ or PUM2 alone. Right lane shows yeast supernatant that expressed DAZ as a positive control. (Center) PUM2 protein coimmunoprecipitated with DAZ protein from primate COS-7 cells. Western analysis showed that PUM2 protein formed a complex with S-tag:DAZ protein captured from COS-7 cells that expressed PUM2 and DAZ. PUM2 protein was not coimmunoprecipitated from supernatant of COS-7 cells when S-beads were incubated with supernatants from cells with PUM2 or DAZ alone. Cell supernatant that expressed PUM2 was included as a positive control (right-most lane in Center). PUM2 protein was coimmunoprecipitated with DAZ from testis extract (Right). Western analysis showed that PUM2 proteins formed a complex with S-tag:DAZ protein when incubated in mouse testis extract. No PUM2 was detected when S-beads were incubated in mouse testis extract that did not contain the S-tag:DAZ fusion protein. Mouse testis extract was included as a positive control (right-most lane in Right). (B) Deletions of the PUM2 protein defined minimal domain of PUM2 required for interaction with DAZ. Eight deletions of PUM2:GAL4 protein were assayed for interaction with intact DAZ in the yeast two-hybrid system. Positions of amino acids on PUM2 constructs are as in GenBank accession no. AF272350. (C) Eleven deletions of DAZ defined minimal region required for PUM2 interaction via the yeast two-hybrid assay. Several constructs truncated the protein near the RRM domain (RNP-2 and -1) and interacted only weakly with PUM2, even though they contain the minimal interaction region. This weak interaction may have been caused by improper folding of DAZ due to disruption of RRM structure. +++, Maximal activity equivalent to that of intact interacting proteins; −, no interaction; ±, weak interaction; +, strong interaction; ++, nearly wild-type interaction.
We narrowed down regions of DAZ and PUM2 that are required for interaction by the yeast two-hybrid system (Fig. 2 B and C). Analysis of truncated PUM2 proteins indicated that DAZ protein interacts with the RNA-binding region of PUM2, which contains an RNA-binding domain with eight Pumilio Fbf (PUF) repeats (23, 24). Constructs that encoded the entire RNA-binding domain (amino acids 471–876 or 532–876) produced PUM2 peptides that interacted with DAZ protein as well as full-length PUM2 (Fig. 2B). In contrast, when residues 795–876, which comprise part of PUF repeat 8 were deleted, interaction with DAZ protein was reduced (Fig. 2B). Deletion analysis of DAZ protein was also used to identify regions of DAZ required for PUM2 interaction. Results indicated that PUM2 interacted with the linker region of DAZ between the RNA-binding DAZ repeat domains (amino acids 124–173; Fig. 2C).
Expression of PUM2 mRNA.
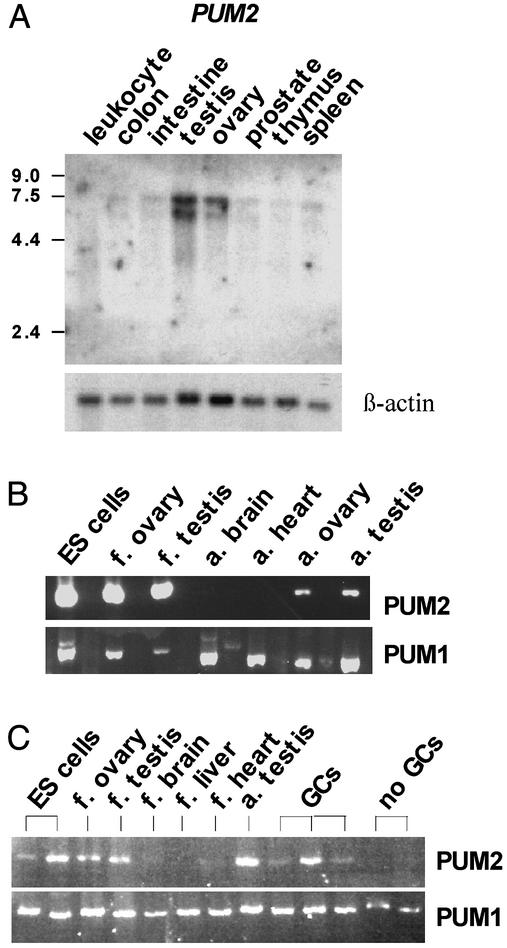
For interaction of DAZ and PUM2 to occur in vivo, the genes that encode them must be expressed in the same cellular/subcellular compartments. We examined expression of PUM2 mRNA and protein and compared it to DAZ. PUM2 mRNA expression was analyzed by both Northern analysis and RT-PCR. As shown by Northern analysis, PUM2 was highly expressed in adult ovary and testis (Fig. 3A). Little expression was detected in other adult tissues (leukocytes, colon, small intestine, prostate, thymus, and spleen) (Fig. 3A). To extend these results, RT-PCR was used to compare mRNA expression of PUM2 with that of PUM1 in human embryonic stem (ES) cells and various fetal and adult tissues (Fig. 3B). RT-PCR analysis indicated that PUM2 was abundantly expressed in ES cells, fetal and adult ovary, and fetal and adult testis; there was little or no expression in other tissues (Fig. 3B). In contrast, PUM1 mRNA was abundant in all tissues (Fig. 3B). Given that expression of PUM2 was greatest in gonads, we asked whether it is expressed in germ cells of the testis, somatic cells or both. To distinguish between these alternatives, expression of PUM2 and PUM1 mRNA was compared in tissue samples from men who had germ cells in their testis biopsies and from men who had no germ cells in their testis. Results showed that PUM2 was expressed most abundantly in testis biopsies with germ cells; variation in expression likely reflects variability in germ cell number and type in individual biopsies (Fig. 3C). PUM2 expression was not observed in biopsies from men who had no germ cells (Fig. 3C). These results suggested that PUM2 was either expressed in germ cells or required germ cells for expression in somatic cells. In contrast, PUM1 was expressed in samples from men with and without germ cells (Fig. 3C).
Figure 3.
Expression of human PUMILIO mRNAs. (A) Northern analysis of adult tissues indicated PUM2 expression in ovary and testis. Two transcripts of ≈5.5 and 6.5 kb were detected. (B) RT-PCR expression analysis of PUM1 and PUM2 in ES cells, fetal ovary and testis, adult brain and heart, and adult ovary and testis. (C) RT-PCR expression analysis of PUM1 and PUM2 in various fetal tissues and from men with normal germ cells (GCs) or no germ cells present in their testis. Germ cell number varied between biopsies.
Localization of PUM2 Protein.
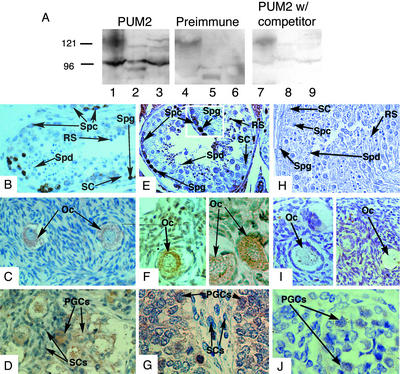
DAZ and DAZL proteins are confined to germ cells (4, 5). We generated antibodies specific to PUM2 to compare localization to that of DAZ and DAZL. Western analysis of mouse ES cell, and human and mouse testis extracts with PUM2 antisera indicated the presence of a dominant protein band of ≈95 kDa (Fig. 4A, lanes 1–3). Evidence that this represented native PUM2 protein is based on the observations that this band was not present on Westerns incubated with PUM2 preimmune sera (lanes 4–6) and was reduced in intensity when PUM2 antisera was preincubated with recombinant PUM2 protein (lanes 7–9). In addition to the dominant ≈95-kDa PUM2 band, we observed two weaker protein bands of higher molecular weight in human and mouse testis and mouse ES cells that were not present in preimmune sera and were competed by PUM2 peptide. These likely correspond to isoforms of PUM2 generated from the same gene on chromosome 2 that correspond to ESTs whose sequence is deposited in the GenBank database (accession no. XM_015812).
Figure 4.
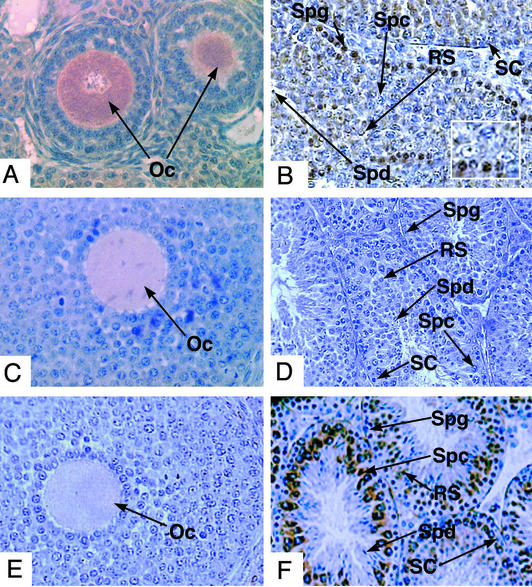
PUM2 expression mirrored that of DAZ and DAZL. (A) Western analysis with PUM2 antisera (lanes 1–3), PUM2 preimmune antisera (lanes 4–6), and PUM2 antisera preincubated with PUM2 protein competitor (lanes 7–9). Cell extracts were from human testis (lanes 1, 4, and 7), mouse testis (lanes 2, 5, and 8), and mouse ES cells (lanes 3, 6, and 9). The major PUM2 protein band was ≈95 kDa in extracts from human testis, mouse testis, and mouse ES cells. Antisera specific to both DAZ and DAZL proteins (4) were used to stain human sections of testis (B), ovary (C), and fetal testis (D). Antisera that recognized a PUM2-specific epitope were used to stain human sections of testis (E) (Inset is a magnified cross section of stained tissue), ovary (F), and fetal testis (G). Preimmune sera were used to stain human sections of adult testis (H), adult ovary (I), and fetal testis (J). Sections B–J were counterstained with hematoxylin/eosin and stained for specific proteins with horseradish peroxidase-conjugated secondary antibodies (brown staining). Magnification: ×200. Spg, spermatogonial cells; Spc, spermatocytes; Spd, spermatids; SC, somatic cells; Oc, oocyte; RS, round spermatid.
PUM2 antisera and antisera specific to DAZ and DAZL proteins (4, 5) were used to compare expression and cellular localization of these proteins. As previously shown (4, 5), both the Y-chromosome encoded DAZ and autosome-encoded DAZL proteins are expressed in germ cells of the human testis; the spermatogonia, early and late spermatocytes and postmeiotic cells (Fig. 4B). In females, only DAZL protein is expressed, in the cytoplasm of oocytes (Fig. 4C). DAZL expression begins early; prenatally in primordial germ cells of fetal testis (Fig. 4D; DAZ is expressed similarly) and fetal ovary (5). The expression of PUM2 mirrors that of DAZ and DAZL:PUM2 protein is most abundant in the cytoplasm and nucleus of spermatogonia but is also present in the cytoplasm of early and late spermatocytes (Fig. 4E) and is present in the cytoplasm of oocytes (Fig. 4F). Expression of PUM2 was also detected in primordial germ cells of fetal testis (Fig. 4G) and fetal ovary (data not shown). As shown, preimmune sera does not cross react with cellular proteins in the germ cells of the testis (Fig. 4H), ovary (Fig. 4I), and fetal testis (Fig. 4J). In addition, use of antisera against PUM1 results in a different pattern of staining: Germ cells, of all stages, as well as somatic cells demonstrate staining (data not shown).
To further examine expression of Pum2 in a mammal that is amenable to staging of germ cell types and confirm expression in spermatogonial stem cells, we examined the expression of Pum2 in mice (Fig. 5). Expression of Pum2 protein in mice mirrored that of humans in cell type and subcellular localization. The Pum2 protein was abundant in the cytoplasm of adult oocytes (Fig. 5A). In the male, Pum2 protein was concentrated in the spermatogonial cells found at the perimeter of spermatogenic tubules of stage I to IX (Fig. 5B Inset; shown is a stage III tubule with A1 spermatogonia near the perimeter). Immunohistochemistry with preimmune PUM2 antisera did not demonstrate significant signal in oocytes or spermatogonia (Fig. 5 C and D). Moreover, this pattern of expression was unique and specific as indicated by the use of antisera against Boule, a close relative of DAZ and DAZL (25). Use of these antisera illustrate the differences in expression pattern between premeiotically expressed DAZ/DAZL/PUM2 proteins and meiotic expression of Boule. The later is not detectable in oocytes (Fig. 5E); it is also absent from spermatogonial stem cells; only spermatocytes contained detectable Boule (Fig. 5F).
Figure 5.
Expression of Pum2 in mouse. Pum2 expression was observed in adult ovary (A) and adult testis (B) (Inset is a magnified cross section of stained tissue). No staining was observed with PUM2 preimmune sera on adult ovary (C) or testis (D). The pattern of PUM2 expression was also compared with that of mouse Boule (25). Boule protein was not present in adult ovary (E) and demonstrates a meiosis-specific expression pattern in adult testis (F). A–F were counterstained with hematoxylin/eosin and stained for specific proteins with horseradish peroxidase-conjugated secondary antibodies (brown staining). Magnification: ×200. Abbreviations are as in Fig. 4.
PUM2 and DAZL Interact on RNA.
Given our observations that PUM2 and DAZ can form a stable complex and colocalize in the germ cell lineage, we addressed whether PUM2 can interact with DAZL protein and form a complex on RNA. We chose to examine interaction with the NRE (Nanos regulatory element) RNA that is repressed by fly Pumilio. In flies, Pumilio interacts with proteins such as Nanos and Brat on the NRE (26) and represses translation of hunchback transcript to establish the embryonic axis (27–29). To determine whether human PUM2 protein can interact with DAZL and bind the NRE sequence as a complex, we first demonstrated that human PUM2 protein could bind to the NRE (Fig. 6A). Then we attached the activation domain of GAL4 to DAZL to assess ability of PUM2 and DAZL to interact and activate transcription of reporter genes, LacZ (Fig. 6B) and HIS3 (data not shown). We found that PUM2 and DAZL formed a stable complex, recognized the NRE RNA, and activated transcription (Fig. 6B).
Discussion
In model organisms, Pumilio proteins are required for germ line maintenance and differentiation (17, 30–34). Germ line stem cells are not maintained and instead differentiate prematurely in Drosophila Pumilio mutants (17). In C. elegans, double mutations in the Pumilio homologs, fbf-1 and fbf-2, result in loss of germ-line stem cell populations (30). It is proposed that FBF proteins may act to control the maintenance of germ-line stem cells, at least partially, by transnational repression of gld-1, a gene that may promote meiotic differentiation (30).
Here, we demonstrate that the human DAZ/DAZL proteins can form a stable complex with human PUM2. This observation potentially ties previous data acquired on the function of Pumilio in the germ cells of other organisms to the function of PUM2 and DAZ/DAZL proteins in human germ cells. Evidence suggests that human PUM2 may play a role similar to that of Pumilio in model organisms: First, the RNA-binding region, required to regulate translation during fly embryogenesis and germ-line development, is 80% identical to that of humans. Moreover, the RNA-binding region of Drosophila Pumilio is the site where cofactors are recruited to Pumilio Fbf (PUF) repeats 7 and 8 and the sequences C-terminal to facilitate Pumilio function as a translational repressor during embryogenesis (24). Our studies indicate that the RNA-binding domain is required for interaction of PUM2 with DAZ and that deletions that remove part of PUF repeat 8, reduced DAZ/PUM2 interaction. Second, PUM2 protein can bind the NRE sequence, the sequence necessary for translational repression of specific transcripts in Drosophila; this suggests that PUM2 may maintain ability to regulate translation. Finally, the pattern of PUM2 expression in embryonic cells and germ cells suggests that it functions in these cell types.
Thus, based on studies on the DAZ gene family and studies in Drosophila and C. elegans that indicate an essential role of Pumilio in germ cell development (17, 30–34), we suggest that PUM2 may function in maintenance of the human germ cell lineage. PUM2 is expressed throughout the development of the female and male germ cell lineages. Perhaps, then, PUM2 functions as a translational regulator in the germ cell lineage in conjunction with DAZ and DAZL proteins. This hypothesis is consistent with recent reports that indicate translational repression by Pumilio homologs may be an ancient and widespread mechanism for maintenance of germ line stem cells (30, 35). The observation that PUM2 message and protein is expressed in embryonic stem cells is also intriguing, although its role in this cell remains to be explored. With identification of conserved embryonic and germ cell components, the stage is set to unravel the function of genes such as PUM2 that may be required for maintenance of cells that give rise to mature germ cells in men and women.
Acknowledgments
We thank Michael Castillo for technical assistance; Anthony Dobson, Susan Fisher, Holly Ingraham, Richard Weiner, Joyce Tung, and Eugene Xu for helpful comments; and Marvin Wickens for three-hybrid resources. This work was supported by the Ford Foundation and the Woodrow Wilson Foundation (F.L.M.), the Polish State Committee for Scientific Research (J.J.), a National Institutes of Health National Research Service Award fellowship (to J.U.), and the Lalor Foundation (M.S.F.), and by grants from the National Institutes of Health (to Ira Herskowitz and R.A.R.P), the Searle Foundation, and the Sandler Family Foundation (to R.A.R.P.).
Abbreviations
- HA
hemagglutinin
- ES
embryonic stem
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos.: PUM2, AF272350 and XM_015812; hQK3, AF272349; BOL, AF272858; DZIP1, AF272347; DZIP2, AF272348; DZIP3, AF279370; DAZL, NM_001351; PUM1, XM_055629; mouse Pum1, NM_030722; mouse Pum2, NM_030723; Xenopus XPum, AB045628; Drosophila Pumilio, X62589).
References
- 1.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, et al. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 2.Reijo R, Alagappan R K, Patrizio P, Page D C. Lancet. 1996;347:1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- 3.Pryor J L, Kent-First M, Muallem A, Van Bergen A H, Nolten W E, Meisner L, Roberts K P. N Engl J Med. 1997;336:534–539. doi: 10.1056/NEJM199702203360802. [DOI] [PubMed] [Google Scholar]
- 4.Reijo R A, Dorfman D M, Slee R, Renshaw A A, Loughlin K R, Cooke H, Page D C. Biol Reprod. 2000;63:1490–1496. doi: 10.1095/biolreprod63.5.1490. [DOI] [PubMed] [Google Scholar]
- 5.Dorfman D M, Genest D R, Reijo Pera R A. Hum Reprod. 1999;14:2531–2536. doi: 10.1093/humrep/14.10.2531. [DOI] [PubMed] [Google Scholar]
- 6.Houston D W, King M L. Development (Cambridge, UK) 2000;127:447–456. doi: 10.1242/dev.127.3.447. [DOI] [PubMed] [Google Scholar]
- 7.Houston D W, Zhang J, Maines J Z, Wasserman S A, King M L. Development (Cambridge, UK) 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- 8.Ruggiu M, Speed R, Taggart M, McKay S J, Kilanowski F, Saunders P, Dorin J, Cooke H. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 9.Reijo R, Seligman J, Dinulos M B, Jaffe T, Brown L G, Disteche C M, Page D C. Genomics. 1996;35:346–352. doi: 10.1006/geno.1996.0366. [DOI] [PubMed] [Google Scholar]
- 10.Neiderberger C, Agulnik A I, Cho Y, Lamb D, Bishop C E. Mamm Genome. 1997;8:277–278. doi: 10.1007/s003359900409. [DOI] [PubMed] [Google Scholar]
- 11.Slee R, Grimes B, Speed R M, Taggart M, Maguire S M, Ross A, McGill N I, Saunders P T K, Cooke H J. Proc Natl Acad Sci USA. 1999;96:8040–8045. doi: 10.1073/pnas.96.14.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarrey J R. In: Cell and Molecular Biology of the Testis. Desjardins C, Ewing L L, editors. New York: Oxford Univ. Press; 1993. pp. 58–89. [Google Scholar]
- 13.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsui S, Dai T, Warren S T, Salido E C, Yen P H. Biol Reprod. 2000;62:1655–1660. doi: 10.1095/biolreprod62.6.1655. [DOI] [PubMed] [Google Scholar]
- 15.Venables J P, Ruggiu M, Cooke H J. Nucleic Acids Res. 2001;29:2479–2483. doi: 10.1093/nar/29.12.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiao X, Trifillis P, Kiledjian M. Biol Reprod. 2002;66:475–485. doi: 10.1095/biolreprod66.2.475. [DOI] [PubMed] [Google Scholar]
- 17.Forbes A, Lehmann R. Development (Cambridge, UK) 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- 18.Chubb C. J Androl. 1992;13:312–317. [PubMed] [Google Scholar]
- 19.Ebersole T A, Chen Q, Justice M J, Artzt K. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 20.Tsui S, Dai T, Roettger S, Schempp W, Salido E C, Yen P H. Genomics. 2000;65:266–273. doi: 10.1006/geno.2000.6169. [DOI] [PubMed] [Google Scholar]
- 21.Ruggiu M, Cooke H. Gene. 2000;252:119–126. doi: 10.1016/s0378-1119(00)00219-5. [DOI] [PubMed] [Google Scholar]
- 22.Wharton R P, Sonoda J, Lee T, Patterson M, Murata Y. Mol Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Zamore P D, Hall T M T. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 24.Edwards T A, Pyle S E, Wharton R P, Aggarwal A K. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 25.Xu E Y, Moore F L, Pera R A R. Proc Natl Acad Sci USA. 2001;98:7414–7419. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonoda J, Wharton R P. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wreden C, Verrotti A C, Schisa J A, Lieberfarb M E, Strickland S. Development (Cambridge, UK) 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 28.Sonoda J, Wharton R P. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murata Y, Wharton R P. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- 30.Crittenden S L, Bernstein D, Bachorik J, Thompson B, Moulder G, Barstead R, Wickens M, Kimble J. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- 31.Lin H, Spradling A C. Development (Cambridge, UK) 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 32.Parisi M, Lin H. Genetics. 1999;153:235–250. doi: 10.1093/genetics/153.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- 34.Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Nat Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- 35.Wickens M, Bernstein D S, Kimble J, Parker R. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]