Abstract
We have sequenced the genome of the intracellular symbiont Buchnera aphidicola from the aphid Baizongia pistacea. This strain diverged 80–150 million years ago from the common ancestor of two previously sequenced Buchnera strains. Here, a field-collected, nonclonal sample of insects was used as source material for laboratory procedures. As a consequence, the genome assembly unveiled intrapopulational variation, consisting of ≈1,200 polymorphic sites. Comparison of the 618-kb (kbp) genome with the two other Buchnera genomes revealed a nearly perfect gene-order conservation, indicating that the onset of genomic stasis coincided closely with establishment of the symbiosis with aphids, ≈200 million years ago. Extensive genome reduction also predates the synchronous diversification of Buchnera and its host; but, at a slower rate, gene loss continues among the extant lineages. A computational study of protein folding predicts that proteins in Buchnera, as well as proteins of other intracellular bacteria, are generally characterized by smaller folding efficiency compared with proteins of free living bacteria. These and other degenerative genomic features are discussed in light of compensatory processes and theoretical predictions on the long-term evolutionary fate of symbionts like Buchnera.
Various bacteria have traded their free-living lifestyle for a permanent physical association with eukaryotic hosts. Both symbiotic and parasitic outcomes of such transitions are associated with genome size reduction in the bacterial partner (1). Bacterial symbioses are widespread among insects, in which they are considered key to several specialized feeding behaviors and to their diversification at large (2). A model system is the obligate association between aphids and their maternally transmitted, intracellular symbiont Buchnera aphidicola (3). The bacteria support the exploitation of the poor diet of plant-phloem sap by aphids through supplementation of deficient nutrients, primarily essential amino acids (4). Buchnera evolved from an enterobacterial-like ancestor 200–250 million years ago (5), and some of its lineages have the smallest prokaryotic genomes reported to date (6).
Here, we present the complete genome sequence of B. aphidicola from the aphid Baizongia pistaciae (BBp) and a comparison with the previously sequenced strains from Acyrthosiphon pisum (BAp) (7) and Schizaphis graminum (BSg) (8). Phylogenetic studies have shown that the lineages leading to BBp and to the common ancestor of BAp and BSg diverged 80–150 million years ago, representing the evolutionarily most basal branching among modern Buchnera (5). Our selection of the Buchnera strain from B. pistaciae for comparative analysis will therefore yield a most comprehensive view of genome evolution in Buchnera and of the process of genome reduction during bacterial lifestyle transition in general.
In contrast to A. pisum and S. graminum, B. pistaciae has a complex life cycle and has never been cultured in the laboratory. Buchnera itself is considered unculturable (3, 4). Insects were therefore collected from the field for isolation of the bacteria, and thus for a direct application of the whole-genome shotgun sequencing method (9) to an environmental DNA sample.
Materials and Methods
Sequencing and Assembly.
B. pistaciae was collected from galls from a natural population on Pistacia trees. Buchnera isolation and subsequent bacterial DNA preparations were performed on samples pooled from 5–20 galls. Because the parthenogenetic offspring within a gall of B. pistaciae originates from a single fertilized egg, DNA preparations were expected to contain up to 20 distinct genotypes. B. aphidicola was purified as reported (6). DNA was isolated using a Qiagen (Valencia, CA) genomic DNA isolation kit and by quantitative hybridization estimated to contain 55% Buchnera DNA. A small-insert library (1.6–2.0 kb) was generated by mechanical shearing (sonication) of genomic DNA and cloning into pUC18 following a two-step ligation method (10) and using pulsed-field gel electrophoresis for all fragment isolations. Random shotgun sequencing was performed to an 8.9-fold sequence coverage with 8,405 Buchnera-derived sequences (average read length, 652 nt). This required gathering of a total of 15,777 sequences.
Genome Analysis.
Genome assembly and editing were done with Phred, Phrap, and Consed (www.phrap.org). Final assembly resulted in a single contig of the Buchnera chromosome and one small plasmid. Additional sequence reactions were performed to close 42 single-stranded gaps. Coding regions were identified using glimmer (11) and genemark (12), and transfer RNAs were predicted by TRNASCAN-SE (13). blast searches (14) were used to compare sequences of predicted genes and intergenic regions against other Buchnera genomes and the NCBI nonredundant protein and nucleotide databases, and aided the identification of structural RNAs and pseudogenes. Sequence, annotation, and detailed methodology can be found in Supporting Genome Annotation Methods, which is published as supporting information on the PNAS web site, www.pnas.org, as well as at www.pdg.cnb.uam.es/fabascal/Buch_ORFand_www. Potential single-nucleotide polymorphisms (SNPs) in the assembly were automatically identified with Consed, using a Phred quality score-threshold of 30 for high-quality discrepancies. All polymorphisms were checked manually to exclude sequencing errors.
Protein Folding Predictions.
The normalized energy gap α is defined as the minimal value of the relative energy gap divided by the structural distance between the predicted optimal fold and an ensemble of alternative configurations. The predicted optimal fold was identified by aligning the query sequence with all structures in a database representative of the entire PDB and containing nearly 1,200 protein chains. We then chose the alignment corresponding to the minimal effective energy, calculated as indicated by Bastolla and colleagues (15, 16). To reduce the complexity of the search, gaps where preassigned at the positions where they are known to occur from the alignment between the query sequence and the sequence of one of the representative structures. Apart from gap placement, the sequence information was not used for the sake of structure prediction. In all cases studied, the predicted optimal sequence coincided with the representative structure of the protein family to which the query sequence belongs. The remaining alternative structural alignments, typically some hundreds of thousands per structure, were used to estimate the energy gap.
The following representative structures of protein families were present in our database (PDB ID codes are in parentheses, www.rcsb.org/pdb/): epsilon subunit of F1F0-ATP Synthase, atpe_ecoli (1aqt); panthetheine-phosphate adenyltransferase, coad_ecoli (1b6t); d-ala d-ala ligase, ddlb_ecoli (1iov); phosphocarrier protein H, pthp_ecoli (1opd), pthp_strfe (1ptf); Rnase H, rnh_ecoli (2rn2), rnh_theth (1ril); Rnase P, rnpa_bacsu (1a6f), rnpa_staau (1d6t); triosephosphate isomerase, tpis_ecoli (1tre), tpis_bacst (1btm), tpis_trybb (1tpd), tpis_leime (1amk), tpis_yeast (7tim); thioredoxin I, thio_ecoli (2trx), thi2_anasp (1thx); thioredoxin reductase, trxb_ecoli (1tde); tryptophan synthase alpha chain, trpa_salty (1a50); substrate binding domain of DnaK in complex with substrate peptide, dnak_ecoli (1dkz). The following species were compared: Buchnera strains from aphid hosts B. pistaciae, S. graminum, and A. pisum; obligate parasites Borrelia burgdorferi, Chlamydia pneumoniae, Chlamydia trachomatis, Mycobacterium leprae, Mycoplasma capricolum, Mycoplasma genitalium, Mycoplasma pneumoniae, Rickettsia prowazekii, Treponema pallidum, Ureaplasma parvum; free-living bacteria Bacillus halodurans, Bacillus megaterium, Bacillus subtilis, Campylobacter jejuni, Caulobacter crescentus, Deinococcus radiodurans, Escherichia coli, Eubacterium acidaminophilum, Haemophilus influenzae, Helicobacter pylori, Lactococcus lactis, Listeria monocytogenes, Mesorhizobium loti, Mycobacterium smegmatis, Mycobacterium tuberculosis, Neisseria meningitidis, Pasteurella multocida, Pseudomonas aeruginosa, Pseudomonas putida, Salmonella typhimurium, Staphylococcus aureus, Streptococcus pyogenes, Streptomyces coelicolor, Synechocystis sp., Vibrio cholerae, Xylella fastidiosa, and Zymomonas mobilis.
Results and Discussion
General Features of the Genome.
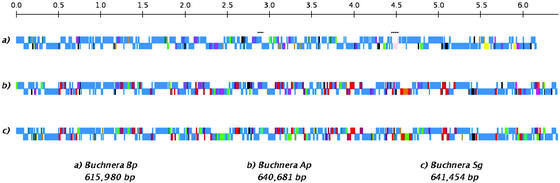
The genome of BBp has a consensus size of 617,838 bp with an average G+C content of 25.3%, and is composed of a 615,980-bp chromosome and a 2,399-bp plasmid (Fig. 1, Table 1; see Fig. 5, which is published as supporting information on the PNAS web site). The putative origin of replication, in the absence of a diagnostic cluster of DnaA boxes, was determined by GC-skew analysis and mapped to a position identical to those in other Buchnera and enterobacterial genomes (7, 8). We identified 544 putative genes and nine pseudogenes, all of which had database matches, and 491 (89%) of which were assigned a function. The functional set includes 507 protein-coding genes, one split ribosomal RNA operon, two structural RNAs, and 32 tRNAs specifying all 20 amino acids (Table 1).
Figure 1.
Comparison of the linearized Buchnera genomes from B. pistaciae, A. pisum, and S. graminum. Nucleotide numbering, marked in 100 kb, starts from the respective origins of replication. Genes above and below lines are transcribed from forward and reverse strands, respectively. Blue, genes present in all genomes; black, RNAs; light green, pseudogenes; pink, inversions; orange, genes unique among the three genomes; red, genes absent only in BBp; dark green, genes absent only in BAp; violet, genes absent only in BSg; yellow, genes chromosomally encoded in BBp and plasmid encoded in BAp and BSg. Hatched lines highlight the inversions in BBp. A map of the 2.4-kb plasmid of BBp is presented in Fig. 5.
Table 1.
Comparison of Buchnera genomes
| Feature | BBp | BAp | BSg |
|---|---|---|---|
| Genome size, bp | 618,379 | 652,095 | 653,001 |
| G+C content, % | 25.3 | 26.3 | 26.2 |
| Genes (plus pseudogenes)* | 553 | 618 | 628 |
| Coding content, % | 83.6 | 88.1 | 83.4 |
| Chromosome | |||
| Size, bp | 615,980 | 640,681 | 641,454 |
| Protein-coding genes | 504 | 560 | 545 |
| Pseudogenes | 9 | 13 | 38 |
| tRNAs | 32 | 32 | 32 |
| rRNAs | 3 | 3 | 3 |
| Other RNAs | 2 | 2 | 2 |
| Plasmids | |||
| Number† | 1 | 2 | 2 |
| Total size, bp‡ | 2,399 | 11,414 | 11,547 |
| Protein-coding genes | 3 | 9 | 9 |
Intrapopulational Variation.
The assembly unveiled putative intrapopulational variation at 1,168 sites, including 72 length polymorphisms (indels) and 1,096 dimorphic SNPs (Table 2). For the identification of polymorphisms, we used a Phred quality-score threshold of 30 for high-quality discrepancies, which corresponds to a probability of 1 in ≈3,000 that a discrepancy can be attributed to a base calling error. All SNPs were found to be dimorphic, with variants being represented on average by 6.4 reads of the “majority variant” and by 2.5 reads of the “minority variant.” The majority of polymorphisms (67%) was evidenced by more than one discrepant read per site. Although we cannot exclude the possibility that a small fraction of the observed variation was introduced through sequencing errors, it is obvious that the distribution of polymorphisms is entirely consistent with expectations based on natural, rather than experimentally introduced, variation (Table 2). Firstly, intergenic regions and pseudogenes were significantly more variable in both classes of polymorphisms than coding regions, whereas pseudogenes in turn were significantly more variable than intergenic regions (binomial test, P < 0.001). Secondly, SNPs showed a strong prevalence of transitions over transversions (5.3), a well known feature of biological sequence evolution that cannot be attributed to sequencing artifacts. The higher frequency of SNPs in pseudogenes (G+C; 23.2%) can partly be attributed to a more recent relaxation of selection against A+T mutational bias compared with intergenic regions (G+C; 15.4%). Likewise, recent relaxation of selection against deletional bias explains the significantly highest frequency of indels in pseudogenes (binomial test, P < 0.001).
Table 2.
BBp genome assembly polymorphisms
| Total | Frequency* | |
|---|---|---|
| Polymorphisms | 1,168 | 1.90E-03 |
| Indels | 72 | 1.17E-04 |
| Average size, bp | 1.8 | NA |
| Intergenic regions | 52 | 5.12E-04 |
| Pseudogenes | 8 | 1.02E-03 |
| RNAs | 1 | 1.30E-04 |
| ORFs | 11 | 2.20E-05 |
| SNPs | 1,096 | 1.78E-03 |
| Transitions | 921 | 1.09E-03 |
| Transversions | 175 | 5.71E-03 |
| Ratio | 5.3 | NA |
| Intergenic regions | 283 | 2.79E-03 |
| Pseudogenes | 47 | 5.99E-03 |
| RNAs | 2 | 2.61E-04 |
| ORFs | 764 | 1.54E-03 |
| Synonymous | 480 | 3.15E-03 |
| Nonsynonymous | 284 | 7.93E-04 |
NA, not applicable.
Frequency of occurrence per site in respective class.
Five of a total of 13 polymorphisms that affected the reading frame of protein-coding genes were potentially deleterious and may represent genuine snapshots of gene inactivation. Indels ranged in size from 1–14 bp (average, 1.8 bp) and resulted in a variable genome size range of 128 bp. The majority of indels (86%) appeared to be the result of slipped-strand mispairing involving A/T stretches.
Assuming a minimum of two distinct genotypes to explain the observed variation, we estimate that SNPs in coding regions occur at maximal frequencies of 3.2 × 10−3 as synonymous substitutions per site (Ks) and of 7.2 × 10−4 as nonsynonymous substitutions per site (Ka). The Ks/Ka ratio of 4.0 is smaller than previous average multilocus estimates for pairs of Buchnera strains (5.1–7.0; ref. 17). The lower Ks/Ka ratio may reflect fractions of both recent mutations within the population, as well as of mutations that occurred within the present generation of predominantly larval aphids and were not yet purged by selection. In general, lower Ks/Ka ratios in Buchnera proteins than in those of free-living bacteria are caused by an accelerated rate of nonsynonymous substitutions and are a signature of increased fixation of mildly deleterious mutations (18).
Genome Comparison.
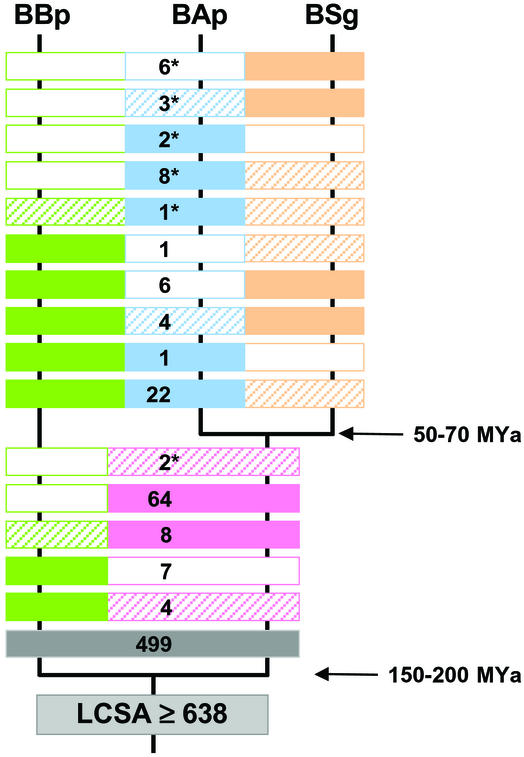
A total of 638 genes have been identified in the genomes of BBp, BAp, and BSg (in this figure we count the gene ribD, which is present as a split gene in BAp and BSg, as a single gene). This set represents a most parsimonious reconstruction of the gene content of the last common, symbiotic ancestor (LCSA) of the three Buchnera lineages (Fig. 2; and see Table 3, which is published as supporting information on the PNAS web site). The perfect chromosomal synteny previously described for BAp and BSg (8) was found to extend to the BBp genome (Fig. 1) and hence to the phylogenetic root of Buchnera. Genomic stasis must thus have set in soon after the establishment of the LCSA, which makes Buchnera an ≈200 million-year-old, enterobacterial “gene-order fossil.” We identified only four minor rearrangements in BBp relative to BAp and BSg (Fig. 1), including two inversions (one covering genes ygfZ-prfB-lysS-lysA-lgt-thyA and one covering the single gene pyrF) and two translocations (involving the resident plasmids and the essential amino acid biosynthesis genes trpEG and leuABCD).
Figure 2.
Numerical comparison of the gene content of Buchnera BBp, BAp, and BSg genomes. Solid boxes, genes present; hatched boxes, pseudogenes; open boxes, genes absent. Asterisked numbers denote genes that were putatively lost or inactivated in parallel in independent lineages. Arrows indicate estimated divergence times (5).
Putatively functional copies of 499 genes (78% of the LCSA's gene repertoire) are present in all three genomes. A set of 139 genes (22%) accounts for differences in gene content among the species and includes genes that were entirely lost from one or more lineages, as well as genes that have lost functionality but that are still recognizable as pseudogenes (7, 8). As expected on the basis of phylogenetic relationships, the difference in functional gene content between BBp on the one hand and BAp and BSg on the other hand is substantially larger than the difference between BAp and BSg. The latter pair differs in the presence or functionality of 54 genes, and they collectively differ in 85 genes from BBp. On top of this, BBp shows 25 and 41 gene content differences with BAp and BSg, respectively (Fig. 2).
Using the parsimony criterion and the established phylogenetic relationships, a minimum of 22 genes (3.4%) can be inferred to have undergone parallel evolution. This low level of homoplasy means that the true gene content of the LCSA must have been close to our minimal estimate of 638 genes. A striking example of parallel evolution is found among the set of eight genes that were completely lost in BBp and which are present as pseudogenes in BSg, whereas functional copies are retained in BAp (fourth bar from top in Fig. 2). Six of these genes occur in two clusters on the chromosome (cysDGNHI and cysQ) and are involved in assimilatory sulfate reduction and cysteine biosynthesis.
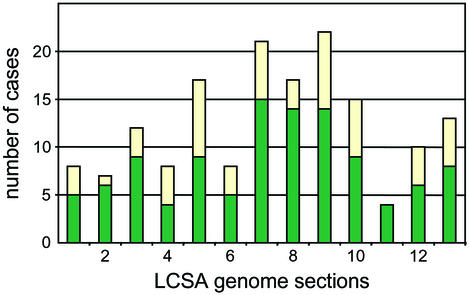
Horizontal gene acquisition in chromosomes (1, 8) has not occurred in Buchnera and the observed synteny therefore facilitates a most parsimonious reconstruction of the history of gene loss from the three genomes since their divergence from the LCSA. Our results indicate that most losses occurred through inactivation and disintegration of individual genes (123 of a total of 163 events). The minority of cases in which two or more adjacent genes were lost includes one large segment in BBp that covers eleven genes (nlpD, ψygbB, ψygbP, ygbQ, and cysC, -D, -G, -N, -H, -I, and -J). Overall, this pattern supports the hypothesis that ongoing gene loss in Buchnera proceeds predominantly through dispersed inactivation of single genes (19). Most unexpectedly, however, we observed a significant clustering (χ2 test, P < 0.002) of gene loss and inactivation toward the region containing the terminus of replication in the LCSA chromosome (Fig. 3). This suggests that gene inactivation and genome shrinkage proceed fastest from within this region in Buchnera, either as a result of weaker selection on genes in the proximity of the terminus or as a deleterious effect of an impaired replication machinery (see below).
Figure 3.
Relation between number of events of gene loss and pseudogene formation from the LCSA genome and genomic position of affected genes. The 635 chromosomally encoded genes of the reconstructed LCSA genome were numbered from the putative origin of replication onwards and arranged into 13 bins of ≈50 genes. Green bars represent total numbers per bin of deletion events from all three Buchnera genomes. Yellow bars represent total numbers per bin of pseudogene formation.
The reconstructed gene repertoire of the LCSA shows a massive reduction in numbers of genes across all functional categories relative to the closely related E. coli K-12 genome (see Fig. 6A, which is published as supporting information on the PNAS web site). Only four genes (yba2, -3, -4, and aroQ) are absent from the latter. Most strongly affected are regulatory and transport functions, whereas translation and metabolism of nucleotides and amino acids show more moderate shrinkage. Both the disproportionate loss of regulation and conservation of a minimal set of information processing functions are features Buchnera shares with other symbionts and obligate parasites (20–23). Extensive loss of transport capabilities and conservation of gene content for essential amino acid biosynthetic pathways, on the other hand, are confirmed to be prime characteristics of the aphid symbiont (7). It is possible that some of the functions lost in Buchnera are compensated for by the host (7).
The integrated view of metabolism, cellular processes, and information processing in Buchnera previously given on the basis of the BAp genome (7) essentially applies to the LCSA, which resembles BAp's gene content most closely. Ongoing gene loss in the lineages leading to BBp, BAp, and BSg has occurred in nearly all functional categories (see Fig. 6B). Several of the observed differences between the genomes are likely to be implicated in host-specific properties of life cycle dynamics or plant utilization. Examples include the loss in BBp of genes for various cofactors and vitamins, the loss of the biotin biosynthesis genes in BAp and BSg, and the convergent losses in the sulfate reduction and cysteine biosynthesis pathways in BBp and BSg. One exception to the general conservation of essential amino acid biosynthetic pathways in Buchnera is the loss of the ornithine pathway in BBp (argA, -EBC, and -D), with which it lacks the capacity to synthesize the essential amino acid arginine. Possibly a consequence of the absence of this pathway is the loss of several, physically unlinked genes involved in pyrimidine (pyrC, -D, and -I) and spermidine (speDE) metabolism.
Replication, Recombination, and Repair.
Perhaps the most extraordinary impact of genome reduction in the LCSA is seen in the complement of genes for replication, recombination, and repair (see Table 3). Relative to the LCSA, BBp, BAp, and BSg underwent further losses in this category of seven, two, and six genes, respectively. Homologous recombination pathways were already lost entirely in the LCSA, a condition that, together with the lack of repeated sequences in Buchnera (8), may explain the observed synteny of the sequenced genomes. We postulate that retention of recBCD in all genomes, in the absence of recA, may serve a general exonuclease repair function as a substitute for recombinational repair.
The MutHSL methyl-directed mismatch repair system is intact in BBp, but degenerate in BAp and BSg, notably through loss of the endonuclease component mutH. In view of the functioning of this pathway in the closely related species E. coli (24), retention of the system is puzzling, given that there is no methylation pathway present in Buchnera. BBp has lost the oxidative damage repair gene mutT, which is responsible for the removal of 8-oxo-dGTP before misincorporation into DNA. Of the six nucleoprotein encoding genes present in the LCSA and BAp, only DNA-binding protein HU-α (hupA) and the single-stranded DNA-binding protein (ssb) are retained in BBp. In addition to transcriptional regulation and chromosome structure, these proteins play an auxiliary role during replication and their absence in BBp may imply chromosomal and replicative instability. Additional losses in BBp associated with the replication machinery include type I topoisomerase activity (topA) and genes for primosomal proteins PriA and DnaT. The latter seem to imply the absence of a reassembly pathway for dissociating replisomes, which occur, for instance, at blocked replication forks, and are further indications of a decreased efficiency of replication in BBp.
Detailed analysis of the replication genes in the three Buchnera genomes and in the recently published genome of the related symbiont Wigglesworthia of the tsetse fly (20) revealed remarkable truncations of the genes dnaX and polA, which encode subunits of DNA polymerase III holoenzyme (Pol III-HE) and DNA polymerase I (Pol I), respectively. All bacterial genomes sequenced to date carry an orthologue of dnaX that encodes either the τ subunit or both the τ and γ subunits of Pol III-HE, τ as a full-length translational product and γ as a shorter product arising from ribosomal frameshifting or transcriptional slippage of τ (25). Subunit τ functions in dimerization of Pol III-HE and is implicated in the coordination of leading- and lagging-strand replication (26). The truncation of dnaX in Buchnera and Wigglesworthia has resulted in a complete loss of τ, a condition unique among bacteria and one likely to decrease the efficiency, accuracy, and processivity of Pol III-HE (27–29). Further attenuation of Pol III-HE integrity compared with E. coli is suggested by the absence of genes encoding the θ, χ, and ψ subunits in Buchnera genomes. The truncation of polA in Buchnera and Wigglesworthia implies the loss of both the polymerase and 3′ to 5′ exonuclease (proofreading) domains of DNA polymerase I, leaving only the 5′ to 3′ exonuclease domain intact. This suggests that Pol I is retained for its role in the removal of RNA primers in replication, and that Pol III is the sole DNA polymerase active in both symbionts. In summary, Buchnera appears to rely on an exceptionally reduced replication, recombination, and repair machinery, an attribute that itself constitutes a major agent of genome evolution. As such, it must have contributed to accelerated sequence evolution and gene inactivation in Buchnera.
Protein Stability.
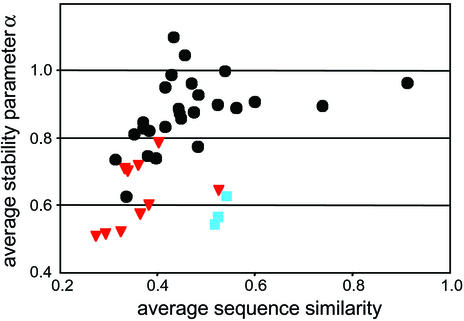
One expected manifestation of genome-wide accumulation of mildly deleterious mutations is a decrease in the thermodynamic stability of encoded macromolecules. To assess the extent of this effect in Buchnera proteins, we compared thermodynamic parameters in 11 protein families for a set of 40 bacterial species, including the 3 Buchnera strains and 10 obligate intracellular parasites. A requisite for efficient protein folding is a smooth free-energy landscape (30). A quantitative measure of this property can be obtained through the normalized energy gap (15, 31, 32). The normalized energy gap was estimated for each protein sequence by using an effective energy function (15, 16) and a set of nearly 1,200 protein structures, representative of the entire PDB and containing the 3D structure of at least one member of each protein family. The predicted native structure coincided in all cases with one of the known structures in the same family. In Fig. 4, we plotted the estimated energy gap α for each species, averaged over the 4–10 proteins available for that species, against the average sequence similarity to the reference structure of each protein. The correlation between these two quantities is an irrelevant artifact of our method, because the larger the sequence similarity, the better the predicted native structure will resemble the real one. Nevertheless, Fig. 4 clearly shows that the estimated energy gap for Buchnera proteins is much smaller than one would expect on the basis of sequence similarity alone. Moreover, this result holds not only for the average gap, but also for the gap of all individual proteins. A strikingly similar pattern is observed for other bacteria with obligate intracellular lifestyles.
Figure 4.
Relation between average protein stability parameter α for 11 proteins in 40 bacterial species and their average sequence similarity to the reference structure of each protein. Bacterial lifestyles are indicated as follows: black dots, free-living bacteria and facultative pathogens; red triangles, obligate parasites; blue squares, Buchnera strains (see Materials and Methods for protein families and species included).
A decreased energy gap is expected to reduce the efficiency of protein folding and to cause problems of presence of intermediate states in the folding process, with consequently slow folding, limited stability of the native state, and possible misfolding and aggregation. One protein family examined is the chaperone DnaK, which belongs to a class of proteins that assist in the folding and refolding of other proteins. Chaperones are abundantly expressed and evolutionarily among the most conserved proteins in intracellular bacteria. Interestingly, the normalized energy gap of DnaK is large, not only in free-living bacteria but, as an exception to the overall pattern, also in bacteria with an intracellular lifestyle. This suggests that DnaK is subject to a relatively strong selective pressure in intracellular bacteria. A recent simulation of Buchnera population dynamics, using experimental populations of E. coli undergoing intense genetic drift, demonstrated that genome-wide deleterious mutational effects could be buffered by constitutive overexpression of the molecular chaperone GroELS (33). In light of the present demonstration of decreased protein stability in Buchnera and a previous finding of its abundant expression of GroELS (34), these data support the hypothesis that GroELS functions in a compensatory mechanism to the accumulation of destabilizing amino acid substitutions in Buchnera proteins (18).
Conclusions
Following the hypothesis that the presymbiotic ancestor of Buchnera had a large, enterobacterial-like genome containing at least 1,800–2,400 genes (19, 35), our comparative analysis of three Buchnera strains has indicated that most of the reduction in genome size (65–74%) occurred soon after the establishment of the symbiosis but before the diversification of the major lineages of extant aphids. Since their divergence from the LCSA, the genomes of BAp, BSg, and BBp underwent further reductions of 5–15%. This pattern is consistent with the view that genome size reduction associated with bacterial lifestyle transition proceeds at an exponentially decreasing pace (36). From our analysis, a view of degenerate, rather than adaptive, genome evolution in Buchnera emerges. The symptoms include mutational bias, erosion of regulatory systems (regulatory genes, promoters, Shine Dalgarno sequences, transcription attenuators, and DnaA boxes), accumulation of mutations affecting protein stability, ongoing pseudogene formation and gene loss across all functional categories, a bias of gene inactivation toward the terminus of replication, and, most importantly given their exacerbating effect on mutational rates, a continued reduction of the arsenal of repair systems and of the replication machinery.
Genetic isolation and small effective population size are major determinants of degenerate genome evolution in Buchnera (18). In general, such populations are subject to increased genetic drift and reduced efficacy of selection and, as a result, show an irreversible accumulation of mildly deleterious mutations and a progressive loss of fitness. The predicted long-term evolutionary outcome of this process, known as Muller's ratchet, are mutational meltdown and population extinction (37). The time scale of such a fate for symbionts like Buchnera will depend on a number of factors, including the strength of compensatory processes such as the stabilizing effect of chaperones on cellular proteins (33), strengths of selection on both host and symbiont, and the exact range of important population genetic parameters like effective population size (38). The varying extents of genome reduction revealed by the present analysis and the recent finding of Buchnera strains with extremely reduced genomes (6) seem to further indicate that prolonged genomic stasis is unsustainable and a symptom of genome degeneracy. At the level of aphid hosts, the adverse effects of genomic exhaustion in Buchnera may also be overcome by its replacement with novel symbiotic bacteria. Indeed, a number of recent studies of facultative symbionts support the generality of a “replacement hypothesis” in insect symbioses (39–41).
Tamas et al. (8) inferred from the ≈50 million years of genomic stasis between BAp and BSg that the ecological diversification of aphids cannot be attributed to the genetic diversity of Buchnera. In our extension of the comparison to genomes that diverged over a period three to four times longer, approaching the origin of symbiotic Buchnera, we also observe that diversity in terms of gene content between basal branchings boils down to “less” rather than “different.” This is a reinforcement of the conclusions of Tamas et al. (8), and one that demotes Buchnera to a provider of only a narrow range of essential nutrients, the requirements for which are apparently very similar among aphids, despite their feeding on an evolutionarily wide range of plant species. Seen from a different angle, however, one can also emphasize that, armed with Buchnera, aphids met a necessary and sufficient condition for their early radiation in the niche of plant-sap feeding.
Supplementary Material
Acknowledgments
We thank J. Pérez-Mercader for vision and support, and R. Gross and F. González-Candelas for reading the manuscript. This work was supported by Fondo Social Europeo, European Union; Fondo Social Europeo para el Desarrollo Regional, European Union; Comunidad Autónoma de Madrid, Spain; Comunidad Autónoma de València, Spain; Instituto Nacional de Técnica Aeroespacial, Spain; and Ministerio de Ciencia y Tecnología, Spain (Grants 2FD1997-1006 and BFM2000-1383).
Abbreviations
- BBp
complete genome sequence of Buchnera aphidicola from Baizongia pistaciae
- BAp
previously sequenced strain of B. aphidicola from Acyrthosiphon pisum
- BSg
previously sequenced strain of B. aphidicola from Schizaphis graminum
- LCSA
last common, symbiotic ancestor
- SNPs
single-nucleotide polymorphisms
Footnotes
References
- 1.Ochman H, Moran N A. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 2.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. New York: Interscience; 1965. pp. 297–332. [Google Scholar]
- 3.Baumann P, Baumann L, Lai C Y, Rouhbakhsh D, Moran N A, Clark M A. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 4.Douglas A E. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 5.Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc London Ser B. 1993;253:167–171. [Google Scholar]
- 6.Gil R, Sabater-Munoz B, Latorre A, Silva F J, Moya A. Proc Natl Acad Sci USA. 2002;99:4454–4458. doi: 10.1073/pnas.062067299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 8.Tamas I, Klasson L, Canback B, Naslund A K, Eriksson A S, Wernegreen J J, Sandstrom J P, Moran N A, Andersson S G. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 9.Fraser C M, Fleischmann R D. Electrophoresis. 1997;18:1207–1216. doi: 10.1002/elps.1150180803. [DOI] [PubMed] [Google Scholar]
- 10.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Delcher A L, Harmon D, Kasif S, White O, Salzberg S L. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borodovsky M, McIninch J. Comput Chem. 1993;17:123–133. [Google Scholar]
- 13.Lowe T M, Eddy S R. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bastolla U, Vendruscolo M, Knapp E W. Proc Natl Acad Sci USA. 2000;97:3977–3981. doi: 10.1073/pnas.97.8.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bastolla U, Farwer J, Knapp E W, Vendruscolo M. Proteins. 2001;44:79–96. doi: 10.1002/prot.1075. [DOI] [PubMed] [Google Scholar]
- 17.Clark M A, Moran N A, Baumann P. Mol Biol Evol. 1999;16:1586–1598. doi: 10.1093/oxfordjournals.molbev.a026071. [DOI] [PubMed] [Google Scholar]
- 18.Moran N A. Proc Natl Acad Sci USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silva F J, Latorre A, Moya A. Trends Genet. 2001;17:615–618. doi: 10.1016/s0168-9525(01)02483-0. [DOI] [PubMed] [Google Scholar]
- 20.Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. Nat Genet. 2002;32:402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- 21.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, et al. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 22.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 23.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 24.Rupp W D. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol. Press; 1996. pp. 2277–2294. [Google Scholar]
- 25.Marians K J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol. Press; 1996. pp. 749–764. [Google Scholar]
- 26.Kim S, Dallmann H G, McHenry C S, Marians K J. J Biol Chem. 1996;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Xu L, Sandler S J, Marians K J. Proc Natl Acad Sci USA. 1999;96:3552–3555. doi: 10.1073/pnas.96.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Dallmann H G, McHenry C S, Marians K J. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 29.Dallmann H G, Kim S, Pritchard A E, Marians K J, McHenry C S. J Biol Chem. 2000;75:15512–15519. doi: 10.1074/jbc.M909257199. [DOI] [PubMed] [Google Scholar]
- 30.Bryngelson J D, Wolynes P G. Proc Natl Acad Sci USA. 1987;84:7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutin A M, Abkevich V I, Shakhnovich E I. Proc Natl Acad Sci USA. 1995;92:1282–1286. doi: 10.1073/pnas.92.5.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bastolla U, Roman H E, Vendruscolo M. J Theor Biol. 1999;200:49–64. doi: 10.1006/jtbi.1999.0975. [DOI] [PubMed] [Google Scholar]
- 33.Fares M A, Ruiz-González M X, Moya A, Elena S F, Barrio E. Nature. 2002;417:398. doi: 10.1038/417398a. [DOI] [PubMed] [Google Scholar]
- 34.Baumann P, Baumann L, Clark M A. Curr Microbiol. 1996;32:279–285. doi: 10.1007/s002849900016. [DOI] [PubMed] [Google Scholar]
- 35. Moran, N. A. & Mira, A. (2001) Genome Biol.2, research0054.1–12. [DOI] [PMC free article] [PubMed]
- 36.Moran N A. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 37.Lynch M, Bürger R, Butcher D, Gabriel W. J Hered. 1993;84:339–344. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 38.Rispe C, Moran N A. Am Nat. 2000;156:425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- 39.Fukatsu T, Ishikawa H. J Insect Physiol. 1992;38:765–773. [Google Scholar]
- 40.Moran N A, Baumann P. Trends Ecol Evol. 1994;9:15–20. doi: 10.1016/0169-5347(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 41.von Dohlen C D, Kohler S, Alsop S T, McManus W R. Nature. 2001;412:433–436. doi: 10.1038/35086563. [DOI] [PubMed] [Google Scholar]
- 42.Baumann L, Baumann P, Moran N A, Sandstrom J, Thao M L. J Mol Evol. 1999;48:77–85. doi: 10.1007/pl00006447. [DOI] [PubMed] [Google Scholar]
- 43.Lai C Y, Baumann L, Baumann P. Proc Natl Acad Sci USA. 1994;91:3819–3823. doi: 10.1073/pnas.91.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.