Abstract
Identifying the genes responsible for human diseases requires combining information about gene position with clues about biological function. The recent availability of whole-genome data sets of RNA and protein expression provides powerful new sources of functional insight. Here we illustrate how such data sets can expedite disease-gene discovery, by using them to identify the gene causing Leigh syndrome, French-Canadian type (LSFC, Online Mendelian Inheritance in Man no. 220111), a human cytochrome c oxidase deficiency that maps to chromosome 2p16-21. Using four public RNA expression data sets, we assigned to all human genes a “score” reflecting their similarity in RNA-expression profiles to known mitochondrial genes. Using a large survey of organellar proteomics, we similarly classified human genes according to the likelihood of their protein product being associated with the mitochondrion. By intersecting this information with the relevant genomic region, we identified a single clear candidate gene, LRPPRC. Resequencing identified two mutations on two independent haplotypes, providing definitive genetic proof that LRPPRC indeed causes LSFC. LRPPRC encodes an mRNA-binding protein likely involved with mtDNA transcript processing, suggesting an additional mechanism of mitochondrial pathophysiology. Similar strategies to integrate diverse genomic information can be applied likewise to other disease pathways and will become increasingly powerful with the growing wealth of diverse, functional genomics data.
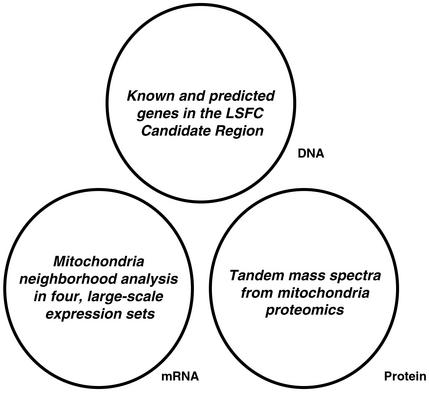
Positional cloning of human disease genes remains a challenging task. It has been facilitated by the availability of the draft sequence of the human genome (1) but remains difficult because genetic mapping provides only limited recombination resolution, especially for complex disorders. Sifting through many poorly characterized genes continues to be formidable. Traditionally, basic gene properties such as tissue expression patterns and protein domain analysis have been used to guide the selection of candidate genes. Ideally, geneticists would be equipped with richer sources of structural and functional information. Fortunately, genomics has sparked the creation of vast databases of biological information including genome sequence, genetic variation, RNA expression, protein-interaction networks, and others. These large-scale experimental data sets provide a wealth of experimental observations that, in principle, can be used to detect subtler correlations between gene properties and disease properties without the need for patient samples. In the current study, we describe the integration of global information about DNA, mRNA, and protein (Fig. 1) to facilitate disease-gene identification. We report the successful application of this approach to a human cytochrome c oxidase (COX) deficiency, Leigh syndrome, French-Canadian type (LSFC).
Figure 1.
DNA, mRNA, and protein data sets that are used in this study.
COX is the terminal complex of the electron-transport chain and serves to transfer reducing equivalents from cytochrome c to molecular oxygen. COX consists of 13 structural subunits, three of which are encoded by mitochondrial DNA (mtDNA). Many accessory proteins are needed for proper subunit assembly and coordination with heme and copper cofactors (2). Of the five clinically distinct autosomal recessive human COX deficiencies that have been described, the genes responsible for four forms have been cloned. Notably, all encode COX assembly factors (SURF1, SCO1, SCO2, and COX10) (2, 3). The fifth form of autosomal recessive COX deficiency is LSFC. It is characterized by Leigh syndrome (a subacute neurodegeneration of the brainstem and basal ganglia) and recurrent episodes of acute and often fatal metabolic acidosis and coma (4). COX activity is most diminished in the liver and brain and to a lesser extent in muscle, fibroblasts, and kidney (5). As a result of a founder effect (6), LSFC is common to the Saguenay-Lac St-Jean region of Quebec (population, ≈300,000), where it has a carrier rate of ≈1 in 23 individuals, with ≈1 in 2,000 live births being affected.
We previously used a genome-wide association study to map LSFC to chromosome 2p16-21 (7). This disease locus does not contain any of the genes encoding known COX structural subunits or assembly factors. The clinical and biochemical features of LSFC suggest that the pathway underlying this disorder is involved in mitochondrial biology. To detect functional relationships between the candidate genes and the putative disease pathway, we designed a strategy to integrate three types of genomic information: DNA sequences of known and predicted human genes, mRNA expression profiles from a wide range of cells and tissues, and tandem MS data from a mitochondria proteomics project (Fig. 1). The integrative approach identifies a single candidate gene, LRPPRC (leucine-rich pentatricopeptide repeat-containing protein), which we show to be the causative gene underlying LSFC.
Methods
Annotation of the LSFC Candidate Region.
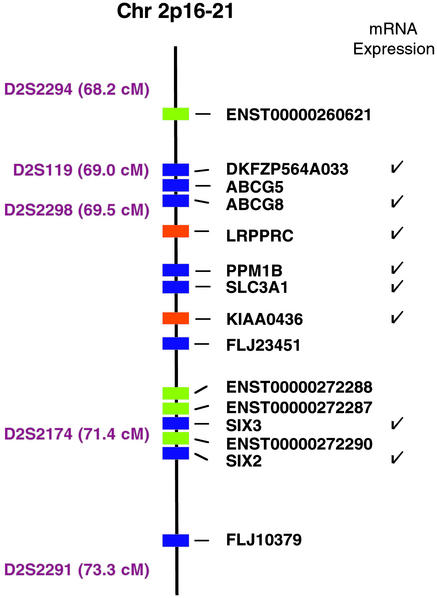
The LSFC candidate region, defined by microsatellite markers D2S2294 (68.2 centimorgans) and D2S2291 (73.3 centimorgans), corresponds to the physical coordinates chr2:46994838–48992238 (Human Genome, August 2001, hg8 assembly). We downloaded annotation tables using the University of California (Santa Cruz) genome browser, http://genome.ucsc.edu/. This region contains 9 RefSeq genes, 19 Ensembl transcripts, 32 human mRNAs, and 33 Genscan predictions. We collapsed these genes and transcripts and found that there were 15 distinct, nonoverlapping genes with RefSeq, Ensembl transcript, or human mRNA support (shown in Fig. 2) and an additional 15 nonoverlapping Genscan predictions.
Figure 2.
Physical map of the LSFC candidate region (Human Genome, August 2001, chr2:46994838–48992238). Microsatellite markers and genetic distances are shown to the left of the chromosome map. Genes with varying levels of annotation support are shown with different colors (RefSeq gene, blue; Ensembl gene, green; human mRNA, orange). An additional 15 computationally predicted genes lie within this region but are not shown. Genes represented in mRNA expression sets are indicated with a check to the right of the gene names.
Mapping Microarray Data Sets to the Genome.
We used a Perl wrapper in conjunction with the BLAST-like alignment tool (BLAT) to align all consensus probe sequences to the August 2001 assembly of the human genome (8). All sequence alignments within the LSFC candidate region then were confirmed by using a higher stringency translated DNA BLAT procedure. Only top-ranking alignments within the LSFC candidate region were then evaluated for overlap with the genome annotations. Any such probe with strong experimental support (known gene, human mRNA, or Ensembl transcript) was reannotated and shown in Fig. 2. Alignment statistics and probe annotations are available at http://www-genome.wi.mit.edu/mpg/lsfc/.
Mitochondria Neighborhood Analysis.
We used four large-scale, publicly available mRNA expression data sets: expression set 1, the normal samples from a Whitehead Institute cancer-classification project (14 normal human tissues; 16,063 genes/ESTs, 393 mitochondrial) (9); expression set 2, the RIKEN (Japan) Expression Array Database data set (49 embryonic/adult mouse tissues; 14,561 genes/ESTs, 290 mitochondrial) (10); expression set 3, the Genomics Institute of the Novartis Research Foundation (San Diego) expression atlas for human (25 human cell lines; 12,600 genes, 356 mitochondrial); and expression set 4, the Genomics Institute of the Novartis Research Foundation expression atlas for mouse (46 adult mouse tissues; 10,043 genes/ESTs, 345 mitochondrial) (11).
The expression data sets were downloaded from the web sites of each institution (Whitehead Institute, http://www-genome.wi.mit.edu/mpr/gcm.html; RIKEN, http://read.gsc.riken.go.jp/; and Genomics Institute of the Novartis Research Foundation, http://expression.gnf.org/). We also downloaded the consensus FASTA files corresponding to the HG-U95Av2, MG-U74Av2, HU6800, and Hu35KsubA probe sets (www.affymetrix.com/, Affymetrix, Santa Clara, CA), as well as the “19K” sequences from the RIKEN READ database (http://read.gsc.riken.go.jp/).
To identify oligonucleotide and cDNA sequences represented on the expression chips that correspond to known mitochondrial genes, we performed a BLASTX search of probe-set consensus sequences against the entries in MITOP (12). A human or mouse probe meeting a BLASTX threshold E < e−30 or E < e−15, respectively, was annotated as mitochondrial. For expression sets 1, 3, and 4 (Affymetrix oligonucleotide arrays), all average differences below 20 were clipped, and then the gene vectors were centered to 0 and normalized to a variance of 1. Expression set 2, which was normalized already, was used in the downloaded format.
The mitochondria neighborhood index, NR, is defined as the number of mitochondrial genes that lie within the query gene's R nearest expression neighbors. For a given query, G, all genes represented in the expression set are first ordered according to the Euclidean distance of their normalized expression pattern across all experimental conditions. Of the R genes with expression patterns that are most similar to G, NR(G) is the number of genes that are annotated as mitochondrial. Normalized expression sets, mitochondrial annotations, and neighborhood analysis results are available at http://www-genome.wi.mit.edu/mpg/lsfc/.
Purification of Mitochondria.
Human HepG2 cells were cultured in DMEM supplemented with FCS. Cells (≈1 × 109) were harvested, and mitochondria were isolated as described (13). The crude mitochondrial preparation then was loaded onto a discontinuous gradient consisting of 4 ml of 30% (vol/vol) Percoll and 4 ml of 70% Percoll centrifuged at 20,000 rpm for 40 min at 4°C in a Sorvall TH641 swinging bucket rotor. The mitochondria collected from the 30–70% interface were washed in isolation buffer with 1 mg/ml BSA. Purity of the preparation was confirmed by Western blot analysis by using an antibody against cytochrome c (1:1,000 dilution, Molecular Probes) and against calreticulin (1:20,000, Calbiochem), an endoplasmic reticulum marker. We also performed transmission electron microscopy of the purified mitochondria and confirmed that the preparation was free of contamination by other organelles.
Tandem MS of Mitochondrial Proteins.
Mitochondria were solubilized in 6 M guanidine hydrochloride containing 20 mM Tris⋅HCl, pH 8.5, and 50 mM DTT for 30 min. Proteins were alkylated with 300 mM iodoacetamide for 1 h, and the resulting mixture was applied to a Superdex 200 HR (Amersham Pharmacia) column equilibrated with 6 M guanidine hydrochloride in 100 mM NH4HCO3. The proteins were eluted at a flow rate of 0.4 ml/min. Samples (500 μl) were collected and then digested with 2.5 μg of endoproteinase Lys-C for 18 h. After 5-fold dilution with 100 mM NH4HCO3, the digestion was continued in the presence of 2.5 μg of trypsin for 6 h. Finally, the peptides were desalted on solid-phase extraction cartridges (Oasis, Waters) and freeze-dried for later use in liquid chromatography MS/MS. For liquid chromatography MS/MS, an Agilent 1100 HPLC system was coupled directly to a QSTAR Pulsar quadrupole time-of-flight mass spectrometer (PE/MDS Sciex, Toronto) equipped with a nanoelectrospray ion source (Protana Engineering, Odense, Denmark). The samples were loaded onto a C18 PepMap nano-precolumn (LC Packings, San Francisco) and eluted into the mass spectrometer through a 75-μm-i.d. fused-silica emitter (New Objective, Cambridge, MA) packed with 3.5-μm Zorbax C18 reverse-phase material (Agilent, Palo Alto, CA). Solvent A was 0.4% acetic acid/0.005% heptafluorobutyric acid in water; solvent B was 90% acetonitrile/0.4% acetic acid/0.005% heptafluorobutyric acid in water.
Mapping Tandem Mass Spectra to the LSFC Candidate Region.
MS/MS spectra were searched against a custom nucleotide database of 93 entries consisting of all RefSeq genes, human mRNAs, Ensembl transcripts, and Genscan transcripts within the LSFC candidate region. Data sets were searched by using the MASCOT software (Matrix Sciences, London). Peptides achieving a MASCOT peptide score of >25 and confirmed by manual inspection were deemed significant.
Mutation Screening and Genotyping.
Screening of DNA from LSFC patients and Centre d'Etude du Polymorphisme Humain (Paris) controls for mutations occurred at the Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research and at the Montreal Genome Centre. PCR products were designed to span all exons and exon–intron boundaries. The PCR primers used by each group are available at http://www-genome.wi.mit.edu/mpg/lsfc/ and www.genome.mcgill.ca/. DNA was sequenced for all 38 exons of LRPPRC for both strands by using the ThermoSequenase BigDye direct cycle-sequencing kit (PE Applied BioSystems). Mutations and polymorphisms in LRPPRC were corroborated at both sites.
After mutation discovery, genotyping was performed in the remaining patients, parents, and healthy controls. Genotyping of the exon 9 mutation was performed by resequencing, as described above. Genotyping of the exon 35 mutation was performed by primer extension of PCR products and detection by matrix-assisted laser desorption ionization time-of-flight MS. PCR (5′-AGCGGATAACTCAGAAACCTTCACTTACTG-3′ and 5′-AGCGGATAACGGAACAACAACAAAATCGGG-3′) and homogeneous MassExtend (5′-AATAATGTTTTAATTTTTAGAGAT-3′) primers were designed by using SpectroDesigner (Sequenom, San Diego). PCR products were purified by using the shrimp alkaline phosphatase method and extended by addition of the homogeneous MassExtend primer as per the Sequenom MassArray protocol. All traces were inspected visually by two observers.
Recently, an autosomal recessive syndrome of cystinuria was reported (14) and found to be caused by an ≈179-kb deletion in 2p16 that includes at least three genes (SLC3A1, PPM1B, and KIAA0436). Because this syndrome included some clinical features of mitochondrial encephalomyopathies, we screened LSFC DNA for mutations in these three genes. No mutations were identified.
Results
Identifying Known and Predicted Genes Within the LSFC Candidate Region.
We previously used a genome-wide association study to map LSFC to chromosome 2p16-21 and identified a common ancestral haplotype found in all patients (7). Individual recombination events allowed us to refine the location of the LSFC gene to a 2.4-centimorgan region (flanked by microsatellite markers D2S119 and D2S2174; Fig. 2). To be conservative, in the present study we chose not to rely on single crossover events and therefore examined the 5.1-centimorgan region flanked by two recombinants, which we term the LSFC candidate region (Fig. 2).
We systematically analyzed the LSFC candidate region and curated a comprehensive list of the known and predicted transcripts. We identified a total of 15 distinct, nonoverlapping genes with strong experimental support (from the RefSeq, Ensembl transcript, or human cDNA collections; Fig. 2). Nine of these genes (from the RefSeq collection) have well established gene structures, whereas the remaining six have structures inferred from computational predictions and mRNA overlap. In addition, the region contains an additional 15 nonoverlapping, de novo computational gene predictions (Genscan) (15).
Neighborhood Analysis of Large-Scale mRNA Expression Data Sets.
Next, because functionally related genes tend to be transcriptionally coregulated (16), we sought to systematically identify genes that exhibit mRNA expression patterns resembling those of known mitochondrial genes. We began by examining a set of ≈300 well studied, nuclear-encoded genes known to encode proteins localized to the mitochondrion (12). These genes, particularly those involved in oxidative phosphorylation, tend to be coregulated in yeast, as shown by microarray experiments (16). We hypothesized that mouse and human mitochondrial genes are also transcriptionally coregulated, and that genes of unknown function that are coregulated with this group may encode polypeptides targeted to this organelle, or perhaps nonmitochondrial proteins that are needed for its proper biogenesis. To search for such coregulated genes, we created a measure, NR, which we refer to as the mitochondria neighborhood index (Fig. 3A). For a given gene, G, we identify the R neighboring genes with the most similar expression pattern (using an appropriate measure of similarity, see Methods), and we then define NR(G) as the number of known mitochondrial genes that lie among these R genes. For example, if the 100 nearest expression neighbors of a given gene G include 20 that encode mitochondrial proteins, then N100(G) = 20. We examined four large, publicly available microarray data sets that profile the expression of ≈10,000–16,000 genes across 134 mouse and human tissue samples and calculated NR for all genes in each data set. In this way, we were able to organize all human genes according to their similarity in expression space to known mitochondrial genes. As predicted, genes encoding known mitochondrial proteins tended to have proportionately higher values of NR compared with random genes (Fig. 3B), demonstrating that they are coregulated in these data sets and validating the mitochondria neighborhood index.
Figure 3.
Evaluating mRNA expression neighborhoods for enrichment in mitochondrial genes. (A) Schematic illustration of the mitochondria neighborhood index. The coordinate of each gene (circle) is defined by its expression vector in an mRNA microarray experiment. Genes are close to one another if they have similar expression profiles (based on an appropriate distance metric, see Methods). The mitochondria neighborhood index, NR(G), is defined as the number of known mitochondrial genes (orange circles) among the R nearest neighbors of the query gene, G (blue circle). In this cartoon, N10 = 5 because there are five mitochondrial genes within the query's 10 nearest-neighboring genes. (B) Distribution of N100 values. The blue histogram shows the distribution of N100 for all genes, and the red histogram plots N100 for known mitochondrial genes, in expression set 4. *, the histogram bin containing LRPPRC (see text and Table 1).
Mitochondrial Proteomics.
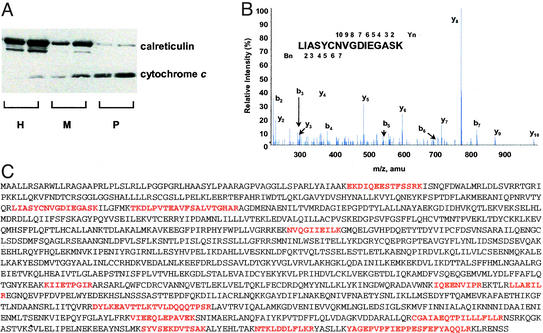
Next, we used proteomics data from an ongoing effort to systematically characterize the protein constituents of this organelle. Briefly, we purified mitochondria from human HepG2 cells using differential centrifugation and confirmed the purity of the preparation by immunoblot analysis (Fig. 4A) and electron microscopy (data not shown). The mitochondrial proteins were size-fractionated and digested with trypsin, and the resulting peptides were subjected to analysis by tandem MS/MS (17). We acquired high-quality MS/MS spectra (Fig. 4B), each corresponding to a mitochondrial peptide. The mass spectra can be correlated to fragment masses predicted by the in silico digest of a given nucleotide or protein sequence. Thus, organelle proteomics provides a tool for determining whether a query sequence may have given rise to an observed mitochondrial peptide.
Figure 4.
Organelle proteomics. (A) Western blot of human HepG2 homogenate (H), crude mitochondrial fraction (M), and Percoll-purified mitochondria (P) probed with an antibody against cytochrome c, a marker for mitochondria, and calreticulin, a marker for contamination by endoplasmic reticulum. Two different loading volumes (5 and 10 μl) were used for each sample. (B) Representative tandem mass spectrum showing y-ion and b-ion series along with the deduced peptide sequence. (C) The predicted LRPPRC (GenBank accession no. XP_031527.3) amino acid sequence with high-scoring peptides, identified by organelle proteomics, marked in red.
Integration of DNA, RNA, and Protein Data Sets Pinpoints a Single Gene.
We combined the DNA, mRNA, and protein data sets (Fig. 1) with the goal of identifying the LSFC gene. First, we surveyed the literature and carefully reviewed the annotations of the 15 experimentally supported transcripts in the LSFC candidate region to search for evidence of any involvement in mitochondrial biology. However, none were known previously to be involved with mitochondrial function. Next, we sought to determine whether any of the LSFC candidate genes exhibit transcriptional coregulation with known mitochondrial genes. For this purpose, we were limited to those LSFC candidate genes (Fig. 2) that were represented in publicly available microarray experiments. One of these genes, LRPPRC, had a strikingly high mitochondrial neighborhood index, NR, across the four large microarray data sets (Table 1), comparable to that of known mitochondrial genes. The NR of LRPPRC was particularly prominent in expression set 4, where the well characterized mitochondrial genes themselves exhibit tightest coregulation (Fig. 3B). We then used the proteomic data set to determine whether any of the DNA sequences in the LSFC candidate region may have given rise to the observed mitochondrial peptide fragment spectra. For this purpose, we were able to test all genes (both known or experimentally supported genes and ab initio gene predictions). We found that all 12 high-scoring mitochondrial peptides that localized to the LSFC candidate region could be accounted for by a single gene, LRPPRC (Fig. 4C). Thus, two complementary approaches (mRNA neighborhood analysis and organelle proteomics) point to the same gene, LRPPRC, which thus emerges as our top-ranking candidate gene for LSFC.
Table 1.
Summary of mitochondria neighborhood analysis
| Gene | Set 1
|
Set 2
|
Set 3
|
Set 4
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N100, 2.4* | N250, 6.1 | N500, 12.2 | N100, 2 | N250, 5 | N500, 10 | N100, 2.8 | N250, 7 | N500, 14.1 | N100, 3.4 | N250, 8.6 | N500, 17.2 | |
| DKFZP564A033 | 0 | 1 | 1 | 0 | 2 | 3 | 1 | 5 | 7 | |||
| 1 | 1 | 4 | 2 | 4 | 7 | |||||||
| 2 | 4 | 6 | ||||||||||
| ABCG8 | 0 | 0 | 3 | |||||||||
| LRPPRC | 5 | 20† | 34† | 4 | 8 | 14 | 8 | 13 | 17 | 52† | 74† | 76† |
| PPM1B | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 3 | 6 | 6 | 10 | 11 |
| 0 | 1 | 3 | 2 | 4 | 5 | 8 | 11 | 15 | ||||
| 5 | 9 | 12 | ||||||||||
| SLC3A1 | 8† | 10 | 15 | 1 | 5 | 9 | 7 | 23† | 27 | |||
| 3 | 3 | 6 | ||||||||||
| KIAA0436 | 1 | 6 | 14 | 2 | 4 | 8 | ||||||
| SIX3 | 2 | 4 | 6 | 0 | 4 | 4 | ||||||
| 1 | 2 | 5 | ||||||||||
| SIX2 | 2 | 3 | 8 | |||||||||
Mitochondria neighborhood index, NR, is shown for each query gene (shown in first column) across four different expression sets using three different values of R (R = 100, 250, and 500). E(NR) is the expected index assuming a binomial distribution of mitochondrial genes across the entire expression set. Boldface items denote the largest entry in a column. Note that some genes are represented more than once in an expression set, and the NR value for each instance is shown on a separate row. See Methods for a description of expression sets.
NR, E(NR).
† An index significantly greater than E(NR) (P < 0.001).
Mutations in LRPPRC Cause LSFC.
LRPPRC (GenBank XM_031527) encodes a poorly understood, 130-kDa protein initially identified on the basis of lectin affinity (18). Our proteomics data are supportive of a 38-exon gene structure for LRPPRC, with a total genomic length of ≈100 kb and a predicted transcript length of 5,098 bp.
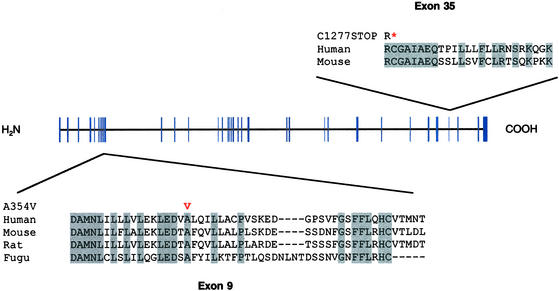
To prove that LRPPRC is responsible for LSFC, we sequenced the regions of all 38 exons in two patients, one parent, and one unrelated control. We identified a single base change (nucleotide 1,119 C → T transition) in exon 9 predicting a missense A354V change at a residue conserved in human, mouse, rat, and Fugu (Fig. 5). We then genotyped the remaining patients, and of 22 patients total, 21 were homozygous for the A354V mutation, and one was heterozygous. As expected, the A354V mutation was heterozygous in 31 of the 32 parents tested but is not present in 175 unrelated controls.
Figure 5.
Mutations identified in LRPPRC. LRPPRC has 38 exons (blue) predicted to encode a 1,394-aa protein. The amino acid sequence corresponding to exons 9 and 35 are shown as well as the aligned sequences from mouse, rat, and Fugu. The exon 9 missense mutation, A354V, and the exon 35 truncation, C1277STOP, are shown in red. Conserved residues are shaded in gray. *, a stop codon.
The only patient (COX015-35) that was heterozygous for the A354V amino acid change was, in fact, heterozygous for the founder-risk haplotype (data not shown). Because LSFC is an autosomal recessive disorder, we reasoned that he must harbor a second, distinct mutation and therefore screened all 38 exons in this patient. We identified an 8-nt deletion in exon 35 resulting in a premature stop at amino acid 1,277 (Fig. 5). Therefore, this patient is a compound heterozygote. The exon 35 deletion was found in this patient and his mother but not in any of 350 unrelated controls. The identification of two mutations in LRPPRC provides definitive genetic proof that LRPPRC is responsible for LSFC.
Discussion
In the present study we used an integrative genomics approach to identify the gene causing a human COX deficiency.
The genes previously identified as causing COX deficiencies are all believed to be involved with subunit assembly of the COX complex (SURF1) and prosthetic group addition (COX10, SCO1, and SCO2) (2, 3). At present, the precise role of LRPPRC is not known (19). Clues to its function come from recent studies of heteronuclear ribonuclear proteins, which comprise a family of polypeptides that bind to pre-mRNA and mRNA (20). These proteins are involved in the maturation and trafficking of mRNA. A recent report found LRPPRC to be bound to polyadenylated mRNA in a shuttling complex with heteronuclear ribonuclear protein K (20), which in separate studies has been shown to bind mtDNA-encoded mRNA (21). Furthermore, the reciprocal best match of LRPPRC in yeast is Pet309, a mitochondrial protein needed for the proper splicing and translational initiation of mtDNA-encoded COX mRNA (22, 23). The sequence similarity is too weak (pairwise BLASTP E = 0.037) to be reliable on its own. However, the fact that these genes are reciprocal best matches, are both implicated in mRNA processing, and are both connected by mutational evidence to mitochondrial biology suggests that LRPPRC and pet309 are true homologues. It further suggests that LRPPRC encodes an mRNA-binding protein involved in the processing and trafficking of mtDNA-encoded transcripts. Taken together, previous reports and our current findings suggest that LRPPRC participates in mRNA processing both in the nucleus and the mitochondrion. Clearly, functional studies are needed to test this hypothesis, decipher the mechanisms that target this protein to different compartments, and understand the consequences of the mutations identified in this study.
Defective processing of mtDNA-encoded mRNA represents a mechanism in mitochondrial pathophysiology that has not been previously described. Our findings should help advance our fundamental understanding of human mitochondrial translational control, an area that is poorly understood.
The identification of LRPPRC mutations will be beneficial in the Saguenay-Lac St-Jean region, where LSFC is common and associated with high infantile and childhood mortality. Our findings will enable carrier testing and provide improved prenatal diagnostic options to members of this community.
More generally, this project illustrates the power of combining whole-genome experimental data sets. We started with a 2.0-megabase region of the genome containing 30 known or predicted genes, and by combining functional data of mRNA expression and subcellular localization, we were able immediately to pinpoint the causative gene for LSFC. The approach is powerful, detects subtle gene properties, and obviates the need for patient biopsies. As improved gene annotation, higher-density mRNA expression sets, and more comprehensive protein-interaction maps become available, it should be possible to apply such integrative analysis more broadly. Moreover, other global views of gene function such as high-throughput physiology and systematic RNA interference-based disruptions will provide orthogonal information that can add more power to the approach.
Although the present project focused on a mitochondrial disease, the strategy can be tailored to other disorders in which an underlying pathway or subcellular compartment has been directly or indirectly implicated. For example, studies of diabetes mellitus could “rank” genes by similarity of RNA and protein expression to known genes involved in pathways related to the disease (such as free fatty acid metabolism and insulin signaling). Such an approach could identify new genes not known previously to be involved in the disease but transcriptionally coregulated with or physically interacting with members of these pathways.
Any single functional genomics measure may suffer from incomplete coverage, imperfect sensitivity, or low specificity, but the combination of data from cross-platform technologies can spotlight candidate genes with increased confidence. In this regard, the integrative analysis of functional genomics data holds promise for accelerating disease-gene discovery.
Acknowledgments
We are grateful to Ken Dewar, Mark Daly, James Engert, John Higgins, Joseph Lehar, Pablo Tamayo, Marc De Braekeleer, Caroline Beard, and Jean Larochelle for valuable discussions; Carole Dore, Jean Chang, April Cooke, Donna Sinnett, and Jacek R. Wisniewski for expert technical contributions; and Ben Gewurz, Joel Hirschhorn, David Altshuler, and members of the E.S.L. laboratory for comments on the manuscript. We express our heartfelt gratitude to the patients and their families for ongoing cooperation and support. This work was supported by grants from L'Association de L'Acidose Lactique du Saguenay-Lac St-Jean (to B.R., T.J.H., and J.D.R.), Canadian Genetics Disease Network (to B.R.), Genome Quebec (to T.J.H.), and Canadian Institutes of Health Research (to R.S., T.J.H., and B.R.), a Howard Hughes Medical Institute postdoctoral fellowship (to V.K.M.), and a grant from Affymetrix/Bristol-Myers Squibb/Millennium (to E.S.L.).
Abbreviations
- COX
cytochrome c oxidase
- LSFC
Leigh syndrome, French-Canadian type
- LRPPRC
leucine-rich pentatricopeptide repeat-containing protein
References
- 1.Lander E S, Linton L M, Birren B, Nussbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Shoubridge E A. Am J Med Genet. 2001;106:46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- 3.Robinson B H. Pediatr Res. 2000;48:581–585. doi: 10.1203/00006450-200011000-00004. [DOI] [PubMed] [Google Scholar]
- 4.Morin C, Mitchell G, Larochelle J, Lambert M, Ogier H, Robinson B H, De Braekeleer M. Am J Hum Genet. 1993;53:488–496. [PMC free article] [PubMed] [Google Scholar]
- 5.Merante F, Petrova-Benedict R, MacKay N, Mitchell G, Lambert M, Morin C, De Braekeleer M, Laframboise R, Gagne R, Robinson B H. Am J Hum Genet. 1993;53:481–487. [PMC free article] [PubMed] [Google Scholar]
- 6.De Braekeleer M, Dao T N. Hum Biol. 1994;66:205–223. [PubMed] [Google Scholar]
- 7.Lee N, Daly M J, Delmonte T, Lander E S, Xu F, Hudson T J, Mitchell G A, Morin C C, Robinson B H, Rioux J D. Am J Hum Genet. 2001;68:397–409. doi: 10.1086/318197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kent W J. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang C H, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov J P, et al. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miki R, Kadota K, Bono H, Mizuno Y, Tomaru Y, Carninci P, Itoh M, Shibata K, Kawai J, Konno H, et al. Proc Natl Acad Sci USA. 2001;98:2199–2204. doi: 10.1073/pnas.041605498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su A I, Cooke M P, Ching K A, Hakak Y, Walker J R, Wiltshire T, Orth A P, Vega R G, Sapinoso L M, Moqrich A, et al. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharfe C, Zaccaria P, Hoertnagel K, Jaksch M, Klopstock T, Lill R, Prokisch H, Gerbitz K D, Mewes H W, Meitinger T. Nucleic Acids Res. 1999;27:153–155. doi: 10.1093/nar/27.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mootha V K, Wei M C, Buttle K F, Scorrano L, Panoutsakopoulou V, Mannella C A, Korsmeyer S J. EMBO J. 2001;20:661–671. doi: 10.1093/emboj/20.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parvari R, Brodyansky I, Elpeleg O, Moses S, Landau D, Hershkovitz E. Am J Hum Genet. 2001;69:869–875. doi: 10.1086/323624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burge C, Karlin S. J Mol Biol. 1997;268:78–84. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 16.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 17.Pandey A, Mann M. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 18.Hou J, Wang F, McKeehan W L. In Vitro Cell Dev Biol Anim. 1994;30:111–114. doi: 10.1007/BF02631402. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, McKeehan W L. Genomics. 2002;79:124–136. doi: 10.1006/geno.2001.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mili S, Shu H J, Zhao Y, Pinol-Roma S. Mol Cell Biol. 2001;21:7307–7319. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostrowski J, Wyrwicz L, Rychlewski L, Bomsztyk K. J Biol Chem. 2002;277:6303–6310. doi: 10.1074/jbc.M110267200. [DOI] [PubMed] [Google Scholar]
- 22.Manthey G M, Przybyla-Zawislak B, McEwen J E. Eur J Biochem. 1998;255:156–161. doi: 10.1046/j.1432-1327.1998.2550156.x. [DOI] [PubMed] [Google Scholar]
- 23.Manthey G M, McEwen J E. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]