Abstract
Transplantation immunity based on the recognition of MHC molecules is well described in vertebrates. Vertebrates, however, do not undergo transplantation reaction naturally. The phylogenetically closest group in which transplantation reactions can occur is the Urochordata. Therefore, these animals occupy a key position for understanding the evolution of the vertebrate immune system. When screening for genes differentially expressed during allorecognition in Botryllus schlosseri, we isolated a gene coding for a type II transmembrane protein with a C-type lectin-binding domain and close similarity to vertebrates CD94 and NKR-P1. Here we show that the gene, BsCD94-1, is differentially regulated during allorecognition and that a subpopulation of blood cells carries the corresponding receptor on its cell surface. Southern blot analysis with DNA from individual colonies and intronless BsCD94-1 probe reveal variation between individuals at the genomic level. CD94 in vertebrates is one of the markers for natural killer cells and binds to MHC class I molecules. Natural killer cells play a major role in recognition and elimination of allogeneic cells. Their evolutionary origin, however, remained unknown. The results presented here indicate that the elaboration of the vertebrate immune system may have its roots in an ancestral population of cells in the urochordate blood.
The development of natural killer (NK) cells was a critical event in the evolution of the immune system (1). To discriminate between “normal” and virus-infected, tumor, or allogeneic cells, mammalian NK cells monitor MHC class I expression on target cells by means of inhibitory NK cell receptors (NKRs) (2–7). The receptors transmit an inhibitory signal that cancels a program for cytotoxic action previously triggered by contact with the target cell. NKRs belong to two distinct groups of molecules: immunoglobulin-like receptors and C-type lectin receptors such as CD94, NKG2, and NKR-P1 in humans and a group of Ly49 receptors in rodents (1). C-type lectin receptors are type II transmembrane glycoproteins with a C-type lectin domain in the extracellular region. They are known to be a hallmark of surface markers for NK cells (5, 8). Despite their significance, the evolutionary origin of NK cells and their receptors has remained a mystery.
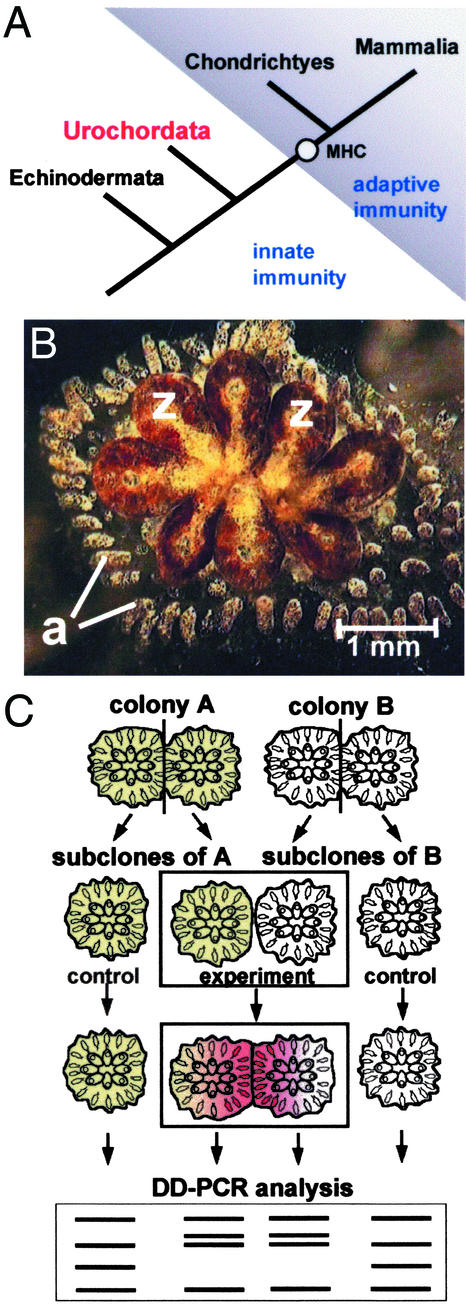
Botryllus schlosseri is a colonial invertebrate chordate that belongs to the earliest branch in the chordate phylum, the subphylum Urochordata (Fig. 1A). A Botryllus colony is composed of numerous zooids that are embedded within the tunic (Fig. 1B). Zooids within a colony commonly form star-shaped clusters around common cloacal apertures. All zooids within a cluster, as all clusters within a colony, are connected one to the other by an extensive network of blood vessels. This common blood system bears sausage-like termini called vascular ampullae, which are found along the periphery of the colony and scattered among the clusters (Fig. 1B). Pairs of colonies that meet naturally or are placed in contact under laboratory conditions either fuse their contacting peripheral ampullae and form an chimeric organism or develop cytotoxic lesions in the contact zone. Rejection appears ≈1 day after contact between nonfusible colonies. According to population genetic studies, allorecognition is controlled by a single fusibility/histocompatibility (Fu/HC) locus with multiple codominantly expressed alleles (9–13). Rejection develops between colonies that do not share alleles of the Fu/HC locus (9–11, 14). The extensive polymorphism resembles the polymorphism of the vertebrate MHC (11, 14, 15). The molecular mechanisms controlling urochordate allorecognition are not known (16). Conspicuous cellular participants in botryllid rejection reactions are the morula cells (MCs), which accumulate at the tips of interacting ampullae, infiltrate the tunic matrix through the ampullar epithelium (17) and cause the formation of lesions called points of rejection. The role of other ascidian blood cells in allogeneic response is unclear. However, in vitro cytotoxic activity against mammalian targets has been described for some granulocytes in the solitary urochordate Ciona intestinalis (18). Here we applied a nonbiased screening strategy to isolate genes differentially expressed during B. schlosseri allorecognition. As a result, we have identified a CD94-related transmembrane receptor protein, a specific cell surface marker of vertebrate NK cells, expressed on the surface of a subpopulation of Botryllus blood cells.
Figure 1.
(A) Phylogeny and the evolution of adaptive immunity. B. schlosseri belongs to the subphylum Urochordata (Tunicata). None of the components of the adaptive immune system have been found yet below the chondrichthyes. (B) A B. schlosseri colony. a, ampullae; z, zooid. A colony consisting of eight zooids is ≈4 mm in diameter. (C) Outline of the experimental approach used in DD PCR nonbiased screening for self/nonself recognition genes.
Materials and Methods
Animals and Colony Allorecognition Assay.
Colonies of B. schlosseri were obtained from the Haifa laboratory and Helgoland Biological Station (Helgoland, Germany). For the colony allorecognition assay (15), a pair of subclones, made of two incompatible colonies (containing 8–20 zooids each), were juxtaposed on a glass slide so that animals grew toward each other (see Fig. 1C). At various time points (24–72 h) after contact of the extending ampullae, the interacting subclones were separated and total RNA was extracted. For control, total RNA was also extracted from naive colonies of the same genotype. The control subclones were grown under conditions identical to those for the experimental colonies but were not brought into contact with any other colony.
Differential Display (DD) PCR and Isolation of BsCD94-1.
To monitor gene expression during allorecognition, total RNA was isolated from individual colonies and subjected to the described nonradioactive DD PCR (19). BsCD94-1 cDNA was isolated by using primers T(12)GC and OPA-9 (Operon Technologies, Alameda, CA). The full-length cDNA sequence was obtained by 5′ and 3′ RACE PCR.
Molecular Techniques.
Nucleic acid isolation, cDNA cloning, and DNA sequence analysis were carried out by following standard procedures. For a positive control in RT-PCR, we cloned a part of the Botryllus β-actin gene. For Southern blot analysis, 10–15 μg of genomic DNA was digested with restriction endonucleases. DNA was separated on a 0.7% agarose gel and transferred onto a nylon membrane (Hybond N+, Amersham Pharmacia). The hybridization probe of 305 bp corresponding to nucleotides 295–600 of the full-length cDNA (Fig. 2A) was PCR-amplified (5′ primer, TGCGGAAAATGCACGACGTGATCG; 3′ primer, GGCCAATGCGGTCAACTTATCGTC), gel-purified, and labeled with [α-32P]dCTP. The corresponding sequence isolated with the same primer pair from genomic DNA was identical to the cDNA sequence and contained no introns. Southern hybridization was performed in the hybridization solution [6× SSC (standard saline citrate, 1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), 0.5% SDS, 5× Denhardt's solution, and 0.5% blocking reagent] at 65°C overnight. Stringent washing was performed in 0.1× SSC/0.1% SDS at 65°C.
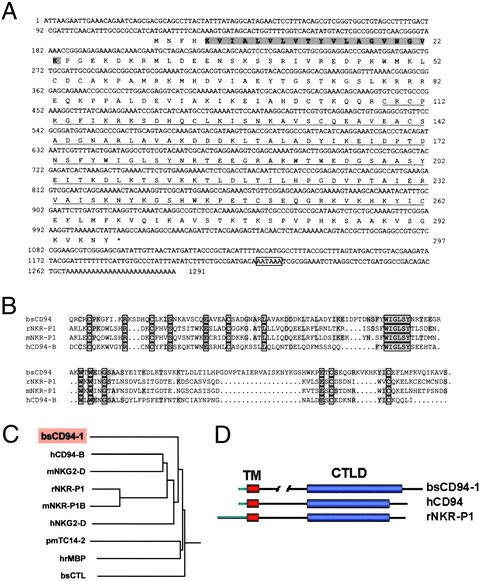
Figure 2.
BsCD94-1 cDNA sequence and alignment with most closely related receptors. (A) Nucleotide sequence and deduced amino acid sequence of BsCD94-1. The transmembrane region is shaded. The C-type lectin-binding domain (CTLD) is underlined. A putative polyadenylation signal is boxed. (B) Amino acid sequence comparison of BsCD94-1 CTLD region with the corresponding regions in rat NKR-P1, mouse NKR-P1, and human CD94-B. (C) Homology tree of BsCD94-1 and related CTLD molecules. BsCD94-1 groups together with transmembrane receptors of NK cells. It is distinct from urochordate soluble lectins such as Botryllus C-type lectin (bsCTL), Halocynthia roretzi mannose-binding protein (hrMBP), and lectin pmTC14-2 from Polyandrocarpa mysaciensis. (D) Schematic diagram depicting the structural similarities among Botryllus BsCD94-1, human CD94, and rat NKR-P1, showing the short cytoplasmic domain of BsCD94-1. TM, transmembrane region.
Preparation of the BsCD94-1 Antiserum.
The antiserum to BsCD94-1 was generated by injecting mice (BALB/c) with a recombinant BsCD94-1 protein according to standard procedures. Recombinant BsCD94-1 was produced by cloning the cDNA fragment corresponding to amino acids 60–297 (Fig. 2A) in vector pCR T7/NT-TOPO (Invitrogen). Escherichia coli cells were transformed and induced by isopropyl β-d-thiogalactoside (IPTG), following the manufacturer's manual. Recombinant protein was purified by using Ni-NTA His-Bind resin (Novagen). Specificity of the serum was examined by Western blot analysis (see Fig. 4) with recombinant protein as well as protein extract from isolated blood cells and from whole tissue. No staining of Botryllus blood cells was observed with preimmune serum.
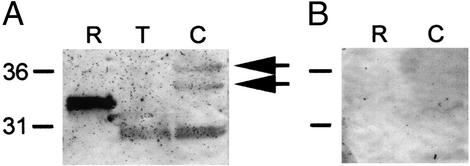
Figure 4.
Antiserum against BsCD94-1 detects specifically the recombinant protein as well as three proteins in Botryllus extract. (A) Western blot using the antiserum and recombinant BsCD94-1 protein (R), protein extract from whole tissue (T), and protein extract from isolated blood cells (C). Equal amounts of extracts were loaded in each lane. Arrows indicate two proteins of approximately expected size in Botryllus blood cells. (B) Western blot with preimmune serum and recombinant protein (R) as well as protein extract from isolated blood cells (C) shows no staining.
Immunofluorescent Staining of Botryllus Colonies and Cells.
Botryllus colonies fixed in 3.7% formaldehyde in sea water on glass slides were washed three times for 30 min in PBBT (1× PBS/1.5% BSA/0.02% Tween 20) at room temperature. Thereafter, antiserum was added to final dilution of 1:500 and incubated for 12 h at 4°C. Colonies were washed three times for 30 min in 1× PBS/0.2% Tween 20. Tetramethylrhodamine B isothiocyanate (TRITC)-conjugated anti-mouse Fab fragments (Sigma) were used for detection at a dilution of 1:5,000 in PBBT. For immunostaining of Botryllus blood cells, cells were isolated from several colonies, washed three times in filtered sea water, and fixed in 4% formaldehyde in sea water. The staining procedure was essentially as described for colonies. To exchange solutions, at the end of every immunostaining step cells were pelleted by centrifugation at 1,500 × g for 10 min.
Results
Nonbiased Screening for Allorecognition-Related Genes Leads to Identification of a CD94/NKR-P1-Related Gene in Botryllus.
To identify genes involved in allorecognition in urochordates, we developed a screening strategy that makes use of the remarkable allorejection process of B. schlosseri and the “colony allorecognition assay” (15). The experimental approach is shown schematically in Fig. 1C. Five independent experiments were performed in parallel. Only cDNAs displaying up- or down-regulation after allocontact in all five experiments were examined further. Of a total of 1,200 analyzed transcripts, 50 (4.2%) were found to be differentially expressed. One of these was represented in a cDNA of ≈600 bases predicted to encode a part of the C-type lectin-binding domain (CTLD) similar to those of receptors of NK cells. Full-length cDNA sequence was obtained by 5′ and 3′ RACE PCR with cDNA from several compatible and incompatible individual colonies. As shown in Fig. 2A, the full-length sequence is 1,291 bases long, corresponds to a 1.3-kb transcript on Northern blots (Fig. 3A), and codes for an ORF of 297 amino acids (Fig. 2A). Full-length cDNAs from two compatible and two incompatible colonies showed only minor changes at the nucleotide level, mostly at the 3′ UTR, which did not alter the predicted protein sequence (data not shown). The predicted protein has a molecular mass of 33.5 kDa. Hydrophobicity analysis revealed a 19-aa transmembrane domain at the N terminus. A blast search comparison of the Botryllus gene indicated the presence of a CTLD with highest similarity to CTLDs of NK receptors NKR-P1, NKG2-D, and CD94 (Fig. 2B) and revealed strong conservation of amino acids known to be of structural and functional relevance. Most prominent among these are a number of conserved cysteines involved in formation of two disulfide bonds as well as two conserved regions (FXWIGL and WXWXDG) described (20) as essential for β-sheet formation (Fig. 2B). Phylogenetic tree analysis (Fig. 2C) shows that the CTLD of the Botryllus protein groups together with CTLDs in vertebrate NK receptors and is distinct from the soluble C lectins described previously in urochordates. Interestingly, and in contrast to proteins from the NKG2 family, the intracellular region of the Botryllus protein is very short, thus resembling the short 7-aa cytoplasmic domain of human CD94 (see Fig. 2D). For that reason we have decided to name the Botryllus gene BsCD94-1.
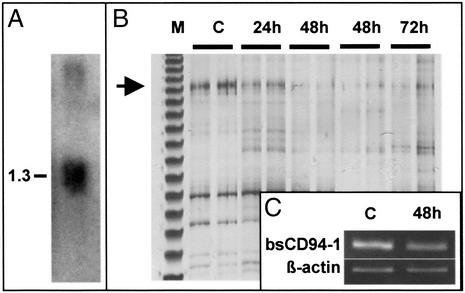
Figure 3.
Expression of BsCD94-1. (A) Northern blot analysis of BsCD94-1. Three micrograms of poly(A)+ RNA was hybridized with the full-length BsCD94-1 cDNA. (B) The DD PCR gel from which the 657-bp cDNA fragment (arrow) of BsCD94-1 was isolated. The transcript is transiently down-regulated after allocontact. M, size marker; C, control colony; 24h, 48h, and 72h, hours after allocontact. (C) RT- PCR analysis of BsCD94-1 mRNA expression in Botryllus cells. C, control; 48h, hours after allocontact. Botryllus β-actin was used as a loading control.
Expression of BsCD94-1.
Northern blot analysis with mRNA from individual colonies revealed the presence of a 1.3-kb BsCD94-1 transcript (Fig. 3A). Because the size of the transcript is similar to the length of the cDNA shown in Fig. 2A, it appears that the deduced amino acid sequence represents a full-length protein. Initial DD PCR screening suggested that the BsCD94-1 gene is transiently down-regulated after allogenic contact (Fig. 3B). The differential expression pattern was confirmed by RT-PCR, using BsCD94-1 sequence-specific primers (Fig. 3C). The quantity of the cDNA template was equalized by using Botryllus β-actin-specific primers. To analyze which tissues or cells expressed the gene, Botryllus blood cells were isolated from whole colonies. When mRNA was extracted from these cells and analyzed by RT-PCR, BsCD94-1 transcripts were detected in blood cells (data not shown).
BsCD94-1 Receptor Is Localized on the Surface of a Subpopulation of Botryllus Blood Cells.
To localize the CD94-related receptor on Botryllus blood cells, we prepared a polyclonal antiserum by immunizing mice with soluble recombinant BsCD94-1 protein (see Materials and Methods). In Western blots (Fig. 4), the serum specifically detected the recombinant BsCD94-1 protein as well as three proteins in Botryllus extracts. In extract prepared from isolated blood cells, two proteins of ≈34 and 36 kDa migrating closely to the expected molecular mass of BsCD94-1 were detected (arrows in Fig. 4A). In addition, the serum recognized a smaller protein of ≈31 kDa. In extract prepared from whole tissue, only the small protein was observed. This protein remains to be identified. Preimmune serum did not stain recombinant protein nor any protein in Botryllus blood cell extract.
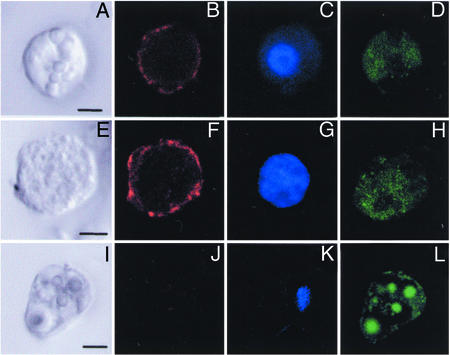
When this antiserum was used in immunocytochemistry, only a subpopulation of blood cells was stained. Immunopositive cells could be divided into three morphologically distinct groups (Fig. 5). The first group (Fig. 5 A–D) shows features of previously described B. schlosseri granulocytes. The nucleus in these cells is relatively small, and the cytoplasm contains small granules. The second group (Fig. 5 E–H) also shows features of granulocytes (21, 22). Morphologically, however, these cells have a large nucleus and a small cytoplasm-to-nucleus ratio, and they are of about 16 μm in diameter. The third group (data not shown) is made up of large cells with a single vacuole and a small kidney-shaped nucleus. These cells in B. schlosseri were previously referred to as signet ring cells; in C. intestinalis they are known as univacuolar refractile granulocytes (22). Optical sectioning by confocal microscopy (Fig. 5 B and F) indicated that in all three groups immunostaining was restricted to the cell surface, confirming the view that BsCD94-1 is a transmembrane protein. Interestingly, Botryllus MCs, characterized by their strong autofluorescent vesicles (Fig. 5L) and known to accumulate in the area of allogeneic contact (17), were not stained by the antiserum (Fig. 5J). Similarly, no antibody staining was observed in the various types of Botryllus pigmented cells (data not shown).
Figure 5.
Surface localization of BsCD94-1 on a subset of Botryllus blood cells by using BsCD94-1 polyclonal antiserum and confocal microscopy. (A, E, and I) Blood cells in differential interference contrast. (B, F, and J) Tetramethylrhodamine B isothiocyanate (TRITC) channel. (C, G, and K) 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei. (D, H, and L) Autofluorescence in FITC channel that allows one to distinguish granulocytes (A and E) from MCs (I).
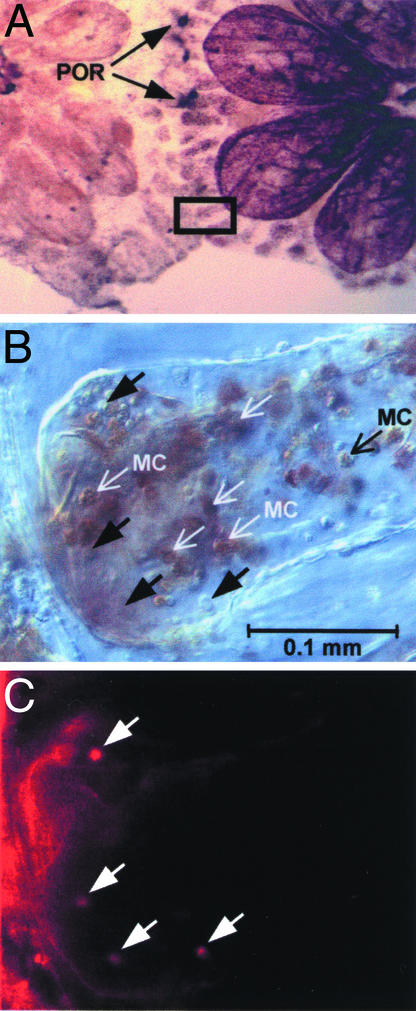
To elucidate the role of the BsCD94-1-positive cells during allorejection and to locate them in situ, we used the colony allorecognition assay. Pairs of incompatible colonies were allowed to contact each other with their ampullae until points of rejections were visible (Fig. 6A). As shown in Fig. 6C, anti-BsCD94-1 staining was restricted to a small population of cells in blood vessels, ampullae, and tunic with no preferential location. While some of them were located close to the contacting zone, others were found more inside the ampullae. In agreement with other observations (17), a large number of MCs accumulated at the tips of the interacting ampullae. Immunopositive cells showed morphological features of the cells described previously (Fig. 5 A–H). As in a single-cell preparation (Fig. 5J), none of the MCs were stained by the BsCD94-1 antiserum. Taken together, these results show that a subpopulation of Botryllus blood cells with morphological features of granulocytes carries the BsCD94-1 transmembrane receptor on their cell surface.
Figure 6.
Whole-mount immunostaining of rejecting colonies. (A) Overview showing contacting ampullae and points of rejection (POR). The boxed area is shown in higher magnification in B and C. (B) Differential interference contrast image of ampullae during allorejection. Note the accumulation of MCs. Black arrows indicate immunopositive non-MCs (indicated by white arrows in C), which show morphological features of cells depicted in Fig. 5 A and E. (C) Few cells throughout the ampullae (white arrows) carry the BsCD94-1 receptor at their surface.
Genomic Organization of BsCD94-1.
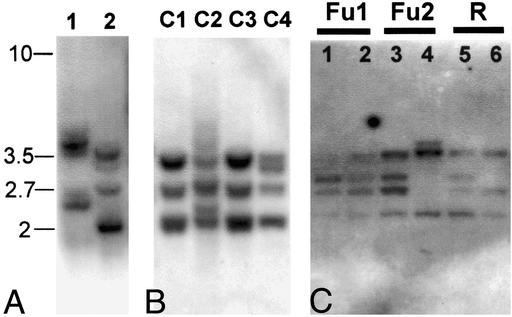
In vertebrates, C-type lectin NK receptors are encoded by a number of related genes that all reside in one genomic region, the NK complex (23). Because the urochordate genome predates the expansion in gene number that occurred in vertebrates (24), it was of interest to determine whether BsCD94-1 was a single gene or member of a gene family. For that purpose we performed Southern blot analysis. Genomic Botryllus DNA was digested with restriction endonucleases and probed with a fragment of BsCD94-1 that did not contain any corresponding restriction site and no intron sequences. Fig. 7A shows the hybridization pattern observed when using genomic DNA pooled from several Botryllus colonies collected in Helgoland. The ClaI as well as the EcoRV digest revealed several distinct fragments hybridizing to the BsCD94-1 probe under highly stringent conditions. The pattern could either be caused by cross-hybridization with closely related genes or be due to the presence of more than one copy of BsCD94-1 in the Botryllus genome.
Figure 7.
Genomic complexity of the BsCD94-1 gene. (A) Southern blot analysis of ClaI (lane 1) and EcoRV (lane 2) -digested DNA pooled from several Botryllus colonies collected at Helgoland. (B) Southern blot analysis of EcoRV-digested DNA from four individuals (C1–C4) collected at Helgoland. (C) Southern blot analysis of EcoRV-digested DNA from six individuals from the laboratory culture at Haifa, Israel, which had been subjected to a colony allorecognition assay before DNA extraction. Individuals 1 and 2 (Fu1) as well as 3 and 4 (Fu2) were able to fuse; individuals 5 and 6 (R) were incompatible and rejected each other.
Unexpectedly, Southern blot analysis using genomic DNA from individual colonies revealed differences in the hybridization pattern in different colonies. Fig. 7B shows the different banding pattern of EcoRV-digested DNA from four individual colonies randomly collected at Helgoland. Colonies 1 and 3 display identical hybridization pattern, whereas colonies 2 and 4 are different. Similar differences could be observed when analyzing ClaI-digested DNA (data not shown). Rehybridization of the filter with the Botryllus β-actin gene as a probe revealed no differences between individuals (data not shown), indicating that the variability is restricted to the BsCD94-1 locus. The variation in the banding pattern among individuals raised the question of whether the genetic variability at the BsCD94-1 locus is involved in determining the ability to fuse and form a chimera. To address this issue, we performed Southern blot analysis with DNA samples from individual B. schlosseri colonies that had been growing in the laboratory in Haifa, Israel, and analyzed for fusibility by the colony-allorecognition assay. Fig. 7C shows the hybridization pattern observed when using EcoRV-digested genomic DNA. Two pairs of compatible individuals (Fu1 and Fu2) and one pair of incompatible individuals (R) were used. The banding pattern varies among individuals and is different from the banding pattern observed in Helgoland by showing additional bands. These observations support the view of genetic diversity at the BsCD94-1 locus among B. schlosseri individual colonies. However, because the banding pattern varies between both compatible and rejecting individuals, the genetic variability at the BsCD94-1 locus is unlikely to be directly involved in determining the ability to fuse.
Discussion
Transplantation immunity and the complex adaptive immune system including MHC and T cell antigen receptor (TCR) molecules are well described in cartilaginous fishes and other vertebrates. None of these organisms, however, undergoes natural transplantation reactions. The phylogenetically closest group that undergoes transplantation reactions naturally is the urochordates. At present, molecules involved in adaptive immunity have not been found outside vertebrates. Urochordates, as the earliest branch in the chordate phylum, occupy a key position for understanding the evolution of the vertebrate immune system (Fig. 1A; refs. 14, 24, and 25). In an nonbiased search for genes differentially expressed after allografting in the urochordate B. schlosseri, we have isolated a homologue of a vertebrate NKR. The results have a number of interesting implications.
First, in vertebrates, CD94 transmembrane receptors are considered specific cell surface markers of NK cells, which are a subset of lymphocytes (5). Our data show that the Botryllus counterpart, BsCD94-1, is also expressed on the surface of a subpopulation of blood cells, implying that Botryllus blood may contain cells functionally related to vertebrate NK cells. Botryllus blood cells display polymorphisms as extensive as those found in vertebrate blood cells and were reported to play important roles in recognition and effector functions involved in colony specificity (17, 26). Despite numerous articles, however, their identification is equivocal. No common terminology has been adopted in classifying them. A major obstacle in analysis of Botryllus blood cell populations is the small amount of blood (about 15 μl) that can be obtained from a single colony. Moreover, problems of identification arise, because presumed different cell types may be different developmental stages of the same cell type. Nothing is known about cell lineages of Botryllus blood cells. Difficulties in correlating ultrastructural studies with observations on living cells add to the confusion about the nature of the cells. For that reason, it was difficult for us to determine the cell-type specificity of cells expressing BsCD94-1. Our studies, however, clearly indicate that the transmembrane receptor is not present on Botryllus MCs or on any of the pigmented cells (see Fig. 5). This is interesting, because MCs have been proposed (17) as the effector cells in Botryllus self/nonself recognition. Our data show that CD94-1-expressing cells are morphologically similar to urochordate granulocytes (18, 22). The extent of structural conservation between the Botryllus BsCD94-1 molecule and the vertebrate orthologs strongly implies functional conservation. We, therefore, propose that B. schlosseri blood cells carrying this receptor are mediators of allorecognition. Because key molecules of vertebrates' NK cells are present in a subpopulation of Botryllus blood cells, these cells may be considered as ancestral NK cells.
Second, the cytoplasmic domain of BsCD94-1 is short (Fig. 2D) and resembles, in this respect, the 7-aa cytoplasmic domain of human CD94. Our results, therefore, raise the question of how signaling is achieved in Botryllus after CD94-1 receptor activation. In human CD94, the signal transduction capacity of the CD94/NKG2 heterodimer is derived from the cytoplasmic domain of the NKG2 partner chain. We propose, therefore, that because of its short cytoplasmic domain, BsCD94-1 should be a heterodimeric receptor that requires a partner chain for becoming a functional NKR. Its identification will require further detailed examination.
Third, at this time the ligand for the Botryllus BsCD94-1 receptor is unknown. Is it similar to that used by vertebrate NK CD94/NKG2/NKR-P1 and, therefore, is the presence of BsCD94-1 in B. schlosseri tied in with the presence of MHC-related proteins? The high degree of structural conservation of the extracellular domain of BsCD94-1 supports the idea that ligands such as nonclassical MHC class I molecules are binding to BsCD94-1.
Fourth, sequencing of the BsCD94-1 coding region indicated no significant sequence differences between individuals. However, Southern blot analysis under highly stringent conditions and an intronless probe revealed a pattern of several genomic fragments that differed among individual colonies. This finding could indicate cross-hybridization of the probe with other closely related genes that may be polymorphic. Thus, not only the CD94-related gene but also the complexity of the family of C-type lectin receptors may be conserved between urochordates and vertebrates. At present, the Southern blot hybridization patterns distinguishing individuals within a population cannot be correlated with the ability to fuse with each other.
Taken together, these results support a model in which a subpopulation of Botryllus blood cells through their BsCD94-1 receptor monitor the cell surfaces for up- and down-regulation of self molecules, much like NK cells in vertebrates. The high degree of structural conservation of the extracellular domain of BsCD94-1, with the possibility that the ligand may belong to the HLA-E group of nonclassical MHC I, provides substance for the proposed notion (11) that “protochordate allorecogniton is controlled by an MHC-like gene system.”
Acknowledgments
We thank Friederike Anton-Erxleben for assistance with confocal microscopy. We also thank the Biologische Anstalt Helgoland for the Botryllus supply, Sandra Pankow for help with blood cell separation, Nicholas Zavazava and Judith Steube for discussion, and Dieter Kabelitz and Silvia Bulfone-Paus for critically reading the manuscript. Part of this work was supported by the United States–Israel Binational Science Foundation, the Israel Science Foundation (456/01), and National Institutes of Health Grant R01-DK54762. This work was supported by the Deutsche Forschungsgemeinschaft. K.K. was supported by a fellowship from the Deutscher Akademischer Austauschdienst.
Abbreviations
- DD
differential display
- NK
natural killer
- MC
morula cell
- NKR
NK cell receptor
- CTLD
C-type lectin-binding domain
Footnotes
References
- 1.Parham P. Immunol Rev. 2001;181:5–289. [Google Scholar]
- 2.Perez-Villar J J, Carretero M, Navarro F, Melero I, Rodriguez A, Bottino C, Moretta A, Lopez-Botet M. J Immunol. 1996;157:5367–5374. [PubMed] [Google Scholar]
- 3.Moretta A, Moretta L. Curr Opin Immunol. 1997;9:694–701. doi: 10.1016/s0952-7915(97)80051-9. [DOI] [PubMed] [Google Scholar]
- 4.Lanier L L. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 5.Braud V M, Allan D S, O'Callaghan C A, Soderstrom K, D'Andrea A, Ogg G S, Lazetic S, Young N T, Bell J I, Phillips J H. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 6.Kärre K. Nature. 1995;267:978–979. doi: 10.1126/science.7863341. [DOI] [PubMed] [Google Scholar]
- 7.Vivier E, Tomasello E, Paul P. Curr Opin Immunol. 2002;14:306–311. doi: 10.1016/s0952-7915(02)00337-0. [DOI] [PubMed] [Google Scholar]
- 8.Biassoni R, Cantoni C, Pende D, Sivori S, Parolini S, Vitale M, Bottino C, Moretta A. Immunol Rev. 2001;181:203–214. doi: 10.1034/j.1600-065x.2001.1810117.x. [DOI] [PubMed] [Google Scholar]
- 9.Oka H, Watanabe H. Proc Jpn Acad Sci. 1957;33:657–659. [Google Scholar]
- 10.Sabbadin A. Am Zool. 1982;22:765–773. [Google Scholar]
- 11.Scofield V L, Schlumpberger L A, West L A, Weissman I L. Nature. 1982;295:488–502. doi: 10.1038/295499a0. [DOI] [PubMed] [Google Scholar]
- 12.Stoner D, Weissman I L. Proc Natl Acad Sci USA. 1996;93:15254–15259. doi: 10.1073/pnas.93.26.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoner D S, Rinkevich B, Weissman I L. Proc Natl Acad Sci USA. 1999;96:9148–9153. doi: 10.1073/pnas.96.16.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito Y, Hirose E, Watanabe H. Int J Dev Biol. 1994;38:237–247. [PubMed] [Google Scholar]
- 15.Rinkevich B, Porat R, Goren M. Proc R Soc London Ser B. 1995;259:319–324. [Google Scholar]
- 16.De Tomaso A W, Saito Y, Ishizuka K J, Palmeri K J, Weissman I L. Genetics. 1998;149:277–287. doi: 10.1093/genetics/149.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rinkevich B, Tertakover S, Gershon H. Mar Biol (Berlin) 1998;131:227–236. [Google Scholar]
- 18.Parrinello N, Cammarata M, Arizza V. Biol Bull. 1996;190:418–425. doi: 10.2307/1543035. [DOI] [PubMed] [Google Scholar]
- 19.Lohmann J, Schickle H P, Bosch T C G. BioTechniques. 1995;18:200–202. [PubMed] [Google Scholar]
- 20.Sawicki M W, Dimasi N, Natarajan K, Wang J, Margulies D H, Mariuzza R A. Immunol Rev. 2001;181:52–65. doi: 10.1034/j.1600-065x.2001.1810104.x. [DOI] [PubMed] [Google Scholar]
- 21.Milanesi C, Burighel P. Acta Zool (Stockholm) 1978;59:135–147. [Google Scholar]
- 22.Parrinello N. In: Invertebrate Immunology. Rinkevich B, Müller W E G, editors. Berlin: Springer; 1996. pp. 190–213. [Google Scholar]
- 23.Hofer E, Sobanov Y, Brostjan C, Lehrach H, Düchler M. Immunol Rev. 2001;181:5–19. doi: 10.1034/j.1600-065x.2001.1810101.x. [DOI] [PubMed] [Google Scholar]
- 24.Corbo J C, Di Gregorio A, Levine M. Cell. 2001;106:535–538. doi: 10.1016/s0092-8674(01)00481-0. [DOI] [PubMed] [Google Scholar]
- 25.Burnet F M. Nature. 1971;232:230–235. doi: 10.1038/232230a0. [DOI] [PubMed] [Google Scholar]
- 26.Schlumpberger J M, Weissman I L, Scofield V L. J Exp Zool. 1984;229:401–411. doi: 10.1002/jez.1402290308. [DOI] [PubMed] [Google Scholar]