Abstract
The diversity and mode of life of microbial eukaryotes in hydrothermal systems is very poorly known. We carried out a molecular survey based on 18S ribosomal RNA genes of eukaryotes present in different hydrothermal niches at the Mid-Atlantic Ridge. These included metal-rich and rare-earth-element-rich hydrothermal sediments of the Rainbow site, fluid–seawater mixing regions, and colonization devices (microcolonizers) containing organic, iron-rich, and porous mineral substrates that were exposed for 15 days to a fluid source. We identified considerable phylogenetic diversity, both at kingdom level and within kinetoplastids and alveolates. None of our sequences affiliates to photosynthesizing lineages, suggesting that we are targeting only autochthonous deep-sea communities. Although sediment harbored most phylogenetic diversity, microcolonizers predominantly contained bodonids and ciliates, indicating that these protists pioneer the colonization process. Given the large variety of divergent lineages detected within the alveolates in deep-sea plankton, hydrothermal sediments, and vents, alveolates seem to dominate the deep ocean in terms of diversity. Compared with data from the Pacific Guaymas basin, some protist lineages seem ubiquitous in hydrothermal areas, whereas others, notably kinetoplastid lineages, very abundant and diverse in our samples, so far have been detected only in Atlantic systems. Unexpectedly, although alvinellid polychaetes are considered endemic of Pacific vents, we detected alvinellid-related sequences at the fluid–seawater interface and in microcolonizers. This finding can boost further studies on deep-sea vent animal biology and biogeography.
Compared with prokaryotes, microbial eukaryotes thriving in extreme environments have rarely been studied. This fact is partly because of the difficulties imposed by classical cultivation approaches. Recent eukaryotic diversity surveys based on 18S rRNA are revealing an unexpected variety of often divergent lineages in different biotopes, including some extreme environments (1–6). The only available molecular survey of microbial eukaryotes from deep-sea vents was recently carried out in hydrothermal sediments from the Pacific Guaymas basin and revealed an important diversity of hitherto unknown lineages (5). Surprisingly, many of these sequences affiliated to typical photosynthesizing groups (such as green algae or diatoms), leading to the conclusion that autochthonous eukaryotes cannot be distinguished from those deposited from the water column (5). There were two objectives in this study. First, we aimed at characterizing the diversity of autochthonous microbial eukaryotes from Mid-Atlantic Ridge hydrothermal systems. Thus, we have carried out a molecular survey of hydrothermal sediment and seawater–fluid interface. These results should constitute a first base for comparison with data from the Pacific systems. To date, whereas prokaryotes seem ubiquitous in different oceanic regions (7), possibly including vent areas, metazoans are subject to a defined biogeographical distribution (8). At present, molecular data on the oceanic distribution of microbial eukaryotes are still negligible. Second, we aimed at having an insight on the in situ microbial colonization process. For this, we analyzed the eukaryotic diversity in experimental microcolonizers containing different substrates that were collected after a 15-day exposure at a fluid emission source. These data also should provide information about the potential specificity of microbes for a particular substrate.
Methods
Sampling.
Samples were taken with the aid of the remotely operated vehicle (ROV) Victor during the French cruise ATOS 2001 to the Mid-Atlantic Ridge hydrothermal area. A sediment core was obtained from Rainbow hydrothermal sediment (36°6′N, 33°11′W, depth 2,264 m). After removal of the ≈1- to 2-mm uppermost layer in a laminar flux chamber, a fraction of the sediment corresponding to the ≈1-cm upper part was frozen in liquid nitrogen until used. Fluid–seawater mixtures were collected from vents by using 0.75-liter titanium bottles at Lucky Strike (37°17′N, 32°16′W, depth 1,695 m) and Rainbow chimneys; they were then filtered sequentially through 5- and 0.2-μm-diameter Millipore filters. Filters then were frozen in lysis buffer (40 mM EDTA/50 mM Tris⋅HCl/0.75 M sucrose) and stored in liquid nitrogen. Sterile microcolonizers were deployed adjacent to a fluid emission at the Tour Eiffel chimney (Lucky Strike site) for 15 days (Fig. 2) and collected in a closed sterile container by the ROV Victor. Microcolonizers consisted of different substrates placed into extensively perforated 50-ml Corning tubes. Two of them consisted of an inert plastic mesh containing, respectively, a meat-based substrate and iron fragments, and a third one was made of basalt and pumice fragments. The container was opened in a laminar flux chamber on board, and the microcolonizers were stored at 4°C in 75% ethanol/2% NaCl. Because of the turbulent mixture of cold deep-sea water and hot hydrothermal fluid at the sampling sites, reliable temperature measurements are unavailable.
Figure 2.
Microcolonizers analyzed in this work being deposited during the ATOS 2001 cruise at a hydrothermal emission (Tour Eiffel chimney, Mid-Atlantic Ridge). Part of the articulated arm of the ROV Victor can be seen on top holding the floater attached to the microcolonizers. Just below the place where the microcolonizers lie, the bottom of the chimney is densely colonized by mussels (Bathymodiolus azoricus). The microcolonizer area appears fuzzy because of the mixing of hydrothermal fluid and seawater.
Electron Microscopy and Sediment Chemical Analysis.
Samples were dehydrated in increasing ethanol concentrations (50%, 70%, 90%, and 100%), critical-point-dried, and gold-coated. Observation was carried out with a JEOL (JSM-840A) scanning electron microscope operated at 17 kV at the Service de Microscopie Electronique de l'Institut Federatif de Recherche de Biologie Intégrative (Paris). Major elements in sediment were analyzed by inductively coupled plasma (ICP)–atomic emission spectrometry; trace elements and rare-earth composition were analyzed by ICP–mass spectrometry by the Service d'Analyze des Roches et Minéraux du Centre National de la Recherche Scientifique (Nancy, France).
DNA Extraction, 18S rDNA Amplification, Cloning, and Sequencing.
Cellular lysis in sediment and microcolonizer substrates was accomplished after several freeze–thaw cycles in liquid nitrogen by overnight incubation at 55°C in a proteinase K/SDS solution. Nucleic acids were then extracted and purified as described (9). Nucleic acids from filters were directly purified after a proteinase K/SDS lysis step by phenol–chloroform and chloroform–isoamyl alcohol extraction. 18S rRNA genes presented in this work were amplified with PCR by using different combinations of the primers 18S-42F (CTCAARGAYTAAGCCATGCA), 18S-82F (GAAACTGCGAATGGCTC), 18S-1498R (CACCTACGGAAACCTTGTTA), and 18S-1520R (CYGCAGGTTCACCTAC). PCRs were performed under the following conditions: 30 cycles (denaturation at 94°C for 15 s, annealing at 55°C for 30 s, extension at 72°C for 2 min) preceded by a 2-min denaturation at 94°C and followed by a 10-min extension at 72°C. rDNA libraries were constructed, by using the TOPO TA Cloning system (Invitrogen), from PCR products coming from different samples and primer combinations. After plating, 25–100 positive transformants per library were screened by PCR amplification of inserts using flanking vector primers. A total of 291 expected-size amplicons from both libraries was partially sequenced (GENOME express, Grenoble, France) with either primer 18S-42F or 18S-82F. From these, four chimeric sequences were detected both by visual inspection of the alignment and by the Ribosomal Database Project CHECK_CHIMERA tool (10). After preliminary phylogenetic analysis (see below), 37 clones representative of the phylogenetic diversity were chosen for complete sequencing. 18S-42F and the internal primers EK-555F (AGTCTGGTGCCAGCAGCCGC) and EK-1269R (AAGAACGGCCATGCACCAC) were used to complete and overlap insert sequences.
Phylogenetic Analyses.
A preliminary phylogenetic analysis of our 291 partial sequences was done by distance methods (neighbor-joining, NJ) by using the program MUST (11), thus allowing the identification of identical or nearly identical sequences and the selection of clones for complete sequencing. Representative clones (n = 37) were completely sequenced, and their sequences were automatically aligned, by using the program BABA (H.P., unpublished work), with 4,575 eukaryotic 18S rDNA sequences retrieved from GenBank (http://ncbi.nlm.nih.gov/). The multiple alignment was then manually edited by using the program ED from the MUST package (11). NJ trees were constructed for the different eukaryotic taxa to choose a representative subset of sequences, avoiding partial, redundant, and fast-evolving ones, for further phylogenetic analyses. Three different subsets of 18S rDNA sequences were selected: one included alveolate sequences (55 sequences), another included representatives of most major eukaryotic groups (55 sequences), and a third included metazoan sequences (46 sequences). Gaps and ambiguously aligned positions were excluded from our analyses, resulting in alignments of 883, 877, and 1,256 positions, respectively. The smaller number of positions used for the first two analyses was because of the inclusion of environmental partial sequences from a molecular survey of eukaryotic diversity in the Guaymas basin (5), which were ≈550 nucleotides shorter than our sequences (the average length was ≈1,200 nucleotides vs. ≈1,750 nucleotides, respectively). The three data sets were subjected to maximum parsimony (MP) and maximum likelihood (ML) analysis by using PAUP* 4b8 (12). By using an MP tree, the parameter values for a general time-reversible model of nucleotide substitution were estimated, with a six-category discrete approximation of a Γ distribution plus invariable sites (GTR + Γ + I model). The ML trees were constructed by using 20 random additions of species and the tree bisection–reconnection branch-swapping option. Bootstrap replicates (n = 100) were performed for all data sets. Alignments, trees, and list of species used are available upon request.
Results and Discussion
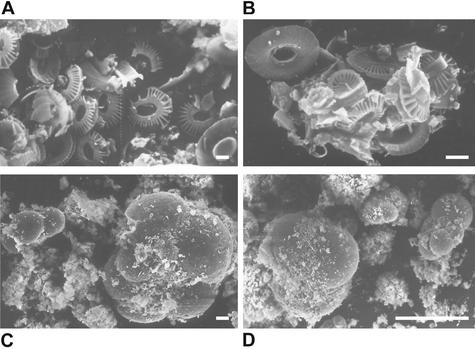
Our samples were collected from the Rainbow and Lucky Strike sites, which are close to the Azores archipelago (see Methods). The Rainbow sediment we studied was particularly interesting because of its very high metal content. The acidic (pH ≈2.8) and hot (≈365°C) Rainbow vent fluids are unique, being enriched in H2, methane, Mn, Fe, Co, Ni, Cu, Zn, Ag, Cd, Cs, Pb, Y, and rare-earth elements (13), thus influencing sediment composition. Sediments beneath the plume at distances of 2–25 km from the site are known to be rich in biogenic CaCO3 (up to 80%) and record enrichments of Fe, Cu, Mn, V, P, and As (14). The analysis of major, trace, and rare-earth elements in our Rainbow sediment sample showed that they were very abundant and that, compared with sediments located further below the plume, Fe, Cu, Mn, V, P, and As content was considerably higher (Tables 1 and 2). Eukaryotes thriving there must consequently be highly metallo-tolerant. Biogenic CaCO3 accounted for ≈32% of total sediment weight (Tables 1 and 2) and seems to be contributed mostly by very abundant haptophyte coccoliths and foraminifer shells, as revealed by direct observation with scanning electron microscopy (Fig. 1). For the colonization experiment, different (organic, iron-rich, mineral) substrates were deployed at a fluid source above a mussel colony (Fig. 2) and recovered after 15 days (see Methods). Although temperature was not recorded over time, microcolonizers were clearly exposed to the vent fluid (Fig. 2). These were at least occasionally exposed to very high temperatures, which is attested to by the fact that part of the 190°C-resistant microcolonizer tubes was partially burnt. This finding leaves open the possibility that some of the eukaryotic lineages detected were thermophilic.
Table 1.
Chemical composition of the Rainbow sediment studied in this work
| Major elements | Rainbow sediment, % | Sediment along Rainbow plume, %* |
|---|---|---|
| SiO2 | 2.34 | |
| Al2O3 | 0.61 | 0.56/0.53/0.46/0.67 |
| Fe2O3 | 4.46 | 1.47/1.53/1.52/1.94 |
| MnO | 0.24 | 0.05/0.06/0.08/0.1 |
| MgO | 0.95 | |
| CaO | 31.30 | |
| Na2O | 1.17 | |
| K2O | 0.07 | |
| TiO2 | 0.05 | 0.05/0.05/0.05/0.06 |
| P2O5 | 0.54 | 0.07/0.07/0.08/0.09 |
| CO2 tot | 32.56 | 82.9/86.2/83.4/86.7 |
| C org | 0.14 | 0.39/nd/0.0/nd |
| S tot | 0.31 |
nd, not determined.
Values from four sediment cores (0.5 cm from surface) taken at 2, 5, 10, and 25 km (2,540, 2,322, 2,363, and 2,519 m deep, respectively) from the Rainbow site following the vent plume. Data are from ref. 14.
Table 2.
Trace elements and rare-earth elements
| Element | Rainbow sediment, ppm | Sediment along Rainbow plume, ppm* | Seawater, ppm† | Element | Rainbow sediment, ppm | Sediment along Rainbow plume, ppm* | Seawater, ppm† |
|---|---|---|---|---|---|---|---|
| As | 53.0 | 25/25/33/39 | 0.0026 | Nb | 1.92 | 0.000015 | |
| Ba | 227 | 0.021 | Nd‡ | 5.99 | 0.0000028 | ||
| Be | <d.l. | 0.0000006 | Ni | 69.6 | 12/18/12/14 | 0.0066 | |
| Bi | 2.91 | 0.00002 | Pb | 13.0 | 0.00003 | ||
| Cd | <d.l. | 0.00011 | Pr‡ | 1.52 | 0.00000064 | ||
| Ce‡ | 10.6 | 0.0000012 | Rb | 2.81 | 0.120 | ||
| Co | 76.2 | 0.00039 | Sb | 0.45 | 0.00033 | ||
| Cr | 87.6 | 21/21/15/8 | 0.0002 | Sm‡ | 1.23 | 0.00000045 | |
| Cs | <d.l. | 0.0003 | Sn | 1.34 | 0.00081 | ||
| Cu | 3625 | 394/334/142/110 | 0.0009 | Sr | 1170 | 8.1 | |
| Dy‡ | 1.19 | 0.00000091 | Ta | 0.14 | <0.0000025 | ||
| Er‡ | 0.623 | 0.00000087 | Tb‡ | 0.200 | 0.00000014 | ||
| Eu‡ | 0.479 | 0.0000013 | Th | 0.87 | 0.0000004 | ||
| Ga | 1.14 | 0.00003 | Tm‡ | 0.098 | 0.00000017 | ||
| Gd‡ | 1.30 | 0.0000007 | U | 0.84 | 0.0033 | ||
| Ge | 0.23 | 0.00006 | V | 126 | 88/92/103/115 | 0.0019 | |
| Hf | 0.27 | <0.000008 | W | 0.52 | <0.000001 | ||
| Ho‡ | 0.240 | 0.00000022 | Y | 9.09 | 0.000013 | ||
| In | 0.34 | <d.l. | Yb‡ | 0.555 | 0.00000082 | ||
| La‡ | 8.51 | 0.0000029 | Zn | 215 | 31/30/21/22 | 0.005 | |
| Lu‡ | 0.085 | 0.00000015 | Zr | 15.2 | 0.000026 | ||
| Mo | 3.03 | 0.01 |
<d.l., below detection limit.
Values from four sediment cores (0.5 cm from surface) taken at 2, 5, 10, and 25 km (2,540, 2,322, 2,363, and 2,519 m deep, respectively) from the Rainbow site following the vent plume. Data are from ref. 14.
Seawater at 3.5% salinity (24).
Rare-earth element.
Figure 1.
Scanning electron microscopy photographs of Rainbow sediment showing coccoliths and foraminifer shells. [Bars = 1 μm (A and B), 10 μm (C), and 100 μm (D).]
After nucleic acid extraction, amplification, and cloning of 18S rRNA genes from the different samples, we partially sequenced ≈300 insert clones. From these, 37 representative clones were selected for complete sequencing, and the sequences aligned with 4,575 additional 18S rRNA gene sequences for phylogenetic reconstruction (see Methods). We selected three subsets of sequences to make compatible time-consuming phylogenetic analyses with a good taxonomic representation of (all and closest relatives to our sequences) eukaryotes, thus minimizing taxonomic sampling-dependent reconstruction artifacts. These subsets cover the whole eukaryotic diversity at a kingdom level, the alveolates and the metazoans (Figs. 3–5).
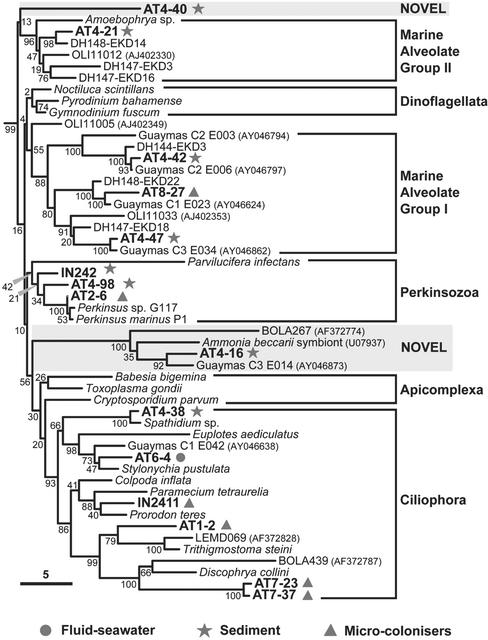
Figure 3.
ML phylogenetic tree of alveolate 18S rDNA sequences. The tree was rooted by using three heterokont sequences (Hyphochytrium catenoides, Dictyocha speculum, and Nannochloropsis salina, not shown). Mid-Atlantic Ridge environmental clones are symbol-coded depending on the sample type. Sequences representing unknown alveolate groups are highlighted. The scale bar indicates the percentage of substitutions for a unit branch length. Numbers at nodes are bootstrap percentages.
Figure 5.
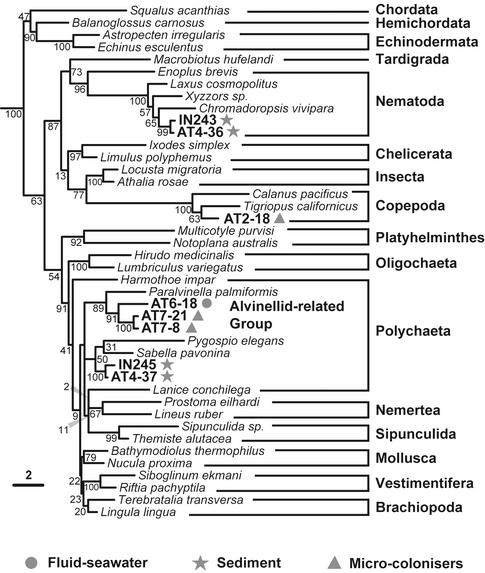
ML phylogenetic tree of metazoan 18S rDNA sequences. The tree was rooted by using two Porifera (Hippospongia communis and Spongilla lacustris) and two Cnidaria (Dendrobrachia paucispina and Craterolophus convolvulus) sequences. Environmental clones are symbol-coded depending on the sample type. The unknown alvinellid-related sequences identified at the Mid-Atlantic Ridge are highlighted. The scale bar indicates the percentage of substitutions for a unit branch length. Numbers at nodes are bootstrap percentages.
By far, the highest level of diversity and phylogenetic uniqueness was found in Rainbow sediment. Alveolates were the most abundant lineages in sediment libraries (62%), including ciliates (3%), marine Group I (7%) and II (7%) alveolates, and phylotypes not ascribed to any of the major alveolate groups (45%; Fig. 3). These alveolates were not only abundant in libraries but very diverse. The finding of Group I and II alveolate sequences in hydrothermal sediments supports the idea that they are ubiquitous in the sea (15), being also present in deep-sea Antarctic plankton (1) and surface Atlantic, Pacific, and Mediterranean waters (2, 3). Group I sequences are particularly diverse in hydrothermal sediments, because many Guaymas clones from the Pacific (5) also cluster in this group (Fig. 3). We identified an additional alveolate group that is particularly interesting. It clusters one sequence from the database, possibly corresponding to a symbiont of the foraminifer Ammonia beccarii, our sequence AT4–16, the clone C3 E014 from the Guaymas basin (5), and the clone BOLA267 from anoxic sediment in a low tidal flat (6). This group is probably related to gregarines, a family of fast-evolving parasitic apicomplexa (16). Sequences belonging to this cluster were relatively frequent in our sediment libraries (14%). The branch basal to this group is long and the statistical support is strong (100%), indicating that this phylogenetic lineage is robust. This strong support contrasts with the poor resolution among the alveolate lineages (low bootstraps and uncertain branching order; Fig. 3). Also, previously uncharacterized alveolates represented by IN242 and AT4–98 were placed at the base of the Perkinsozoa, breaking the basal branch of this group of marine parasites.
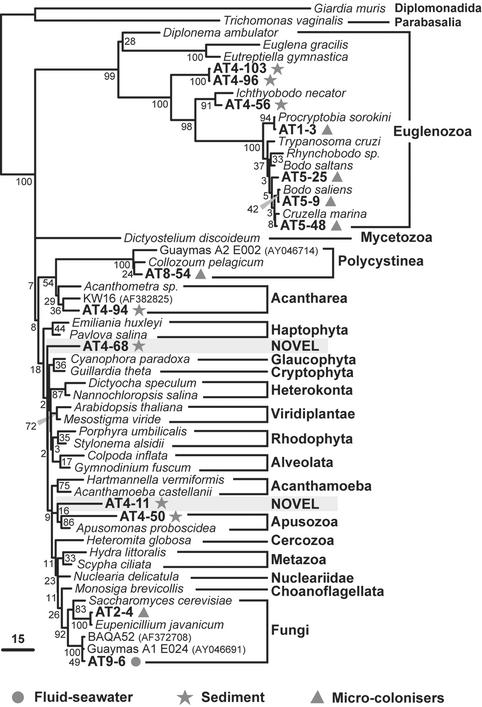
In addition to the large variety of sediment alveolates, we identified very divergent lineages that could correspond to hitherto unknown eukaryotic kingdoms. This is the case of AT4–68 and AT4–11 (Fig. 4). AT4–50 also branches very deeply in the apical region of the eukaryotic tree but is consistently associated with Apusomonas sp. (bootstrap 86%), heterotrophic gliding flagellates of difficult phylogenetic ascription (17). A very divergent group of sequences (AT4–96 and AT4–103) appears at the base of the Kinetoplastida, also breaking the otherwise very long branch of this group. The fact that the most basal kinetoplastid lineages are found in sediment may indicate that these are anaerobic or microaerophilic lineages. We also detected a relatively basal kinetoplastid sequence related to the genus Ichthyobodo (Fig. 4). In addition to kinetoplastids, we identified an acantharean sequence (AT4–94) and several metazoan sequences. All animal sequences detected corresponded either to nematodes or to a cluster apparently distantly related either to brachiopods or to polychaetes (IN245 and AT4–37; Fig. 5). Because the position of the latter is very unstable in different phylogenetic analyses (data not shown), it could correspond either to a previously uncharacterized metazoan group or to a divergent but described lineage for which there are no sequences available in databases. Despite the extensive presence of haptophyte coccoliths attesting to a sedimentary process involving the remains of photosynthetic organisms from the photic zone (Fig. 1), none of the sediment sequences ascribed to photosynthesizing lineages. This result indicates that their DNA is degraded before deposition at the 2,264-m deep Rainbow site.
Figure 4.
Unrooted ML phylogenetic tree of major eukaryotic groups based on 18S rDNA sequences. Mid-Atlantic Ridge environmental clones are symbol-coded depending on the sample type. Clones representing possibly unknown kingdom-level groups are highlighted. The scale bar indicates the percentage of substitutions for a unit branch length. Numbers at nodes are bootstrap percentages.
The molecular analyses of the three different microcolonizer substrates (organic, iron-rich, and mineral) showed no significant differences of eukaryotic diversity among them. This finding agrees with the absence of significant differences in prokaryotic diversity on the same substrates (S. Duperron, P.L.-G., and D.M., unpublished results). We identified a large diversity of eukaryotes including bodonids (Kinetoplastida), different alveolates (ciliates, Perkinsozoa, Group I), metazoa, fungi, and radiolaria (Figs. 3–5). By far, the most abundant phylotypes in the six 18S rDNA libraries analyzed were bodonids (48%) and ciliates (36%). Bodonids are small, free-living, flagellate predators phylogenetically related to the well known parasitic kinetoplastids such as Trypanosoma spp.; bodonid sequences were very diverse (Fig. 4). Ciliate sequences were also very diverse, with some lineages being quite divergent within this group, such as AT1–2 or the cluster AT7–23 + AT7–37 (Fig. 3). These seem more or less distantly related to sequences LEMD and BOLA coming from anoxic environments (6). This result would tend to support a view of a microaerophilic nature for these eukaryotes. Indeed, vent surroundings are poor in oxygen, because fluids are very reducing and oxygen has lower solubility at higher temperatures. In addition to bodonid and ciliate predators, we observed some typical saprophytes, such as fungi (Fig. 4) and one Perkinsus-like sequence (Fig. 3). Perkinsus species parasitize mussels, among other species. Because the microcolonizers were deposited above a colony of B. azoricus (Fig. 2), the finding of Perkinsus phylotypes here could indicate that these deep-sea mussels may be infected by these parasites, similarly to their coastal relatives (18). The large diversity of predatory and parasitic protists found suggests that these organisms can exert a demographic control in vent populations. Some mussels' mortalities attributed to bacterial infections might indeed be the product of protist parasitism.
We also analyzed the eukaryotic diversity in two samples taken at the fluid–seawater interface (7). However, because sample volume was small (0.5–0.75 liters each) and the biomass was low, the results obtained are only indicative. Nevertheless, we detected ciliates, fungi, and metazoans (Figs. 3–5), which correlates well with the diversity found in microcolonizers. Interestingly, with the exception of a few copepod microcolonizer sequences, all metazoan sequences detected in microcolonizers and fluid–seawater interface samples corresponded to polychaetes (possibly larvae or gametes) related to alvinellids (Fig. 5). All described species of alvinellids are endemic of Pacific vents (8). Known alvinellid species build tubes, which are not apparent in Atlantic chimneys. Nevertheless, we cannot exclude the possibility of errant alvinellid-related polychaetes thriving in the Atlantic hydrothermal systems. It cannot be assessed at present whether these have escaped zoologist taxonomists or whether they have been described as deep-sea polychaetes but their 18S rRNA sequences have never been determined. In any case, our results suggest that alvinellids constitute a family larger than that presently recognized and whose ancestors adapted to Pacific and Atlantic deep-sea vents, probably before the separation of the two oceanic provinces.
The results of our study of microbial eukaryotic diversity in Atlantic hydrothermal systems reveal many divergent eukaryotic lineages. None of our sequences affiliates to photosynthesizing lineages, in sharp contrast to the data obtained from the Pacific Guaymas basin (5). This result points to the existence of an autochthonous deep-sea vent community that is not masked by photosynthetic-depositing organisms. Nevertheless, we cannot exclude the possibility that many of the detected lineages correspond simply to deep-sea eukaryotes not specifically associated with hydrothermal vents. To discriminate which lineages are endemic to hydrothermal systems, a comprehensive protist survey of nonhydrothermal deep-sea sediment and plankton will be required. Another striking fact is that, whereas some eukaryotic lineages appear distributed in the Atlantic and Pacific vent areas, kinetoplastid sequences have been detected so far only in the Atlantic region. Kinetoplastids are one of the more represented groups in our samples; bodonids are diverse in fluid–seawater interface and microcolonizers, and early branching kinetoplastids are diverse in sediment. Whether this reflects a true biogeographic difference or a particular substrate specificity (e.g., Rainbow sediments and fluids are exceedingly metal-rich), or whether it is just that the eukaryotic diversity has not been completely described both in Pacific and Atlantic hydrothermal systems cannot be answered at present. Nevertheless, given that kinetoplastid sequences are so abundant and diverse in our samples, the fact that not a single kinetoplastid phylotype had been identified in Pacific samples, for which a total of 276 sequences was analyzed (5), might indicate a different geographic distribution or, at the least, that kinetoplastids have a higher representation in Atlantic systems. In the latter sense, one bodonid species, Rhynchomonas nasuta, was isolated from Pacific vents (19).
At the Atlantic sites studied, whereas the hydrothermal sediment seems to be a reservoir of phylogenetic diversity, including kingdom-level groups, basal kinetoplastids, alveolates, or even animals (e.g., cluster IN245 and AT4–37), substrates exposed to the fluid–seawater interface are colonized mainly by diverse, small, possibly microaerophilic bodonids and ciliates in only 2 weeks. Therefore, these mobile predators pioneer the colonization process. Whether these eukaryotes are thermophilic cannot be assessed yet, but they certainly are adapted to the highly reduced and metal-enriched hydrothermal fluid. Eukaryotes thriving in Rainbow sediment are likely anaerobic and clearly metallo-tolerant because, as shown by chemical analysis, this site holds records of high concentrations in several metals and rare-earth elements. Protists isolated from this environment would thus be good models in which to study the adaptation of eukaryotic cells to various extreme conditions (high metal content, pressure, and temperature).
In combination with previous data from deep-sea plankton (1), the amazing variety of alveolates in both sediment and microcolonizers suggests that alveolates dominate, in terms of genetic diversity, the deep ocean. The existence of so many new alveolate lineages with poor branching order resolution could support the idea that they radiated (1, 15), although a much larger alveolate taxonomic sampling would be required to confirm this hypothesis. Given the phylogenetic uniqueness we found, and given that hydrothermal systems have been present on Earth since early times (20), these are ideal target environments in which to look for eukaryotes predating the mitochondrial acquisition, if they exist (21), that have retained primitive characters. Their potential discovery would allow for the testing of different models on eukaryotic origin (22, 23).
Acknowledgments
We thank Magali Zbinden for efficient sample processing on board during the ATOS cruise, the ATOS chief scientist, Pierre Marie Sarradin, and the crew of the oceanographic ship Atalante. The campaign was made possible by the Institut Français de Recherche pour l'Exploitation de la Mer and European program VENTOX Grant EVK3 CT 1999-00003. This work was financed by the Centre National de la Recherche Scientifique–Institut National des Sciences de l'Univers, Programme Géomicrobiologie des Environnements Extrêmes.
Abbreviations
- MP
maximum parsimony
- ML
maximum likelihood
Footnotes
References
- 1.López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- 2.Moon-van der Staay S Y, De Wachter R, Vaulot D. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 3.Díez B, Pedrós-Alió C, Massana R. Appl Environ Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaral Zettler L A, Gomez F, Zettler E, Keenan B G, Amils R, Sogin M L. Nature. 2002;417:137. doi: 10.1038/417137a. [DOI] [PubMed] [Google Scholar]
- 5.Edgcomb V P, Kysela D T, Teske A, De Vera Gomez A, Sogin M L. Proc Natl Acad Sci USA. 2002;99:7658–7662. doi: 10.1073/pnas.062186399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson S C, Pace N R. Proc Natl Acad Sci USA. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong E F. Curr Opin Microbiol. 2001;4:290–295. doi: 10.1016/s1369-5274(00)00205-8. [DOI] [PubMed] [Google Scholar]
- 8.Van Dover C L, German C R, Speer K G, Parson L M, Vrijenhoek R C. Science. 2002;295:1253–1257. doi: 10.1126/science.1067361. [DOI] [PubMed] [Google Scholar]
- 9.López-García P, Gaill F, Moreira D. Environ Microbiol. 2002;4:204–215. doi: 10.1046/j.1462-2920.2002.00286.x. [DOI] [PubMed] [Google Scholar]
- 10.Maidak B L, Cole J R, Lilburn T G, Parker C T, Saxman P R, Farris R J, Garrity G M, Olsen G J, Schmidt T M, Tiedje J M. Nucleic Acids Res. 2001;29:173–174. doi: 10.1093/nar/29.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philippe H. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer; 2000. , Version 4.0. [Google Scholar]
- 13.Douville E, Charlou J L, Oelkers E H, Bienvenu P, Jove Colon C F, Donval J P, Fouquet Y, Prieur D, Appriou P. Chem Geol. 2002;184:37–48. [Google Scholar]
- 14.Cave R R, German C R, Thomson J, Nesbitt R W. Geochim Cosmochim Acta. 2002;66:1905–1923. [Google Scholar]
- 15.Moreira D, López-García P. Trends Microbiol. 2002;10:31–38. doi: 10.1016/s0966-842x(01)02257-0. [DOI] [PubMed] [Google Scholar]
- 16. Leander, B. S., Clopton, R. E. & Keeling, P. J. (2003) Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 17.Cavalier-Smith T, Chao E E. Proc R Soc London Ser B. 1995;261:1–6. [Google Scholar]
- 18.Perkins F O. In: The Illustrated Guide to the Protozoa. Lee J J, Leedale G F, Bradbury P, editors. Vol. 1. Lawrence, Kans.: Soc. Protozoologists; 2000. pp. 200–202. [Google Scholar]
- 19.Atkins M S, Teske A P, Anderson O R. J Eukaryotic Microbiol. 2000;47:400–411. doi: 10.1111/j.1550-7408.2000.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 20.Nisbet E G, Sleep N H. Nature. 2001;409:1083–1091. doi: 10.1038/35059210. [DOI] [PubMed] [Google Scholar]
- 21.Philippe H, Germot A, Moreira D. Curr Opin Genet Dev. 2000;10:596–601. doi: 10.1016/s0959-437x(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 22.López-García P, Moreira D. Trends Biochem Sci. 1999;24:88–93. doi: 10.1016/s0968-0004(98)01342-5. [DOI] [PubMed] [Google Scholar]
- 23.Forterre P, Philippe H. BioEssays. 1999;21:871–879. doi: 10.1002/(SICI)1521-1878(199910)21:10<871::AID-BIES10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]