Abstract
Phosphoinositides are critical regulators of ion channel and transporter activity. Defects in interactions of inwardly rectifying potassium (Kir) channels with phosphoinositides lead to disease. ATP-sensitive K+ channels (KATP) are unique among Kir channels in that they serve as metabolic sensors, inhibited by ATP while stimulated by long-chain (LC) acyl-CoA. Here we show that KATP are the least specific Kir channels in their activation by phosphoinositides and we demonstrate that LC acyl-CoA activation of these channels depends on their low phosphoinositide specificity. We provide a systematic characterization of phosphoinositide specificity of the entire Kir channel family expressed in Xenopus oocytes and identify molecular determinants of such specificity. We show that mutations in the Kir2.1 channel decreasing phosphoinositide specificity allow activation by LC acyl-CoA. Our data demonstrate that differences in phosphoinositide specificity determine the modulation of Kir channel activity by distinct regulatory lipids.
Phosphoinositides are important signaling molecules, playing critical roles in processes as diverse as vesicular transport, calcium signaling, growth factor signaling, organization of the cytoskeleton, and regulation of ion channel activity (1, 2). There are multiple isomers of biologically active phosphoinositides in the plasma membrane. Phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is the precursor of the two classical second messengers, inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). Phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] and PI(3,4)P2, the products of phosphoinositide 3-kinase (PI3K), are important signaling molecules in many growth factor signal transduction pathways.
Kir channels are important regulators of cell excitability in the nervous system and they also play important roles in processes as diverse as the regulation of the heart rate, pancreatic insulin release, and K+ secretion in the kidney (3–5). A common feature of these channels, which has emerged recently, is their activation by phosphoinositides (6–14). It was recently shown that defects in channel–phosphoinositide interactions lead to disease (14). Several reports have suggested a relatively low phosphoinositide selectivity of certain Kir channels. GIRK (Kir3.x) channels (13, 15), and ROMK1 (Kir1.1) channels (16) were reported to be activated by PI(4,5)P2, PI(3,4,5)P3, and PI(3,4)P2, and to a lesser extent by PI(4)P (13). KATP channels were shown to be activated not only by PI(4,5)P2, but also by PI(3,4,5)P3 (7) and PI(3,4)P2 (17). When applied at higher concentrations, PI(4)P (7, 10, 11), phosphatidylinositol (PI) (11), and phosphatidic acid (18) could activate these channels. Without knowledge of the local concentrations of these anionic phospholipids in the proximity of the channels, their relative importance in the physiological regulation of KATP channels remains unclear.
The metabolic intermediate long-chain (LC) acyl-coenzyme A (CoA) is a negatively charged lipid of physiological importance. It activates several PKC isoforms and plays a role in membrane trafficking and gene transcription (19, 20). LC acyl-CoA has been shown to activate Kir6.2 channels, the pore-forming subunits of KATP channels that serve as cellular metabolic sensors (21–23). Because cellular levels of LC acyl-CoA increase during ischemia (24), they may contribute to the metabolic activation of KATP channels (22). Activation of KATP channels by LC acyl-CoA has also been suggested to play a role in the physiological oscillatory insulin secretion (20), as well as in pathophysiological states. Examples are type 2 diabetes mellitus or obesity, where the glucose sensitivity of the pancreatic β cell is decreased because of the higher levels of circulating free fatty acids. In such cases, the consequential increase of intracellular LC acyl-CoA activates KATP channels (21, 23). It has been suggested that the mechanism of activation of KATP channels by LC acyl-CoA is different from that of activation by PI(4,5)P2 (21, 22).
In this study we report that LC acyl-CoA activation of KATP channels correlates with their low specificity of activation by different phosphoinositides. None of the other Kir channels tested was activated by LC acyl-CoA. Unlike Kir6.2, all other Kir channels were activated preferentially by PI(4,5)P2 over PI(3,4,5)P3, and PI(3,4)P2. Mutants of Kir2.1 that showed low phosphoinositide specificity could in fact be activated by LC acyl-CoA, suggesting that LC acyl-CoA may stimulate Kir channels through a nonspecific phosphoinositide binding site.
Methods
Materials.
Dioctanoyl (diC8) forms of PI(4,5)P2, PI(3,4,5)P3, and PI(3,4)P2 were purchased from Echelon (www.echelon-inc.com). They were dissolved in water to make 2.5 mM stock solutions, which were divided into aliquots and stored at −70°C. Further dilutions were made in bath solution on the day of the experiment. LC arachidonyl-stearoyl (AASt) PI(4,5)P2 was purchased from Roche Molecular Biochemicals (Indianapolis). Aqueous stock and working solutions were prepared and sonicated as described (13, 25). Oleoyl-CoA was purchased from Sigma and was dissolved in water at a concentration of 5 mM. Other materials were purchased from Sigma.
Molecular Biology and Expression in Xenopus Oocytes.
Kir2.2 and Kir4.1 were cloned from a human brain cDNA library (QUICK-Clone, CLONTECH) with a PCR-based approach. Human Kir3.4 and Kir3.1 were cloned previously in our laboratory (26). Other Kir cDNA clones were gifts from various investigators (see Acknowledgments). We subcloned most of the cDNA constructs into pGEMHE vector (27) or its modified version, pGEMSH, to obtain optimal expression in Xenopus oocytes. Point mutants were produced with a QuikChange mutagenesis kit (Stratagene). Mutations were confirmed by DNA sequencing (Cornell University, Ithaca, NY). Oocyte preparation and the details of complementary RNA (cRNA) preparation and injection are described in refs. 25 and 26. cRNAs of the various Kir channels and mutants were injected in the range of 0.5–10 ng per oocyte depending on the expression level of the given construct.
Electrophysiology.
Patch-clamp experiments were performed in the inside-out macropatch configuration as described (13), by using an EPC-7 patch clamp amplifier. PCLAMP 6.01 software (Axon Instruments, Foster City, CA) was used for data acquisition and analysis. In most measurements we used a ramp protocol from −80 to +80 mV, with a holding potential of 0 mV. This protocol was repeated once per second throughout the measurement. On the figures we show for clarity the time course of the current at −80 mV. In some measurements the membrane potential was held constant at −80 mV, with a sampling rate of 30 Hz. The pipette solution contained 96 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 1 mM NaCl, and 10 mM Hepes, pH 7.4. The bath solution contained 96 mM KCl, 5 mM EGTA, and 10 mM Hepes, pH 7.4. Experiments with Kir3.4 and Kir3.1 channels were performed in bath solution containing 20 mM Na+ to promote their activation by different PIP2 analogs (6).
Computer Software.
A computer program developed in our laboratory was used to identify candidate amino acid residues important for the specificity of channel activation by phosphoinositides. The program scans a multiple sequence alignment and evaluates correlation between a specific functional index of proteins, such as the specificity of channel activation by phosphoinositides, and physicochemical properties of residues at that position. Strong correlation raises the possibility that the residues at the position are important for the specific function of proteins. In the current version of the program, correlation between each of five types of physicochemical properties, hydrophobicity, charge, volume, aromaticity, and the existence of a β-carbon in the amino acid side chain, was calculated at each position. A more detailed description of the program can be found on our web site, http://inka.mssm.edu/∼diomedes.
Statistics.
Data are shown as the mean ± SEM, representing 3–12 measurements performed on two or more batches of oocytes. Statistical significance was calculated by using a t test.
Results
Phosphoinositide and Oleoyl-CoA Activation of KATP Channels.
The activation of KATP channels by phosphoinositides has been reported to be nonspecific, depending only on the number of charges in the lipid head-group (7, 10, 11, 17). LC acyl-CoA activates native cardiac (22) and pancreatic (23) KATP channels, as well as recombinant Kir6.2Δ36 channels (21). Kir6.2Δ36 refers to the C-terminal truncation mutant of Kir6.2 that renders these channels active as homomers (28), by removing the 3-aa motif (RKR; ref. 29) that serves as an endoplasmic reticulum (ER) retention signal. As can be appreciated from their structures, LC acyl-CoA (Fig. 1A) and PI(4,5)P2 (Fig. 1B) exhibit some degree of similarity, as they both possess three phosphates in their respective head-groups and they contain one or two long acyl chains. To examine whether LC acyl-CoA and various phosphoinositides exert their effects through a common mechanism, we examined the effect of their sequential applications to recombinant KATP channels (Fig. 1).
Figure 1.
LC acyl-CoA and phosphoinositide activation of KATP channels. (A) Schematic of oleoyl-CoA. (B) Schematic of AASt PI(4,5)P2. (C–F) Inside-out macropatch measurements on recombinant KATP channels expressed in Xenopus oocytes were performed as described in Methods. Currents at −80 mV are shown on all traces after rundown of channel activity. Development of inward current is shown by a downward deflection on the curves. Zero current is shown by a dashed line. (C Left) Representative trace for Kir6.2 channels coexpressed with SUR2A. The applications of 25 μM diC8 PI(4,5)P2, PI(3,4,5)P3, and PI(3,4)P2 are shown by the horizontal lines before and after the application of 10 μM oleoyl-CoA. (Right) Statistical summary of the data (n = 3). The data were normalized to the current level induced by PI(4,5)P2 (100%). (D Left) Representative trace on Kir 6.2-SUR2A channels. Oleoyl-CoA (10 μM) is applied before and after the application of 5 μM AASt PI(4,5)P2. The length of the first application of oleoyl-CoA, also labeled with an arrow, was 10 s. Short pulses of 25 μM diC8 PI(4,5)P2 were applied throughout the experiment as shown by the horizontal lines. (Right) Summary of the data from three similar experiments. (E) Kir6.2Δ36 channels: 25 μM diC8 PI(4,5)P2 is applied repetitively, and the applications of 3 and 10 μM oleoyl-CoA are indicated by horizontal lines. (F) Kir6.2Δ36 channels: the applications of diC8 phosphoinositides (25 μM) and 10 μM oleoyl-CoA are shown by horizontal lines.
We used water-soluble, short acyl chain, diC8 phosphoinositides to activate KATP channels (13, 25). The reversible effect of the water-soluble diC8 PIP2 makes it possible to test several phosphoinositide analogs in the same patch and evaluate their relative effectiveness. Even though diC8 phosphoinositides are water soluble, they are likely to partition into the patch membrane to activate Kir channels, because the shorter chain analog diC4 PI(4,5)P2 has failed to activate Kir2.1 and Kir3.4/3.1 channels (13). The reversible effect of diC8 phosphoinositides is in sharp contrast with the slower but practically irreversible activation by the long-chain PIP2 analogs, such as the naturally occurring AASt PI(4,5)P2, (13, 25).
Fig. 1 C–F shows representative ion current measurements in the inside-out macropatch configuration at −80 mV. We show traces after current rundown, which is caused by the gradual hydrolysis of PI(4,5)P2 by lipid phosphatases in the patch membrane (9). diC8 PIP2 analogs (25 μM), repeatedly applied to inside-out patches of Kir6.2-sulfonylurea receptor (SUR)2A (Fig. 1 C and D), and Kir6.2Δ36 (Fig. 1 E and F), activated the channels in a reversible manner. The effectiveness of PI(4,5)P2, PI(3,4)P2, and PI(3,4,5)P3 was very similar, consistent with previous reports (7, 10, 17). Oleoyl-CoA (10 μM) also activated Kir6.2-SUR2A channels (Fig. 1 C and D) and Kir6.2Δ36 (Fig. 1 E and F). Subsequent applications of diC8 PIP2 analogs after 10 μM oleoyl-CoA did not cause further activation (Fig. 1 C and F). Similar data were obtained with Kir6.2-SUR1 (data not shown). As the effect of oleoyl-CoA washed out, the effect of the diC8 PI(4,5)P2 was slowly revealed (Fig. 1D). When Kir6.2-SUR2A channels were activated by the natural AASt PI(4,5)P2, oleoyl-CoA did not exert any further effect (Fig. 1D). Similar results were obtained with Kir6.2Δ36 and Kir6.2-SUR1 (data not shown). Partial occlusion of the effect of diC8 PI(4,5)P2 by oleoyl-CoA occurred at submaximal concentrations on Kir6.2Δ36 (Fig. 1E). These data suggest that phosphoinositides and oleoyl-CoA exert their effects on Kir6.2 channels through a common mechanism.
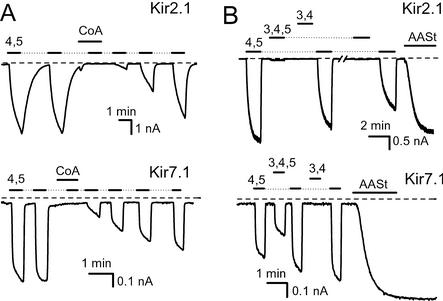
We then asked whether LC acyl-CoA activates other Kir channels. Fig. 2A shows that oleoyl-CoA not only failed to activate recombinant Kir2.1 and Kir7.1 channels, but it in fact inhibited their subsequent activation by diC8 PI(4,5)P2. These results are consistent with previous reports showing that LC acyl-CoA failed to activate cardiac inward rectifiers (Kir2.0; ref. 22) and inhibited recombinant Kir1.1 channels (21). We next asked whether the Kir2.1 and Kir7.1 channels that were not activated by oleoyl-CoA showed higher phosphoinositide specificities than were exhibited by KATP channels. Fig. 2B shows the specificity of phosphoinositide activation of Kir2.1 and Kir7.1 channels. Both these channels showed markedly higher specificity toward PI(4,5)P2 than Kir6.2 channels. Kir2.1 was activated only by PI(4,5)P2, whereas activation by PI(3,4)P2 and PI(3,4,5)P3 was negligible. Kir7.1 was activated by PI(3,4,5)P3, less than by PI(4,5)P2, and not at all by PI(3,4)P2. The specificity profile obtained in these experiments with diC8 phosphoinositides for Kir2.1 channels is similar to that obtained with long acyl chain PIP2 analogs (13), confirming that the results obtained with the diC8 analogs are a good representation of the specificity of Kir channels with native phosphoinositides.
Figure 2.
Reversible activation of Kir channels by diC8 PIP2 analogs and inhibition by oleoyl-CoA. Inside-out macropatch measurements on recombinant Kir channels expressed in Xenopus oocytes were performed as in Fig. 1. (A Upper) Representative trace for Kir2.1 channels. Repetitive applications of 25 μM diC8 PI(4,5)P2 are shown by the horizontal lines before and after the application of 10 μM oleoyl-CoA. (Lower) A similar measurement on Kir7.1 (B Upper) Kir2.1 channels. The applications of 25 μM diC8 PI(4,5)P2, PI(3,4,5)P3, and PI(3,4)P2 are shown by the horizontal lines. At the end of the experiment, 5 μM AASt PI(4,5)P2 was applied. (Lower) A similar measurement on Kir7.1.
Specificity Profile of Kir Channels.
We tested the effect of oleoyl-CoA on several other members of the Kir channel family, which were as follows: Kir1.1, Kir2.2, Kir2.3, Kir4.1, Kir4.2, and Kir3.4* (the homomerically active GIRK4 mutant S143T; ref. 30). We found that none of these channels was activated by 10 μM oleoyl-CoA (data not shown). We then asked whether Kir channels that failed to be activated by LC acyl-CoA exhibited higher phosphoinositide specificity, as was the case with Kir7.1 and Kir2.1.
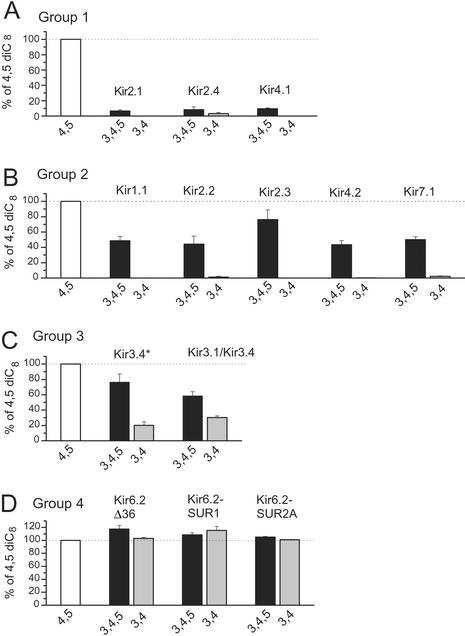
Fig. 3 shows the phosphoinositide-specificity profile of most members of the Kir channel family, expressed as a percentage of the effect of 25 μM diC8 PI(4,5)P2. Based on the diverse specificity of activation by phosphoinositides, we divided Kir channels into four groups. Group 1 contains Kir2.1, Kir2.4, and Kir4.1 (Fig. 3A). These channels show marginal or no activation by PI(3,4)P2, whereas the activation by PI(3,4,5)P3 is <10% of the effect of PI(4,5)P2. Group 2 contains Kir1.1, Kir2.2, Kir2.3, Kir4.2, and Kir7.1 (Fig. 3B). These channels again show marginal or no activation by PI(3,4)P2, but they do show significant activation by PI(3,4,5)P3, which is 40–80% of the effect of PI(4,5)P2. The third group is composed of the Kir3 subfamily members, Kir3.4* homomers, or Kir3.1/Kir3.4 heteromers (Fig. 3C). These channels were activated by PI(3,4)P2 but less so than by PI(3,4,5)P3 or PI(4,5)P2. Group 4 contains Kir6.2, expressed alone or together with SUR1 or SUR2A (Fig. 3D). These channels did not show any preference for PI(4,5)P2; in fact, the activation by PI(3,4,5)P3 or PI(3,4)P2 was slightly higher. In summary, all Kir channels, with the notable exception of Kir6.2, exhibited some preference for PI(4,5)P2 over PI(3,4,5)P3, showing little (Kir3) or no activation by PI(3,4)P2.
Figure 3.
Phosphoinositide specificity profile of the Kir channel family. Inside-out macropatch experiments were performed as shown in Fig. 1. The concentrations of diC8 PI(4,5)P2, PI(3,4)P2, and PI(3,4,5)P3 were 25 μM in all experiments. Data are expressed as a percentage of the activation by diC8 PI(4,5)P2. Because in some patches the effect of PI(4,5)P2 had an increasing or decreasing tendency, diC8 PI(4,5)P2 was applied repeatedly in every experiment. The effects of PI(3,4)P2 and PI(3,4,5)P3 were compared with the average of the following and preceding applications in such cases. A–D show, respectively, groups 1–4 of Kir channels based on the specificity of their activation by phosphoinositides, as described in the text.
We chose to use a single concentration of the diC8 phosphoinositides (25 μM) to compare their relative effects on the various Kir channels, after having performed dose–response measurements with diC8 PI(4,5)P2 for three of the channels shown in Fig. 3. Kir2.1 (14), Kir3.4* (31), and Kir6.2Δ36 gave EC50 activation values of 4.6 ± 0.7 μM, 17.7 ± 1.0 μM, and 62.4 ± 13.9 μM, respectively. diC8 PI(4,5)P2 (25 μM) activated Kir2.1 channels to 75.5 ± 5.2% (n = 7) of the cell attached level, indicating that 25 μM diC8 PI(4,5)P2 activates these channels somewhat less than the natural PI(4,5)P2 present in the oocyte membrane. We did not construct full dose–response curves with PI(3,4)P2 and PI(3,4,5)P3, because their concentrations in biological membranes are much lower than that of PI(4,5)P2 (see Discussion) and 25 μM represents a concentration that is most likely higher than physiological. For this reason, we tested 2.5 μM diC8 PI(3,4,5)P3 on Kir7.1 channels, which gave currents that were 4 ± 1% (n = 4) of those elicited by 25 μM PI(4,5)P2.
Amino Acids Involved in Phosphoinositide Specificity of Kir2.1.
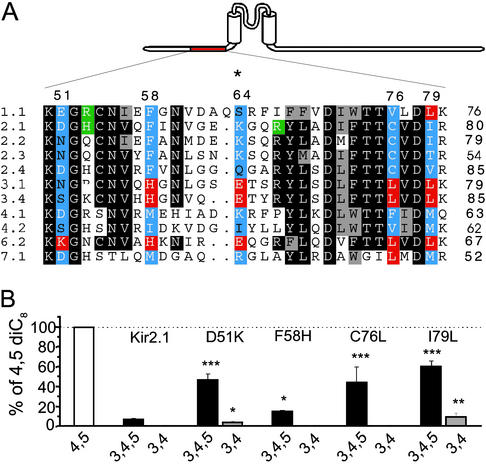
Having characterized the phosphoinositide specificity pattern of Kir channel family members, we searched for amino acid positions where the distribution of the physicochemical properties of specific residues correlated with specificity of activation by phosphoinositides. By careful inspection of the sequence alignment of the Kir channel family, and by using a computer algorithm (see Methods), we identified eight residues in the C terminus and five in the N terminus as potential determinants of specificity. We systematically mutated these amino acids (blue; Fig. 4A) in the background of the highly specific channel Kir2.1 to the corresponding residues (red; Fig. 4A) found in the least specific channel, Kir6.2. Fig. 4B shows that mutation of four N-terminal residues of Kir2.1 to the corresponding residues in Kir6.2 mainly increased sensitivity to PI(3,4,5)P3, compared with the wild-type Kir2.1. Mutation of the fifth predicted residue, K64E, rendered the channels nonfunctional. Similarly, Fig. 5 A and B shows the seven C-terminal residues tested, only two of which had small, but significant, effects on PI(3,4,5)P3 sensitivity. Mutation of an eighth predicted residue, H226S, prevented activation of these channels by diC8 phosphoinositides. Surprisingly, although the C-terminal cytoplasmic portion of Kir channels is longer than the N-terminal, and it contains the majority of PIP2-interacting residues (14, 32), the most dramatic effects were seen with N-terminal mutations.
Figure 4.
Phosphoinositide specificity of N-terminal mutants of Kir2.1. (A) Multiple sequence alignment of the proximal N terminus of Kir channels. Through our computer algorithm, blue residues correlated with specificity and red residues correlated with the lack of specificity. Putative PIP2-interacting residues are green (see ref. 14). Numbers on the top correspond to positions of PIP2-specificity-candidate residues in Kir2.1. The * denotes a nonfunctional mutant (K64E). (B) Phosphoinositide activation of individual mutants in the background of Kir2.1. Experiments were performed and data are displayed as in Fig. 3. Asterisks denote a statistically significant difference from the wild-type Kir2.1: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 5.
Phosphoinositide specificity of the C-terminal and combination Kir2.1 mutants. (A) Multiple sequence alignment of the cytoplasmic C terminus of Kir channels. Only segments containing residues that correlated with phosphoinositide specificity are shown for clarity. Blue residues correlated with specificity and red residues correlated with the lack of specificity. Putative PIP2-interacting residues are green (see refs. 14 and 33). Numbers on the top correspond to positions of PIP2-specificity-candidate residues in Kir2.1. The * denotes a mutant that was not activated by diC8 phosphoinositides (H226S). (B) Phosphoinositide activation of individual mutants in the background of Kir2.1. Experiments were performed and data are displayed as in Fig. 3. (C) Phosphoinositide activation of combination mutants in the background of Kir2.1. Asterisks denote statistically significant differences from the wild-type Kir2.1. *, P < 0.05; ***, P < 0.001.
As none of the N- or C-terminal mutants converted the phosphoinositide specificity of Kir2.1 to that of Kir6.2, we tested the effects of combinations of these mutations on phosphoinositide specificity of Kir2.1 channels. The double-mutant D51K-K185Q (Fig. 5C) was somewhat less specific than either D51K (Fig. 4B) or K185Q alone (Fig. 5B). PI(3,4,5)P3 activated the triple D51K-I79L-K185Q mutant to a similar extent as PI(4,5)P2; however, the activation by PI(3,4)P2 was still <10% of the activation by PI(4,5)P2. The triple D51K-C76L-K185Q and the quadruple D51K-C76L-I79L-K185Q mutants were stimulated by PI(3,4,5)P3 similarly to the triple mutant D51K-I79L-K185Q (Fig. 5C). Additional mutations (F58H, M180F) in these triple- and quadruple-mutant constructs failed to render these channels less specific (data not shown).
Effect of LC Acyl-CoA on Kir2.1 Mutants.
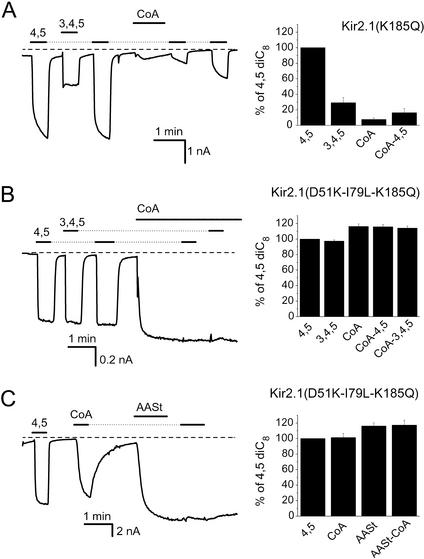
To examine whether the activation of Kir6.2 channels by LC acyl-CoA is because of their unique low phosphoinositide specificity, we tested whether oleoyl-CoA could activate Kir2.1 channel mutants with reduced phosphoinositide specificities. Fig. 6A shows that the K185Q mutant of Kir2.1 channels, showing intermediate phosphoinositide specificity, exhibited only a slight and transient activation by 10 μM oleoyl-CoA, whereas channel activation by diC8 PI(4,5)P2 was still inhibited by the presence of oleoyl-CoA. On oleoyl-CoA washout, the response to diC8 PI(4,5)P2 gradually recovered (Fig. 6A). This result is similar to that obtained with the wild-type Kir2.1 channels (Fig. 2A). The D51K mutant of Kir2.1 was also inhibited by oleoyl-CoA (data not shown). The triple-mutant D51K-I79L-K185Q, the least specific mutant we have produced (see Fig. 5C), on the other hand, was activated by oleoyl-CoA (Fig. 6B). Similar to KATP channels, application of diC8 PI(4,5)P2 or PI(3,4,5)P3 after oleoyl-CoA did not further activate the channels (Fig. 6B). At submaximal concentrations, partial occlusion of the effect of 5 μM diC8 PI(4,5)P2 occurred by 3 μM oleoyl-CoA (data not shown). Oleoyl-CoA application after channel activation by AASt PI(4,5)P2 revealed no further activation (Fig. 6C). These data suggest that oleoyl-CoA and the phosphoinositides acted through a common mechanism. Two other combination mutants of Kir2.1 with altered PIP2 specificity (D51K-C76L-K185Q and D51K-C76L-I79L-K185Q; Fig. 5C) were also activated by oleoyl-CoA (data not shown). In general, oleoyl-CoA activation of Kir2.1 mutants correlated well with the loss of specificity for PI(4,5)P2.
Figure 6.
Effects of oleoyl-CoA and phosphoinositides on mutant Kir2.1 channels. Inside-out macropatch measurements on recombinant Kir channels expressed in Xenopus oocytes were performed as in Fig. 1. The traces show currents at −80 mV after channel rundown. (A) Representative trace for Kir2.1(K185Q) channels. The applications of 25 μM diC8 PI(4,5)P2 and 25 μM diC8 PI(3,4,5)P3 are shown by the horizontal lines before and after the application of 10 μM oleoyl-CoA. (B) Similar measurement as in A with the triple-mutant Kir2.1(D51K-I79L-K185Q) channels. (C) The effect of 10 μM oleoyl-CoA on the triple-mutant of Kir2.1 before and after 5 μM AASt PI(4,5)P2. Twenty-five micromolar diC8 PI(4,5)P2 was applied at the beginning of the experiment.
These data demonstrate that LC acyl-CoA activation of Kir channels depends on their low phosphoinositide specificity.
Discussion
Regulation of the activity of ion channels and transporters by phosphoinositides presents an emerging theme in signal transduction (2). Despite the large number of phosphoinositide-regulated ion transport systems, the question of phosphoinositide specificity has not yet been examined in a systematic manner. Here we determined the specificity profile of the most extensively studied PIP2-regulated ion channel family, the Kir channels. We demonstrate that differences in phosphoinositide specificity of action on Kir channels are functionally important, because they determine the effectiveness of different lipids, such as phosphoinositides and LC acyl-CoA.
The Effect of LC Acyl-CoA Depends on Phosphoinositide Specificity.
KATP channels act as metabolic sensors activated by a rise in LC acyl-CoA (21, 22) in physiological and pathophysiological conditions (20, 22). Despite their differences in biosynthesis, chemical structure, and overall biological roles, the effects of PI(4,5)P2 and LC acyl-CoA on KATP channels share a number of similarities. Both acyl-CoA and PI(4,5)P2 act directly on the Kir6.2 subunit, because they are effective on recombinant Kir6.2Δ36 channels expressed without SUR subunits (21). LC acyl-CoA, like PI(4,5)P2, not only activates run-down Kir6.2 channels but also reduces their ATP sensitivity (21, 22). Finally, even though LC acyl-CoA lacks the glycerol backbone and the inositol ring of PI(4,5)P2, it has one long acyl chain and three phosphates in its head group, similar to PI(4,5)P2 (Fig. 1).
Despite these similarities, two different laboratories concluded that PI(4,5)P2 and LC acyl-CoA act through different mechanisms (21, 22), because other Kir channels (e.g., Kir1.1 and the cardiac inward-rectifiers, Kir2.0) were not activated by LC acyl-CoA, even though they were activated by PI(4,5)P2. Our study strongly suggests that acyl-CoA and phosphoinositides activate KATP channels through a similar mechanism, perhaps acting on the same binding site. In support of this interpretation, KATP channels activated by oleoyl-CoA were insensitive to further activation by PIP2 analogs; conversely, oleoyl-CoA was ineffective after activation of the channel by PI(4,5)P2. Activation by oleoyl-CoA depended on low phosphoinositide specificity of activation, because other wild-type Kir channels that showed higher specificity to PI(4,5)P2 were generally inhibited by oleoyl-CoA. Mutations that reduced the specificity of phosphoinositide activation of Kir2.1 channels rendered them sensitive to LC acyl-CoA activation. The effect of mutations could be to allow LC-CoA to fully bind, or alternatively, to enable the fully bound but not effective LC-CoA to stimulate activity.
PI(4,5)P2 Is the Preferred Phosphoinositide Activator of All Kir Channels Except KATP.
We have shown in this study that Kir channels, other than Kir6.2, were preferentially activated by PI(4,5)P2. This finding may reflect higher binding affinity of PI(4,5)P2, or alternatively, stronger allosteric coupling between phospholipid binding and channel activation. Regardless of the precise mechanism of phosphoinositide specificity in channel activation, it is clear that channel activity is stimulated more efficiently by PI(4,5)P2 than by PI(3,4,5)P3 and PI(3,4)P2.
Bulk measurements of phosphoinositides in biological membranes by biochemical techniques show that PI(3,4)P2 and PI(3,4,5)P3 are barely detectable in resting cells, and their concentration even in stimulated cells is thought to be <10% of that of PI(4,5)P2 (1). Given its much higher concentration in the plasma membrane, PI(4,5)P2 seems to be the major specific physiological activator of almost all Kir channels, even though contributions to Kir channel activity by other phosphoinositides cannot be ruled out, especially in the microenvironment of the channels. KATP channels, on the other hand, may also be regulated by other phosphoinositides as well as other negatively charged lipids.
In conclusion, the only Kir channel that we were able to activate with LC acyl-CoA was Kir6.2. Other Kir channels that showed higher specificity toward PI(4,5)P2 were not activated by LC acyl-CoA. We showed that it is the low phosphoinositide specificity that determines LC acyl-CoA activation, because Kir2.1 mutants with low phosphoinositide specificities could also be activated by LC acyl-CoA. Our data demonstrate that phosphoinositide specificity determines regulation of ion channels by distinct lipid molecules.
Acknowledgments
We thank Dr. X. Yan and H. Walsh for oocyte preparation and Dr. K. W. Chan for help with the Kir6.2 constructs. We thank Dr. Jian Yang and members of the Logothetis laboratory for critical comments on the manuscript. We thank the following colleagues for sharing cDNA clones with us: Dr. L. Y. Jan (University of California, San Francisco) for mouse Kir2.1 and Kir2.3, Dr. A. Karschin (University of Würzburg, Würzburg, Germany) for rat Kir2.4, Dr. C. G. Nichols (Washington University, St. Louis) for mouse Kir4.2, Dr. J. Bryan (Baylor College of Medicine, Houston) for hamster SUR1, Dr. S. Seino (Chiba University, Chiba, Japan) for mouse Kir6.2 and rat SUR2A, and Dr. D. E. Clapham (Children's Hospital, Boston) for human Kir7.1. Finally, we thank Dr. A. Rosenhouse-Dantsker for posting the computer software used in this study on the laboratory web site. This work was supported by National Institutes of Health Grant HL59949 (to D.E.L.). D.E.L. is an Established Investigator of the American Heart Association. T.R. was supported by a postdoctoral fellowship from the Revson Foundation and a National Research Service Award from the National Institutes of Health.
Abbreviations
- Kir
inwardly rectifying K+
- LC acyl-CoA
long-chain acyl-coenzyme A
- diC8
dioctanoyl
- KATP
ATP-sensitive K+ channel
- PI(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PI(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate
AASt, arachidonylstearoyl
- SUR
sulphonylurea receptor
- PIP2
phosphatidylinositol bisphosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fruman D A, Meyers R E, Cantley L C. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Hilgemann D W, Feng S, Nasuhoglu C. Sci STKE. 2001;2001:RE19. doi: 10.1126/stke.2001.111.re19. [DOI] [PubMed] [Google Scholar]
- 3.Nichols C G, Lopatin A N. Annu Rev Physiol. 1997;59:171–191. doi: 10.1146/annurev.physiol.59.1.171. [DOI] [PubMed] [Google Scholar]
- 4.Ashcroft F M, Gribble F M. Diabetologia. 1999;42:903–919. doi: 10.1007/s001250051247. [DOI] [PubMed] [Google Scholar]
- 5.Babenko A P, Aguilar-Bryan L, Bryan J. Annu Rev Physiol. 1998;60:667–687. doi: 10.1146/annurev.physiol.60.1.667. [DOI] [PubMed] [Google Scholar]
- 6.Sui J L, Petit-Jacques J, Logothetis D E. Proc Natl Acad Sci USA. 1998;95:1307–1312. doi: 10.1073/pnas.95.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shyng S L, Nichols C G. Science. 1998;282:1138–1141. doi: 10.1126/science.282.5391.1138. [DOI] [PubMed] [Google Scholar]
- 8.Hilgemann D W, Ball R. Science. 1996;273:956–959. doi: 10.1126/science.273.5277.956. [DOI] [PubMed] [Google Scholar]
- 9.Huang C L, Feng S, Hilgemann D W. Nature. 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 10. Baukrowitz, T., Schulte, U., Oliver, D., Herlitze, S., Krauter, T., Tucker, S. J., Ruppersberg, J. P. & Fakler, B. (1998) Science 1141–1144. [DOI] [PubMed]
- 11.Fan Z, Makielski J C. J Biol Chem. 1997;272:5388–5395. doi: 10.1074/jbc.272.9.5388. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, He C, Yan X, Mirshahi T, Logothetis D E. Nat Cell Biol. 1999;1:183–188. doi: 10.1038/11103. [DOI] [PubMed] [Google Scholar]
- 13.Rohács T, Chen J, Prestwich G D, Logothetis D E. J Biol Chem. 1999;274:36065–36072. doi: 10.1074/jbc.274.51.36065. [DOI] [PubMed] [Google Scholar]
- 14.Lopes C M B, Zhang H, Rohács T, Jin T, Logothetis D E. Neuron. 2002;34:933–944. doi: 10.1016/s0896-6273(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 15.Ho I H M, Murrell-Lagnado R. J Physiol (London) 1999;520:645–651. doi: 10.1111/j.1469-7793.1999.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng W Z, Liou H H, Krishna U M, Falck J R, Huang C L. Am J Physiol. 2002;282:F826–F834. doi: 10.1152/ajprenal.00300.2001. [DOI] [PubMed] [Google Scholar]
- 17.Krauter T, Ruppersberg J P, Baukrowitz T. Mol Pharmacol. 2001;59:1086–1093. [PubMed] [Google Scholar]
- 18.Fan Z, Gao L, Wang W. Am J Physiol. 2003;284:C94–C102. doi: 10.1152/ajpcell.00255.2002. [DOI] [PubMed] [Google Scholar]
- 19.Zammit V A. Biochem J. 1999;343:505–515. [PMC free article] [PubMed] [Google Scholar]
- 20.Deeney J T, Prentki M, Corkey B E. Semin Cell Dev Biol. 2000;11:267–275. doi: 10.1006/scdb.2000.0175. [DOI] [PubMed] [Google Scholar]
- 21.Gribble F M, Proks P, Corkey B E, Ashcroft F M. J Biol Chem. 1998;273:26383–26387. doi: 10.1074/jbc.273.41.26383. [DOI] [PubMed] [Google Scholar]
- 22.Liu G X, Hanley P J, Ray J, Daut J. Circ Res. 2001;88:918–924. doi: 10.1161/hh0901.089881. [DOI] [PubMed] [Google Scholar]
- 23.Larsson O, Deeney J T, Branstrom R, Berggren P O, Corkey B E. J Biol Chem. 1996;271:10623–10626. doi: 10.1074/jbc.271.18.10623. [DOI] [PubMed] [Google Scholar]
- 24.van der Vusse G J, Glatz J F, Stam H C, Reneman R S. Physiol Rev. 1992;72:881–940. doi: 10.1152/physrev.1992.72.4.881. [DOI] [PubMed] [Google Scholar]
- 25.Rohács T, Lopes C M B, Mirshahi T, Jin T, Zhang H, Logothetis D E. Methods Enzymol. 2002;345:71–92. doi: 10.1016/s0076-6879(02)45008-2. [DOI] [PubMed] [Google Scholar]
- 26.Chan K W, Langan M N, Sui J L, Kozak J A, Pabon A, Ladias J A, Logothetis D E. J Gen Physiol. 1996;107:381–397. doi: 10.1085/jgp.107.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liman E R, Tytgat J, Hess P. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 28.Tucker S J, Gribble F M, Zhao C, Trapp S, Ashcroft F M. Nature. 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- 29.Zerangue N, Schwappach B, Jan Y N, Jan L Y. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 30.Vivaudou M, Chan K W, Sui J L, Jan L Y, Reuveny E, Logothetis D E. J Biol Chem. 1997;272:31553–31560. doi: 10.1074/jbc.272.50.31553. [DOI] [PubMed] [Google Scholar]
- 31.Jin T, Peng L, Mirshahi T, Rohács T, Chan K W, Sanchez R, Logothetis D E. Mol Cell. 2002;10:469–481. doi: 10.1016/s1097-2765(02)00659-7. [DOI] [PubMed] [Google Scholar]
- 32.Cukras C A, Jeliazkova I, Nichols C G. J Gen Physiol. 2002;120:437–446. doi: 10.1085/jgp.20028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shyng S L, Cukras C A, Harwood J, Nichols C G. J Gen Physiol. 2000;116:599–608. doi: 10.1085/jgp.116.5.599. [DOI] [PMC free article] [PubMed] [Google Scholar]