Abstract
Alkaloids comprise one of the largest groups of plant secondary metabolites. Berberine, a benzylisoquinoline alkaloid, is preferentially accumulated in the rhizome of Coptis japonica, a ranunculaceous plant, whereas gene expression for berberine biosynthetic enzymes has been observed specifically in root tissues, which suggests that berberine synthesized in the root is transported to the rhizome, where there is high accumulation. We recently isolated a cDNA encoding a multidrug-resistance protein (MDR)-type ATP-binding cassette (ABC) transporter (Cjmdr1) from berberine-producing cultured C. japonica cells, which is highly expressed in the rhizome. Functional analysis of Cjmdr1 by using a Xenopus oocyte expression system showed that CjMDR1 transported berberine in an inward direction, resulting in a higher accumulation of berberine in Cjmdr1-injected oocytes than in the control. Typical inhibitors of ABC proteins, such as vanadate, nifedipine, and glibenclamide, as well as ATP depletion, clearly inhibited this CjMDR1-dependent berberine uptake, suggesting that CjMDR1 functioned as an ABC transporter. Conventional membrane separation methods showed that CjMDR1 was localized in the plasma membrane of C. japonica cells. In situ hybridization indicated that Cjmdr1 mRNA was expressed preferentially in xylem tissues of the rhizome. These findings strongly suggest that CjMDR1 is involved in the translocation of berberine from the root to the rhizome.
Plant cells produce a large variety of alkaloids, which have diverse chemical structures and biological activities. Some of them are used as medicines, such as anticancer drugs, and play an important role in plants as an endogenous biological barrier to protect against pathogens or herbivores because of their strong antimicrobial activities and cytotoxicity. For instance, berberine, a yellow benzylisoquinoline alkaloid, which is used as a bitter stomachic and an antidiarrhetic, shows strong antimicrobial activity toward both Gram-positive and -negative bacteria as well as other microorganisms (1).
In Coptis japonica (Ranunculaceae), a perennial medicinal plant grown in Asian countries, berberine is highly accumulated in the rhizome as the main alkaloid. However, the genes for the biosynthesis of berberine are specifically expressed in the root tissue (2), where only a low level of berberine is detected. This finding suggests that berberine is transported from the root after biosynthesis and is accumulated highly in the rhizome. Cultured C. japonica cells produce berberine and it is accumulated exclusively in the vacuoles (3, 4). Furthermore, exogenous berberine that is added to the culture medium is actively taken up by C. japonica cells (3–5) and is also accumulated in the cell vacuoles (6).
Our previous attempts to identify the transporter of berberine in C. japonica cells showed that the uptake of berberine by C. japonica cells depended on the ATP level, and several inhibitors of P-glycoprotein (gene product of mdr1), a representative ATP-binding cassette (ABC) protein responsible for multiple drug resistance of cancer cells (7), significantly suppressed berberine uptake from the medium (5). These results suggested that an ABC protein was involved in berberine transport in this plant. Based on this idea, we isolated Cjmdr1 (Coptis japonica multidrug resistance 1) as a potential berberine transporter from C. japonica cells by using RT-PCR (8). Cjmdr1 was expressed preferentially in the rhizome, where berberine is highly accumulated compared with other organs (8). Multidrug-resistance protein (MDR) belongs to a subfamily of the ABC superfamily that is widely distributed from prokaryotes to eukaryotes and is involved in the active transport of many divergent compounds in an ATP-dependent manner (9). Based on the results of the Arabidopsis genome project, this plant contains a large number of members of this family, e.g., 129 ORFs in Arabidopsis thaliana (10, 11). However, only a few members have been functionally characterized (12, 13), and the MDR subfamily in particular is even less understood.
In the present paper, we describe a functional analysis of Cjmdr1 by using Xenopus oocytes. We also analyzed the intracellular and cell-specific localization of the gene product of Cjmdr1 to characterize the physiological function of this protein.
Materials and Methods
Cultured Cells.
High berberine-producing cultured C. japonica cells, which were originally induced from the rootlets of C. japonica Makino var. dissecta (Yamabe) Nakai, were maintained as described (14).
Chemicals.
Berberine and other chemicals used in this study were purchased from Wako Pure Chemicals (Osaka) or Nakalai Tesque (Kyoto).
Oocyte Isolation and Injection.
An African clawed frog, Xenopus laevis, was anesthetized by hypothermia (20 min in ice-cold water). Part of one ovary was removed, cut into small portions, and placed in OR2 medium (82.5 mM NaCl/2 mM KCl/1 mM MgCl2/5 mM Hepes–NaOH, pH 7.4). Oocytes were prepared as described (15), and mature stage VI oocytes were injected with Cjmdr1 mRNA (50 nl, 1 ng/nl), or sterile water for control oocytes, by using a micropipette attached to a hydraulic microinjector (Nanoject, Drummond Scientific, Broomall, PA). Glass micropipettes with a tip diameter of 10–30 μm were pulled on a Narishige PE6 pipette puller (Tokyo). Injected oocytes were kept at 18°C for 3 days in ND96 medium (96 mM NaCl/2 mM KCl/1.8 mM MgCl2/1 mM CaCl2/2.5 mM sodium pyruvate/5 mM Hepes–NaOH, pH 7.4), which ensured 90% survival. Damaged oocytes were separated from intact oocytes bearing a clear brown pole animal hemisphere and a distinct equator line.
Expression of CjMDR1 in Xenopus Oocytes.
Cjmdr1 cDNA (4.1 kb) was subcloned in a pCF3 vector derived from the Xenopus oocyte expression vector pBSTA, kindly provided by A. L. Goldin (University of California, Irvine), after slight modifications. The pBSTA vector contains a T7 promoter followed by 5′ noncoding β-globin sequences, a single BglII site for insertion of exogenous DNA, 3′ noncoding β-globin sequences, a poly(A) tail, and a polylinker that allows the plasmid to be linearized before in vitro transcription (16). The addition of 5′ and 3′ noncoding sequences from the β-globin gene greatly increases the expression of exogenous proteins in oocytes (17). In vitro transcription followed by capping of the RNA was performed with the plasmid (1 μg) linearized at the unique site AscI by using the T7 mMESSAGE mMACHINE kit (Ambion, Austin, TX). The template DNA was degraded by DNase I, and Cjmdr1 mRNA was purified on a formaldehyde gel and quantified.
Measurement of Berberine Transport in Oocytes.
All transport experiments were conducted 2–3 days after the injection of oocytes at 18°C. Fluorescence in the oocytes derived from berberine was monitored by using a fluorescence microscope (excitation 420 nm/emission 517 nm). To quantitatively determine the influx of berberine into oocytes, water- and Cjmdr1-injected oocytes (15–20 oocytes per batch) were incubated in the presence of berberine chloride (0.3 or 1 mM) for 1 h unless otherwise specified. In time course experiments, oocytes exposed to 1 mM berberine were sampled periodically (3, 5, 15, 30, and 60 min), lysed in ND96 (pH 5), and lyophilized, and 2 mg of dried material was used for HPLC analysis. Efflux experiments were performed by transferring oocytes loaded with berberine into fresh ND96 solution after careful washing to remove extracellular berberine. Inhibitors were added to the oocyte suspension and incubated for 15 min at 4°C. After 60 min of efflux, the remaining berberine in the oocytes was quantitatively analyzed by HPLC.
The ATP-dependent uptake of berberine in Cjmdr1-injected oocytes was also investigated by depleting ATP from oocytes with a metabolic inhibitor (1 mM potassium cyanide), by suppressing the ATPase activity of the transporter (1 mM vanadate), or by applying classical blockers of ABC transporters (100 μM nifedipine, verapamil, or glibenclamide).
HPLC Conditions.
For HPLC analysis, dried oocytes were suspended in 50% methanol (0.01 M HCl) by vigorous pipetting, and centrifuged at 10,000 × g for 10 min. The supernatant was subjected to HPLC analysis: mobile phase, 50 mM tartaric acid solution containing 10 mM SDS, acetonitrile, and methanol (100:130:33); column, TSK-GEL ODS-80TM (Tosoh, Tokyo; 4.6 mm i.d. × 250 mm); temperature, 40°C; flow rate, 1.2 ml/min; detection, absorbance measured at 260 nm by using a photodiode array detector.
Peptide Antibody Against CjMDR1.
A keyhole limpet hemocyanin conjugate of an oligopeptide of CjMDR1, at position 378 (n-CSYDTSGHKSDDIRGD-c), was injected into a rabbit according to the standard protocol (Sawady Technology, Tokyo). After the sixth boost, antiserum was recovered and used for immunoblot analysis without further purification.
Expression of CjMDR1 in Yeast.
Cjmdr1 cDNA (4.1 kb) was subcloned into yeast expression vector pDR196 (18), kindly provided by W. Frommer (University of Tübingen, Tübingen, Germany), at the NotI site. The resulting plasmid, pDR-CjMDR1, was used to transform strain AD12345678 (yor1Δ, snq2Δ, pdr5Δ, pdr10Δ, pdr11Δ, ycf1Δ, pdr3Δ, pdr15Δ; ref. 19) by the lithium acetate method (20), and then selected by SD medium (−uracil). The yeast transformant was cultured in 100 ml of SD medium (−uracil), and membrane proteins were extracted for the detection of recombinant CjMDR1 as described (21).
Isolation of Plasma Membranes.
Plasma membranes were purified from a microsomal fraction of C. japonica cells by partitioning in an aqueous polymer two-phase system as described (22). The upper phase, in which the plasma membrane was enriched, and the lower phase were used for immunoblot analyses.
For sucrose gradient fractionation, a microsomal pellet was resuspended in a small volume of resuspension buffer and layered onto a noncontinuous gradient containing 10–50% (wt/vol) sucrose in centrifugation buffer (10 mM Tris⋅HCl, pH 7.6/1 mM DTT/1 mM EDTA). The gradient was then centrifuged at 100,000 × g for 2 h and the membranes were collected from the interface between different sucrose concentrations.
Protein Gel Blotting.
For immunoblotting, proteins were denatured in denaturation buffer for 10 min at 50°C, subjected to SDS/7% PAGE, and transferred to an Immobilon poly(vinylidene difluoride) membrane (Millipore). The membrane was treated with BlockAce (Dainippon Pharmaceutical, Osaka) for blocking, and incubated with primary antibodies and secondary horseradish peroxidase-conjugated anti-rabbit IgG antibodies by using standard procedures. The band was visualized by chemiluminescence. Antibodies used for immunodetection were against CjMDR1, vacuolar H+-pyrophosphatase from Arabidopsis (V-PPase), endoplasmic reticulum luminal binding protein (BiP), and plasma membrane H+-ATPase (H+-ATPase).
In Situ Hybridization.
Roots and rhizomes were fixed on ice in freshly prepared solutions of 2% and 4% formaldehyde in 50% ethanol and 10% acetic acid, respectively, for 3 h. The samples were dehydrated through an ethanol series, and embedded with Paraplast plus (Oxford). Then, 10- to 20-μm-thick sections prepared with a microtome (RM 2155, Leica) were hybridized with the digoxigenin-labeled antisense or sense RNA probe of Cjmdr1, which is a 178-bp fragment containing the 3′ untranslated region. In situ hybridization and detection procedures were performed as described (23).
Drug Sensitivity Assay.
The drug sensitivity of yeast transformants was tested by spotting SD cultures (−uracil) onto agar plates containing various compounds. Five microliters of the transformant diluted to same density, OD600 = 0.5, was spotted onto each plate and growth was monitored after incubation of the cells for 44 h at 25°C. The drugs, berberine and cycloheximide, used in this study were dissolved in water, whereas 4-nitroquinoline N-oxide (4-NQO) was dissolved in acetone.
Results
CjMDR1 Functions as a Berberine Influx Pump.
We expressed CjMDR1 in Xenopus oocytes to characterize the function of this ABC protein. Oocytes injected with Cjmdr1 mRNA, or water as a negative control, were placed in ND96 medium containing berberine, washed thoroughly, and then observed under a fluorescence microscope. The Cjmdr1-injected oocytes showed higher fluorescence derived from berberine than the control oocytes, suggesting that CjMDR1 might function as a drug influx pump. This result was unexpected because MDR-type ABC proteins usually function as drug efflux pumps and no eukaryotic ABC transporter has yet been reported, to our knowledge, to be involved in drug influx.
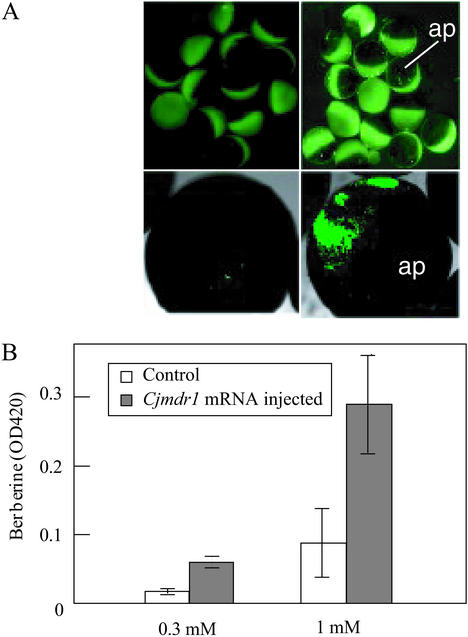
Both microscopic observation (Fig. 1A) and OD420 measurement of berberine accumulation in oocytes (Fig. 1B) showed that the Cjmdr1-injected oocytes had a higher content of berberine than did control oocytes. Confocal microscopic analysis indicated that berberine appeared to be mostly localized in the vegetative pole of the oocytes (Fig. 1A). Because CjMDR1 was suggested to have inward transport activity for berberine, the time course of berberine uptake was monitored quantitatively by HPLC analysis after the lysis of oocytes (Fig. 2). Cjmdr1-injected oocytes showed a constant uptake of berberine from the medium in a time-dependent manner, whereas the berberine level in control oocytes remained nearly unchanged at a low level.
Figure 1.
Berberine accumulation in Cjmdr1-injected oocytes and a negative control. (A) Fluorescent micrographs of Cjmdr1-injected oocytes (Right) and water-injected oocytes (Left). Both oocytes were incubated in the presence of 1 mM berberine in ND96 medium for 1 h. (Lower) Photographs were taken with a confocal microscope. ap, Animal pole. (B) Amounts of berberine accumulated in Cjmdr1-injected or water-injected oocytes, which were incubated in 0.3 or 1.0 mM berberine medium. Amounts of berberine were estimated based on optical density at 420 nm.
Figure 2.
Time course of berberine influx into oocytes injected with Cjmdr1 mRNA. Cjmdr1- and water-injected oocytes were incubated in berberine medium (1 mM) and sampled at the times indicated for transfer into the lysis buffer. The lysate was lyophilized and dissolved in 50% methanol to be analyzed by HPLC with a photodiode array detector. The quantity of berberine was calculated from the peak area.
To examine whether this berberine uptake depended on CjMDR1, the effect of the ATPase inhibitor and the intracellular ATP level was analyzed (Fig. 3A). The uptake of berberine observed in Cjmdr1-injected oocytes without inhibitor was strongly inhibited to the control level by a membrane ATPase inhibitor, vanadate, and similar results were observed on the addition of potassium cyanide, which causes ATP depletion. Effective ABC transporter-inhibitors, verapamil and nifedipine, which also function as Ca2+ channel blockers, inhibited berberine uptake by CjMDR1. Glibenclamide, another type of inhibitor for sulfonylurea receptor and plant ABC transporters, also clearly inhibited berberine uptake (Fig. 3A). The effects of bafilomycin A1 (0.1 μM), a specific V-type ATPase inhibitor, and NH (5 mM), which destroy ΔpH across the membrane, were also tested, but they did not significantly inhibit berberine uptake, i.e., <12% inhibition for both.
(5 mM), which destroy ΔpH across the membrane, were also tested, but they did not significantly inhibit berberine uptake, i.e., <12% inhibition for both.
Figure 3.
Effects of inhibitors commonly used for ABC transporters on berberine uptake in Cjmdr1-injected oocytes. (A) Amounts of berberine taken up in Cjmdr1- and water-injected oocytes after incubation in medium containing 1 mM berberine in the presence of inhibitor. Ber, no addition of inhibitor; potassium cyanide (KCN), 1 mM; vanadate (Vi), 1 mM; nifedipine (Np), 100 μM; verapamil (Vm), 100 μM; glibenclamide (Gb), 100 μM. (B) Berberine retained inside Cjmdr1-injected oocytes incubated in fresh medium after loading with berberine. Inhibitors were added 15 min before the oocytes were transferred from berberine-loading medium to fresh medium without berberine. Vm, 100 μM; Gb, 100 μM.
To characterize CjMDR1 activity, the berberine content in oocytes that were loaded with berberine and then incubated in fresh medium without berberine was quantitatively analyzed (Fig. 3B). Cjmdr1-injected oocytes retained a clearly higher berberine level than the negative control, and this berberine-retaining activity dramatically decreased when cells were treated with the ABC transporter-inhibitors, verapamil and glibenclamide, indicating that berberine was actively retained by CjMDR1.
Kinetic analysis of berberine uptake by Cjmdr1-injected oocytes revealed that the Km value and Vmax were 54.62 ± 5.46 μM and 0.75 ± 0.015 nmol/10 min per mg of protein, respectively (see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). The Km value of CjMDR1 for berberine was similar to, whereas its Vmax was ≈10-fold lower than, that of AtMRP1 for the glutathione conjugate of dinitrophenol (24).
CjMDR1 Is Localized at the Plasma Membrane.
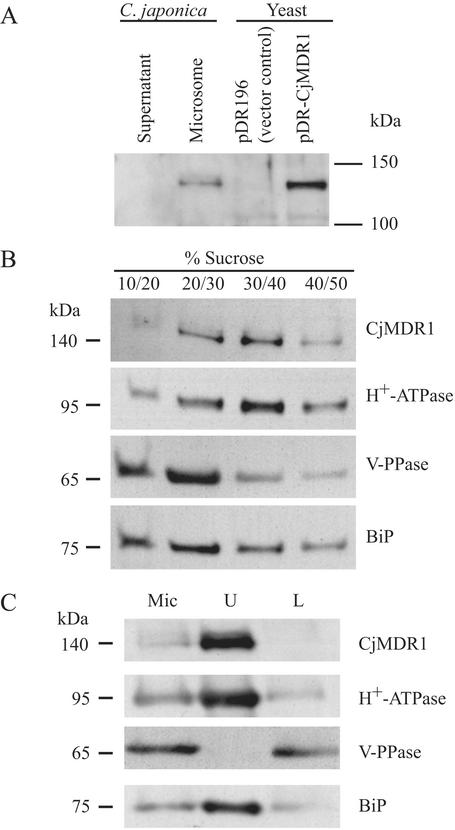
To understand the physiological function of CjMDR1 in planta, we determined its subcellular localization. For this purpose, we prepared polyclonal antibodies against a specific oligopeptide of CjMDR1 as described in Materials and Methods, and the reactivity of these antibodies to CjMDR1 was examined both in a heterologous expression system with yeast and in intact C. japonica membranes (Fig. 4A). In plant membranes, anti-CjMDR1 antibodies detected a band at 140 kDa, whereas no detectable protein was observed in the soluble fraction. A band at 140 kDa was also detected in the membranes of yeast cells transformed with Cjmdr1 cDNA, but was absent in the negative control using the empty vector.
Figure 4.
Immunodetection of CjMDR1 in C. japonica and yeast cells. (A) Soluble and membrane (microsomal fraction) proteins (10 μg per lane) of C. japonica cells and membrane proteins (10 μg per lane) of yeast expressing CjMDR1 or empty vector were separated by SDS/PAGE and blotted onto a poly(vinylidene difluoride) membrane. The membrane was probed with anti-CjMDR1 polyclonal antibodies. (B) Immunodetection of CjMDR1 in C. japonica plasma membranes. Fractionation of total microsomes from C. japonica cells on a noncontinuous sucrose gradient consisting of 10%, 20%, 30%, 40%, and 50% (wt/vol) sucrose. Membrane fractions were collected from the interfaces between different sucrose concentrations. Proteins from each interface were immunodetected with anti-CjMDR1 antibodies. Blots were probed with antibodies raised against CjMDR1, plasma membrane H+-ATPase, V-PPase, and endoplasmic reticulum BiP. (C) Microsomal membranes (Mic) from C. japonica cells were fractionated by the aqueous two-phase partitioning method into an upper phase (U) enriched with plasma membranes and a lower phase (L) containing intracellular membranes. Proteins from each fraction (10 μg per lane) were immunoblotted. The same membrane was reprobed with antibodies against the indicated marker proteins.
Using these antibodies, we determined the subcellular localization of CjMDR1 by two different methods. First, microsomal membranes were fractionated on sucrose density gradients and analyzed by immunoblotting (Fig. 4B). CjMDR1 and the plasma membrane H+-ATPase were cofractionated to give a peak at the interface between 30% and 40% sucrose, whereas membranes containing BiP, a marker of endoplasmic reticulum (25), and V-PPase, a marker of tonoplast membrane, sedimented at positions different from CjMDR1 (Fig. 4B). These results suggested that CjMDR1 was associated with the plasma membrane and not with the tonoplast membrane or endoplasmic reticulum.
The localization of CjMDR1 in the plasma membrane was also supported by an analysis of plasma membranes separated from other intracellular membranes of C. japonica by using the aqueous two-phase partitioning method (22). As shown in Fig. 4C, CjMDR1 was highly enriched in the upper-phase fraction (U) and greatly depleted in the lower-phase fraction (L), which is the same pattern as H+-ATPase. Membrane purity was confirmed by immunodetection of the marker proteins, although a relatively large portion of endoplasmic reticulum marker was also detected in the upper-phase fraction.
In Situ Localization of Cjmdr1 Transcripts.
Northern analysis showed that Cjmdr1 mRNA is highly expressed in the rhizome, where berberine is preferentially accumulated (8). To define the specific cell type in which Cjmdr1 mRNA is accumulated, in situ hybridization experiments were performed by using a digoxigenin-labeled antisense RNA probe prepared from the Cjmdr1 cDNA. The rhizome and the root, which correspond to the sink organ for berberine accumulation and the source organ for berberine biosynthesis, respectively, were sectioned and analyzed (Fig. 5). In the rhizome section, strong signals were observed around the vascular tissue, particularly in the xylem cells (Fig. 5 A–C), with the antisense probe, whereas no detectable signal was seen with the sense probe as a negative control (data not shown). A weak signal could also be detected in the cortex (Fig. 5C). However, almost no signals were detected in root sections with either the antisense or sense probe, although a faint signal was sometimes observed in the vascular tissue (Fig. 5 D and E). These results suggest that an ABC transporter, CjMDR1, might participate in the unloading of berberine in xylem tissues of the rhizome.
Figure 5.
In situ hybridization of Cjmdr1 in the rhizome and root of C. japonica. (A–C) In situ hybridization using a digoxigenin-labeled antisense probe in the rhizome. (D and E) In situ hybridization using a digoxigenin-labeled antisense probe in the root. BF, bast fiber; C, cortex; E, epidermis; M, medulla; Ph, phloem; VB, vascular bundle; X, xylem. (Bars, 500 μm.)
Other Possible Substrates of CjMDR1.
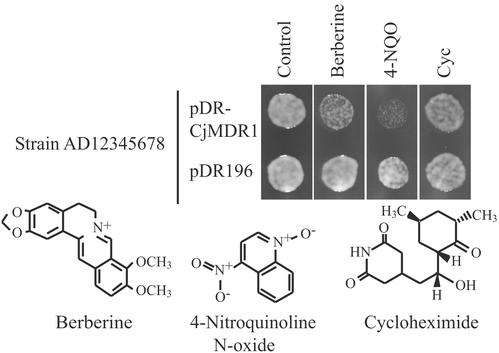
We examined whether CjMDR1 is exclusively specific to berberine by incubating oocytes with other natural compounds, e.g., reticuline, sanguinarine, quinine, and quercetin. However, we did not observe any difference in the uptake of these compounds between Cjmdr1-injected oocytes and the control except for reticuline, a precursor of berberine, whose uptake was slightly higher in Cjmdr1-oocyte only at concentrations as high as 1 mM (data not shown). Moreover, we also investigated the substrate specificity by estimating drug sensitivity in yeast transformants (Fig. 6). CjMDR1 was expressed with a shuttle vector, pDR196, with which a foreign gene is constitutively expressed by PMA1 promoter (18), in yeast Saccharomyces cerevisiae, strain AD12345678 (19). The strain AD12345678 lacks major yeast ABC transporter-encoding genes that confer multidrug resistance. The same yeast strain transformed with the empty vector was used as a negative control. As shown in Fig. 6, the CjMDR1 transformant appeared to be more sensitive to berberine, although the difference was rather small. A larger difference in drug sensitivity between CjMDR1 transformant and the control was found with 4-NQO, a substrate of yeast pleiotropic drug resistance (PDR)-type ABC transporter, SNQ2 (26). These data suggest that 4-NQO was recognized as a substrate of CjMDR1 and taken up into cells, and the sensitivity of yeast was increased. However, cycloheximide, a substrate of the yeast PDR-type ABC transporter, PDR5 (27), showed no clear difference between yeast cells, indicating that CjMDR1 had relatively strong substrate specificity.
Figure 6.
Drug sensitivity of yeast CjMDR1 transformant. Yeast strain AD12345678 was transformed with pDR196 (empty vector) or pDR-CjMDR1. Each transformant was precultured overnight in SD medium (−uracil). The cultures were diluted to OD600 = 0.5. Five microliters was spotted onto an SD medium (−uracil) plate containing berberine (3.5 μM), 4-NQO (0.11 μg/ml), or cycloheximide (Cyc; 0.0175 μg/ml), and incubated for 44 h at 25°C.
Discussion
We isolated Cjmdr1 as a gene encoding a potential berberine transporter from high berberine-producing cells of C. japonica (8), and found that this molecular species is expressed preferentially in the rhizome of the intact plant (8) where the main alkaloid berberine is specifically accumulated, whereas the biosynthetic enzymes we studied are expressed in root tissues (ref. 2; M. Tanaka, N. Izekawa, and F.S., unpublished data. The main root and lateral root should be referred to as the rhizome and root, respectively). In this study, we have provided direct evidence that this MDR-type ABC transporter, CjMDR1, takes up berberine in a heterologous system with Xenopus oocytes (Fig. 2). We also showed that CjMDR1 was expressed in xylem tissue in the rhizome (Fig. 5) and localized in the plasma membrane of C. japonica cells (Fig. 4). All of these findings are consistent with the hypothesis that this ABC protein is involved in the unloading of berberine in the rhizome; i.e., berberine is biosynthesized in root tissues and translocated upward probably through xylem transport, whereas in the rhizome, the berberine molecule is trapped by plasma membrane-localized CjMDR1 and accumulated in the rhizome tissue.
It is not yet clear why C. japonica has specific tissues for berberine biosynthesis and accumulation, but feedback inhibition of biosynthetic enzyme by berberine might be one possible explanation (28). The accumulation of berberine in the rhizome would also be beneficial for the plant, because the rhizome is a sink organ for starch and berberine may protect this tissue from attack by soil pathogens. Elicitor-induced induction of berberine biosynthesis also supports the physiological function of berberine and related alkaloids as phytoalexins (29). The compartmentation of biosynthetic cells/tissues and sink cells/tissues is generally observed for plant secondary metabolites, such as tropane alkaloids, nicotine alkaloids, and so on (30). The Coptis transport system may be a model for understanding such a transport mechanism.
Our investigation also showed that CjMDR1 functioned as an influx pump for berberine. Thus far, all of the mammalian ABC transporters characterized that are localized in the plasma membrane show excretion activity for substrates; e.g., P-glycoprotein and members of the multidrug-resistance-related protein (MRP) subfamily in animal cells efflux anticancer drugs from cytosol. Tonoplast-localized plant ABC transporters such as members of the MRP family export glutathione conjugates of xenobiotics and chlorophyll catabolite from cytosol into the vacuolar matrix. While such transport of CjMDR1 from outside the cells toward the cytosol has often been reported for prokaryotic organisms, we know of no other example for eukaryotic ABC protein. Because this transport activity was clearly observed in both Xenopus oocyte and yeast, we assume that CjMDR1 was sorted and folded actively in the correct orientation.
With regard to substrate specificity, CjMDR1 does not seem to be exclusively specific for the isoquinoline alkaloid berberine, an endogenous substrate in C. japonica, but may also recognize reticuline and 4-NQO, a berberine precursor and a quinoline derivative, respectively (Fig. 6). Sanguinarine, quinine, quercetin, and cycloheximide, which have different chemical structures, do not seem to be recognized by CjMDR1, which suggests that CjMDR1 does not show as broad a substrate specificity as other MDR-type ABC transporters, e.g., P-glycoprotein.
Plant ABC transporters comprise one of the most active research fields in recent years. The Arabidopsis genome project showed that this plant requires a large number of ABC protein superfamily members. Recently, there have been many excellent studies to elucidate the function of multidrug-resistance-related protein subfamily members (24, 31–33), PDR subfamily protein (34), and the unique peroxisome-localized ABC protein (35, 36). However, little is known about the endogenous substrates of these ABC proteins, and the MDR subfamily, such as PMDR1 of potato (37), HvMDR2 of barley (38), and TaMDR1 of wheat (39), is still open for investigation, except for AtPGP1 (40), one of the 22 members in Arabidopsis. The Arabidopsis ABC protein that is most similar to CjMDR1 is AtPGP4, the function of which is still unknown. Our present findings may provide some new clues to clarify their functions and physiological roles.
Supplementary Material
Acknowledgments
We thank Dr. W. Frommer, University of Tübingen (Tübingen, Germany) for the gift of pDR196; Dr. A. L. Goldin, University of California (Irvine) for the gift of pBSTA; Dr. M. A. De Waard, Wageningen University (Wageningen, The Netherlands) for kindly providing the yeast strain; Dr. M. Boutry, Université Catholique de Louvain (Louvain-la-Neuve, Belgium) for providing anti-H+-ATPase antibodies; Dr. N. Koizumi, Nara Institute of Science and Technology (Ikoma Nara, Japan) for providing anti-BiP antibodies; Dr. M. H. Sato, Kyoto University, for anti-V-PPase antibodies; and the Kyoto Botanical Garden of Takeda Chemical Industries for the intact C. japonica plants. This work was supported in part by Grant-in-Aid for Scientific Research 10217206 and Grant of Research for the Future Program 00L01605 from the Ministry of Education, Science, Sports, and Culture of Japan (to K.Y.), and by the Commissariat à l'Energie Atomique (to I.B. and C.F.).
Abbreviations
- ABC
ATP-binding cassette
- MDR
multidrug-resistance protein
- 4-NQO
4-nitroquinoline N-oxide
- PDR
pleiotropic drug resistance
- CjMDR1
Coptis japonica MDR1 protein
- BiP
luminal binding protein
- V-PPase
vacuolar H+-pyrophosphatase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Iwasa K, Nanba H, Lee D U, Kang S I. Planta Med. 1998;64:748–751. doi: 10.1055/s-2006-957572. [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara H, Takeshita N, Terano Y, Fitchen J H, Tsujita T, Katagiri Y, Sato F, Yamada Y. Phytochemistry. 1993;34:949–954. [Google Scholar]
- 3.Sato H, Kobayashi Y, Fukui H, Tabata M. Plant Cell Rep. 1990;9:133–136. doi: 10.1007/BF00232088. [DOI] [PubMed] [Google Scholar]
- 4.Sato F, Takeshita N, Fujiwara H, Katagiri Y, Huan L, Yamada Y. Plant Cell Tissue Organ Cult. 1994;38:249–256. [Google Scholar]
- 5.Sakai K, Shitan N, Sato F, Ueda K, Yazaki K. J Exp Bot. 2002;53:1879–1886. doi: 10.1093/jxb/erf052. [DOI] [PubMed] [Google Scholar]
- 6.Sato H, Tanaka S, Tabata M. Phytochemistry. 1993;34:697–701. [Google Scholar]
- 7.Ueda K, Cardarelli C, Gottesman M M, Pastan I. Proc Natl Acad Sci USA. 1987;84:3004–3008. doi: 10.1073/pnas.84.9.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yazaki K, Shitan N, Takamatsu H, Ueda K, Sato F. J Exp Bot. 2001;52:877–879. doi: 10.1093/jexbot/52.357.877. [DOI] [PubMed] [Google Scholar]
- 9.Higgins C F. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 10.Sánchez-Fernández R, Davies T G, Coleman J O, Rea P A. J Biol Chem. 2001;276:30231–30244. doi: 10.1074/jbc.M103104200. [DOI] [PubMed] [Google Scholar]
- 11.Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu U, Muller-Rober B, Schulz B. Planta. 2002;214:345–355. doi: 10.1007/s004250100661. [DOI] [PubMed] [Google Scholar]
- 12.Rea P A, Li Z-S, Lu Y-P, Drozdowicz Y M, Martinoia E. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:727–760. doi: 10.1146/annurev.arplant.49.1.727. [DOI] [PubMed] [Google Scholar]
- 13.Theodoulou F L. Biochim Biophys Acta. 2000;1465:79–103. doi: 10.1016/s0005-2736(00)00132-2. [DOI] [PubMed] [Google Scholar]
- 14.Sato F, Yamada Y. Phytochemistry. 1984;23:281–285. [Google Scholar]
- 15.Dascal N. CRC Crit Rev Biochem. 1987;22:317–387. doi: 10.3109/10409238709086960. [DOI] [PubMed] [Google Scholar]
- 16.Shih T M, Smith R D, Toro L, Goldin A L. Methods Enzymol. 1998;293:529–556. doi: 10.1016/s0076-6879(98)93032-4. [DOI] [PubMed] [Google Scholar]
- 17.Krieg P A, Melton D A. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer W B. FEBS Lett. 1995;370:264–268. doi: 10.1016/0014-5793(95)00853-2. [DOI] [PubMed] [Google Scholar]
- 19.Decottignies A, Grant A M, Nichols J W, de Wet H, McIntosh D B, Goffeau A. J Biol Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- 20.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yazaki K, Kunihisa M, Fujisaki T, Sato F. J Biol Chem. 2002;277:6240–6246. doi: 10.1074/jbc.M106387200. [DOI] [PubMed] [Google Scholar]
- 22.Larsson C, Widell S, Kjellbom P. Methods Enzymol. 1987;148:558–568. [Google Scholar]
- 23.Yamada S, Katsuhara M, Kelly W B, Michalowski C B, Bohnert H J. Plant Cell. 1995;7:1129–1142. doi: 10.1105/tpc.7.8.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y-P, Li Z-S, Drozdowicz Y M, Hortensteiner S, Martinoia E, Rea P A. Plant Cell. 1998;10:267–282. doi: 10.1105/tpc.10.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas I G. Experientia. 1994;50:1012–1020. doi: 10.1007/BF01923455. [DOI] [PubMed] [Google Scholar]
- 26.Servos J, Haase E, Brendel M. Mol Gen Genet. 1993;236:214–218. doi: 10.1007/BF00277115. [DOI] [PubMed] [Google Scholar]
- 27.Hirata D, Yano K, Miyahara K, Miyakawa T. Curr Genet. 1994;26:285–294. doi: 10.1007/BF00310491. [DOI] [PubMed] [Google Scholar]
- 28.Sato F, Takeshita N, Fitchen J H, Fujiwara H, Yamada Y. Phytochemistry. 1993;32:659–664. [Google Scholar]
- 29.Funk C, Gugler K, Brodelius P. Phytochemistry. 1987;26:401–405. [Google Scholar]
- 30.Hashimoto T, Yamada Y. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:257–285. [Google Scholar]
- 31.Liu G, Sánchez-Fernández R, Li Z-S, Rea P A. J Biol Chem. 2001;276:8648–8656. doi: 10.1074/jbc.M009690200. [DOI] [PubMed] [Google Scholar]
- 32.Tommasini R, Vogt E, Fromenteau M, Hortensteiner S, Matile P, Amrhein N, Martinoia E. Plant J. 1998;13:773–780. doi: 10.1046/j.1365-313x.1998.00076.x. [DOI] [PubMed] [Google Scholar]
- 33.Gaedeke N, Klein M, Kolukisaoglu U, Forestier C, Muller A, Ansorge M, Becker D, Mamnun Y, Kuchler K, Schulz B, et al. EMBO J. 2001;20:1875–1887. doi: 10.1093/emboj/20.8.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jasinski M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M. Plant Cell. 2001;13:1095–1107. [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M. Plant Cell Physiol. 2002;43:1–11. doi: 10.1093/pcp/pcf023. [DOI] [PubMed] [Google Scholar]
- 36.Zolman B K, Silva I D, Bartel B. Plant Physiol. 2001;127:1266–1278. [PMC free article] [PubMed] [Google Scholar]
- 37.Wang W, Takezawa D, Poovaiah B W. Plant Mol Biol. 1996;31:683–687. doi: 10.1007/BF00042240. [DOI] [PubMed] [Google Scholar]
- 38.Davies T G, Theodoulou F L, Hallahan D L, Forde B G. Gene. 1997;199:195–202. doi: 10.1016/s0378-1119(97)00367-3. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki T, Ezaki B, Matsumoto H. Plant Cell Physiol. 2002;43:177–185. doi: 10.1093/pcp/pcf025. [DOI] [PubMed] [Google Scholar]
- 40.Sidler M, Hassa P, Hasan S, Ringli C, Dudler R. Plant Cell. 1998;10:1623–1636. doi: 10.1105/tpc.10.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.