Abstract
The natural history and pathogenic potential of the recently identified TT virus (TTV) are currently a matter of intensive investigation. In an attempt to shed some light on these issues, nasal and blood specimens of 1- to 24-month-old children hospitalized with a clinical diagnosis of acute respiratory disease (ARD) were examined for the presence, load, and genetic characteristics of TTV. The results have indicated that at least in young children, the respiratory tract not only represents a route by which abundant TTV can be shed into the environment but also may be a site of primary infection and continual replication. Although we found no compelling evidence that TTV was the direct cause of ARD in some of the children studied, the average loads of TTV were considerably higher in patients with bronchopneumonia (BP) than in those with milder ARD, raising interesting questions about the pathophysiological significance of TTV at this site. Furthermore, group 4 TTV was detected almost exclusively in children with BP.
TT virus (TTV)—a small, nonenveloped virus with a single-stranded, negative-polarity, circular DNA genome of 3.8 kb—was initially thought to be a circovirus similar to chicken anemia virus, porcine circovirus, and other viruses of animals, but it is now under consideration as the possible type species of an independent virus family (17, 18, 34, 35). The taxonomy of the vast array of related viral agents that have been identified in humans since the first description of TTV by Nishizawa et al. in 1997 (21) is also uncertain. Recently, however, Okamoto and Mayumi (22) have divided TTV isolates into at least four phylogenetic clusters differing by over 40% at the nucleotide level: group 1 has the original TTV isolate as the prototype and includes genotypes 1 to 6; group 2 has PMV as the prototype and comprises genotypes 7, 8, 17, 22, and 23; group 3 has SANBAN and SENV as representative isolates and includes genotypes 9 to 16 and 18 to 20; and group 4 has YONBAN as the prototype and includes genotype 21. Furthermore, while the present report was in preparation, the same group identified several novel genotypes within group 4 TTV as well as members of a proposed fifth group (26). Additional related viruses are more dissimilar due to their smaller genome (2.8 kb) and are presently designated TTV-like minivirus or TLMV (6, 33, 34).
The natural history and pathogenic potential of TTV are currently the subject of active investigation. From the epidemiological standpoint, evidence has accumulated that TTV viremia is extremely common in the general population worldwide, starting from early childhood (1, 5, 10, 26, 27), implying the existence of a very efficient means of transmission, and the observation that infectious TTV is shed in the feces has led to the suggestion that the fecal-oral route is the most likely one (36). With regard to the life cycle in infected hosts, both self-limited and persistent systemic TTV infections have been described (reviewed in reference 5) and chronic TTV viremia has been shown to result from the production and clearance of 1010 to 1011 virions per day (16), indicative of the existence of body sites where the virus is continuously and actively replicated. Furthermore, it has been reported that chronically infected individuals harbor high levels of mature TTV DNA as well as replicative DNA intermediates and mRNA in all body sites examined (23) except the central nervous system (14), thus implying that TTV is polytropic in nature or grows in a cell type(s) that is represented throughout the human body. With regard to pathogenesis, early indications that TTV might cause some forms of cryptogenetic hepatitis have been questioned (7, 20) while attempts to link the virus to other diseases have either been unsuccessful or need confirmation (5, 38), leaving the virus with “orphan” status with regard to clinical disease.
Thus far, the routes of TTV entry into the body, the locations of initial virus amplification before infection becomes systemic, and the clinical consequences of primary infection, if any, have been investigated only to a limited extent. In this study we have approached these issues by examining children with acute respiratory infections (ARD). The results have indicated that the respiratory tract represents not only a route by which abundant TTV can be shed into the environment but also a site of primary infection and continual replication. Although we found no compelling evidence that TTV was the direct cause of ARD in some of the children studied, average burdens of TTV were considerably higher in patients with bronchopneumonia (BP) than in those with milder forms of ARD, raising interesting questions about the pathophysiological significance of this virus. Furthermore, group 4 TTV was detected almost exclusively in children with BP.
MATERIALS AND METHODS
Study subjects, specimens and detection of CRV.
All specimens were from children with ARD admitted to the Department of Pediatrics, University Hospital of Pisa, and were collected on admission day after informed parental consent had been obtained. The duration of clinical symptoms before hospitalization was 6.8 ± 5.3 days. Nasal samples were harvested with sterile alginate swabs that were gently pushed toward the nasopharynx until resistance was encountered and then rotated a few times to collect abundant fluid and cells. Plasma was obtained by centrifuging blood collected under nuclease-free conditions in sodium citrate tubes. Peripheral blood mononuclear cells (PBMC) were obtained by standard Ficoll-Hypaque density gradient centrifugation (ICN Biomedicals, Aurora, Ohio), washed thoroughly, and treated to remove adsorbed extracellular virus as previously described (15). Simultaneous nasal and plasma samples from the 24 children who entered the pilot study (19 boys and 5 girls aged 1 to 24 months [mean, 9.5 months], all negative for hepatitis B virus, hepatitis C virus, and human immunodeficiency virus; no history of blood transfusions or blood product use) were harvested from November 1999 through April 2000. The nasal secretions collected from these children were quantified by weighing the swabs before and after collection (range of weights, 13 to 72 mg; mean ± standard deviation, 42 ± 15 mg), diluted 1:10 (wt/vol) with phosphate-buffered saline (PBS), and clarified at 900 × g at 4°C for 10 min to separate cells and cell debris. Finally the cell-free nasal fluid was stored in aliquots at −20°C until used for DNA extraction. The cell pellets, whose contribution to the total weight of secretions was found to be negligible, were repeatedly washed in abundant PBS, resuspended in 200 μl of PBS, and also stored at −20°C with no examination of their cellular composition. The samples used to expand the population investigated were also from children aged 1 to 24 months and were submitted to our laboratory for routine virological analysis between May 2000 and November 2001. Some swabs from this second group were vortexed in 0.5 ml of sterile saline, and the resulting suspension was stored as above, without further processing. All nasal samples were tested for common respiratory viruses (CRV) by direct immunofluorescence (adenovirus, cytomegalovirus, influenza A and B viruses parainfluenza virus types 1 to 3, and respiratory syncytial virus [RSV]), enzyme immunoassay (RSV), and rapid culture in shell vials (all the above viruses).
TTV detection and quantification.
DNA was extracted from 200-μl samples of all specimens by using the QIAamp DNA minikit (Qiagen, Chatsworth, Calif.) as specified by the manufacturer. The presence and load of TTV in nasal and plasma specimens were determined in a single step by a previously described TaqMan (TM)-PCR assay (14, 27). This assay is based on a highly conserved segment of the viral untranslated region (Table 1) and has the potential for sensitive and specific detection of all the genotypes of TTV present in GenBank at the time of writing. Furthermore, the method does not detect TLMV, as determined by testing TLMV plasmid sequences and also confirmed by the sequence data below. The procedures used for quantification of copy numbers and evaluation of specificity, sensitivity, intra- and interassay precision, and reproducibility have been described previously (27). The lower limit of sensitivity was 1.0 × 103 viral genomes per ml or μg of total DNA for both nasal and blood specimens. The presence of PCR inhibitors in extracted nasal secretions was ruled out by testing several nasal samples spiked with known numbers of TTV DNA copies. The yields of total DNA extracted from PBMC and nasal cells were determined by agarose gel electrophoresis and by comparison with appropriate dilutions of quantitative molecular markers.
TABLE 1.
PCR oligonucleotides used for quantifying and grouping TTV isolates
| Oligonucleotide | Sequence (5′-3′)a | Genome localization | Position (nt)b | PCR product length (bp) |
|---|---|---|---|---|
| Universal TM-PCR | 63 | |||
| AMTS | GTGCCGIAGGTGAGTTTA | UTR | 177-194 | |
| AMTAS | AGCCCGGCCAGTCC | UTR | 226-239 | |
| AMTPTUc | TCAAGGGGCAATTCGGGCT | UTR | 205-223 | |
| Group-specific PCR | ||||
| Group 1 | 191 | |||
| TTV2 | CAGTTAGTGGTGAGCCGAA | ORF1 | 1348-1366 | |
| TTV3 | GACACAGAIATTACAGCCCC | ORF1 | 1126-1145 | |
| TTV5 | ACAGCCCCCAGCATACATCC | ORF1 | 1138-1157 | |
| TTV6 | GTGGTGGCATAGACCGTTAG | ORF1 | 1309-1328 | |
| Group 2 | 259 | |||
| PMV1 | ACAIAGATGTCCCTGGAGTAGA | ORF1 | 1018-1039 | |
| PMV2 | AGAGGCACTGATGTTTACTGGC | ORF1 | 1327-1348 | |
| PMV3 | TGGAGTAGATCGAACGAAGA | ORF1 | 1031-1050 | |
| PMV4 | CTGTAAATAGAGTGGGGGG | ORF1 | 1271-1289 | |
| Group 3 | 255 | |||
| SS1 | GACCARCTAGACCTGGCCAGATA | ORF1 | 1061-1083 | |
| SS2 | GTTTGTGGTGAGCMGAAYGG | ORF1 | 1382-1401 | |
| SS4 | GTACCASTKGTCTWCAAA | ORF1 | 1298-1315 | |
| Group 4 | 273 | |||
| PY1 | CCATTTTGTRCAGCCCGCCA | UTR | 103-122 | |
| PY2 | ACGGCATGACTTTGTGTYCTG | UTR | 389-409 | |
| PY3 | AGCCCGCCAATTTCTGTTC | UTR | 114-132 | |
| PY4 | GGAGAACAGAGCRRGGGAAT | UTR | 367-386 |
I, inosine; R, A or G; M, A or C; Y, C or T; S, G or C; K, G or T; W, A or T.
Nucleotide (nt) positions are according to isolate AB017610 for TM-PCR and group 1-specific PCR and to isolates AF261761, AB028669, and AB038624 for group 2-, 3-, and 4-specific PCR, respectively.
AMTPTU probe was labeled with 6-carboxyfluorescein and 6-carboxytetramethylrhodamine at the 5′ and 3′ ends, respectively, and contained a propynilic chemical group bound to each thymidine.
TTV typing by group-specific PCR assays.
Selected specimens found positive by the TM-PCR assay were amplified by four nested or seminested PCR protocols, using the group-specific primers listed in Table 1. The reactions were performed in a Perkin-Elmer 9600 thermal cycler (Applied Biosystems, Foster City, Calif.), amplified products were analyzed by gel electrophoresis followed by ethidium bromide staining, and the amplicon size was determined by comparison with appropriate molecular size markers. To exclude the presence of carryover contamination, each round of assays included appropriate negative controls added during the steps of DNA extraction and PCR amplification. Specimens were examined at least in duplicate, and if the results of tests of different specimens from a patient differed, the specimens were reextracted and tested again in duplicate. The sensitivity of each group-specific assay was measured by testing serial dilutions of a known concentration of recombinant plasmids having the appropriate segment of viral genome inserted and found to range between 10 and 100 DNA copies.
Sequencing and sequence analysis.
Amplification products from the group-specific PCR assays were directly sequenced using the Big Dye Terminator cycle-sequencing kit (Applied Biosystems). Cycle sequencing was carried out with an automatic DNA sequencer (ABI model 373; Applied Biosystems) and performed in both orientations for confirmation. Nucleotide sequences were aligned with previously published sequences. Multiple alignments were created using CLUSTALW, and phylogenetic relationships were estimated by using DAMBE (version 4.0.59 [37]) and the neighbor-joining method.
Statistical analysis.
Pearson's chi-square test was applied to evaluate the heterogeneity of contingency tables. Differences between means and distributions were evaluated by the two-tailed Student t test and the Wilcoxon rank sum test, respectively. Associations between variables were determined by applying Pearson's correlation coefficient.
Nucleotide sequence accession numbers.
Sequences obtained were submitted to GenBank (accession no. AY093391 to AY093412).
RESULTS
In-depth examination of TTV in 24 paired nasal and blood specimens.
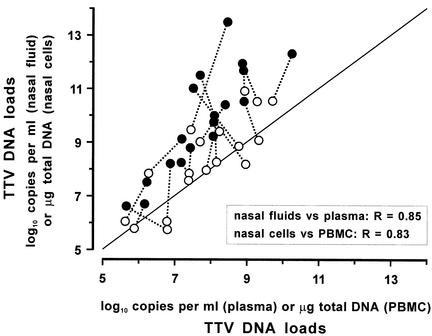
For the pilot study of the nasal and blood specimens, nasal swabs were weighed before and after specimen collection to permit precise comparisons of viral load with those in plasma, and nasal fluids and cells in the swabs were processed separately and invariably tested in parallel with the corresponding plasma and PBMC. Universal TM-PCR analysis detected TTV in 18 of the 24 children studied, and in each case all specimens were concordantly positive or negative. As shown in Fig. 1, the levels of TTV found in individual patients varied widely but the loads in the four specimens from each patient were clearly correlated. Importantly, TTV DNA copy numbers per milliliter in nasal fluids constantly exceed those in the corresponding plasma samples, in 15 cases by over 1 log10 unit and in one by as many as 5 log10 units (mean difference, 2 log10 units; P < 0.001). TTV DNA copy numbers per microgram of total DNA associated with nasal cells were also often but not constantly greater than in the corresponding PBMC, and the differences were less pronounced (mean difference, 0.5 log10 unit, P = 0.02).
FIG. 1.
TTV loads in paired nasal and blood samples from the 18 children in the pilot study that were found virus positive in both specimens. •, TTV DNA copy numbers in nasal fluid versus plasma; ○, TTV DNA copy numbers in total DNA from nasal cells versus PBMC. Broken lines connect the data for each individual patient. Most points lie above the diagonal line, indicating higher loads in nasal samples than in blood samples.
The 18 pairs of nasal fluid and plasma TTV isolates were then characterized by grouping and sequencing. Groups were determined by reamplifying the clinical specimens with four specifically designed group-specific PCR assays targeted either to the viral ORF1 (groups 1 to 3) or UTR (group 4). Of the nasal specimens, two were not amplified by any such reaction and remained unclassified although they again reacted TTV positive when retested by universal TM-PCR. The others yielded either one (11 specimens), two (3 specimens), or three (2 specimens) positive reactions, indicating that the respiratory tracts of five children contained more than one TTV group. Group 3 was the most heavily represented (14 samples), followed by groups 1 (6 samples) and 4 (3 samples). Characterization of plasma TTV yielded results identical to those for the nasal fluid TTV in 13 cases. The others showed different forms of discordance, which were also demonstrated by the results of examining the corresponding PBMC and nasal cells, although the presence of an unclassified TTV in one of the two sites was common (Table 2).
TABLE 2.
Discrepant results obtained by grouping contemporaneous TTV-positive nasal and blood samples
| Patient | Sexa/age (mo) | Clinical diagnosis | Group of TTV in:
|
|||
|---|---|---|---|---|---|---|
| Nasal fluid | Nasal cells | Plasma | PBMC | |||
| 576 | M/16 | BP | 3, 4 | 3, 4 | 3 | 3 |
| 4999 | F/8 | Bronchiolitis | 3 | 3 | NTb | NT |
| 5174 | M/5 | Bronchiolitis | 3 | 3 | 1, 3 | 1, 3 |
| 5210 | F/10 | BP | 3 | 3 | NT | NT |
| 5575 | M/9 | BP | NT | NT | 1 | NT |
M, male; F, female.
NT, not typeable.
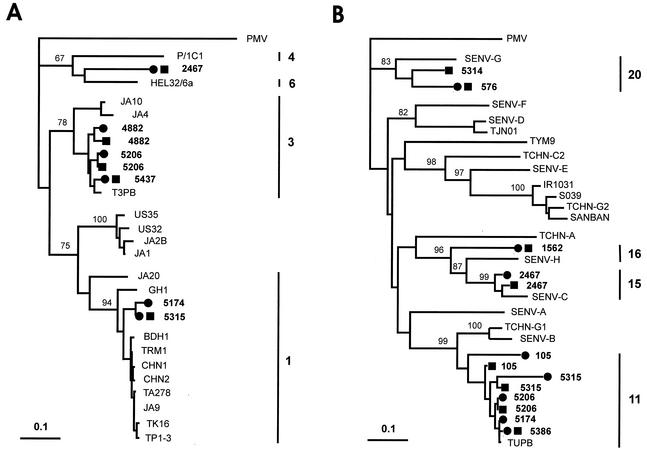
Amplicons from 32 of 45 group-specific PCR assays were sequenced, and in each case the original classification by the PCR assay was confirmed by sequence analysis (data not shown). Twenty-seven sequences were in the ORF1 region and could be used for a meaningful comparison of paired nasal and plasma sequences as well as for classification of the viral isolates into genotypes. Of 12 paired nucleotide sequences examined, 6 were identical; the others diverged between 1 and 16%. Figure 2 shows the phylogenetic trees constructed by using such sequences together with representative GenBank sequences. Group 1 sequences (interpatient nucleotide homologies between 55 and 97%) clustered within genotypes 3 (five sequences), 1 (two sequences), and 6 (one sequence). Group 3 sequences (interpatient nucleotide homologies between 54 and 99%) fell into genotypes 11 (eight sequences), 20 (two sequences), 15 (two sequences), and 16 (one sequence). The additional five amplicons sequenced, being derived from group 4-specific PCR assays, were less informative due to high conservation of the untranslated region targeted and showed a maximum interpatient divergence of 3% (data not shown).
FIG. 2.
Phylogenetic analysis of 27 TTV isolates from the pilot study. (A) Tree based on the 151-bp ORF1 segment amplified by the group 1-specific PCR. (B) Tree based on the 214-bp ORF1 segment amplified by the group 3-specific PCR. Both segments had primer nucleotides subtracted. Nasal (▪) and plasma (•) isolates from the present study are in boldface type. GenBank sequences are indicated by the isolate name. The PMV isolate was used as the outgroup. Bootstrap values above 67% of 1,000 replicates are shown at branch points, to show the significance of the grouping. Bars represent the number of nucleotide substitutions per site. Bars with numbers to the right of each tree indicate the TTV genotypes within which the sequences identified in this study clustered (the others are omitted for simplicity).
Correlation between TTV loads and ARD severity in a large group of children
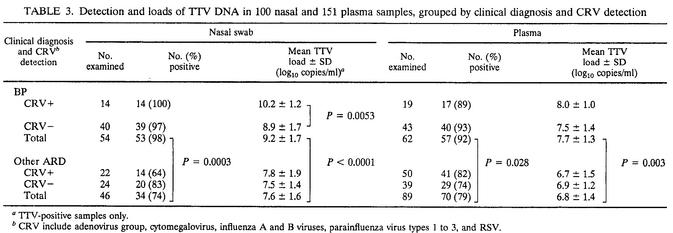
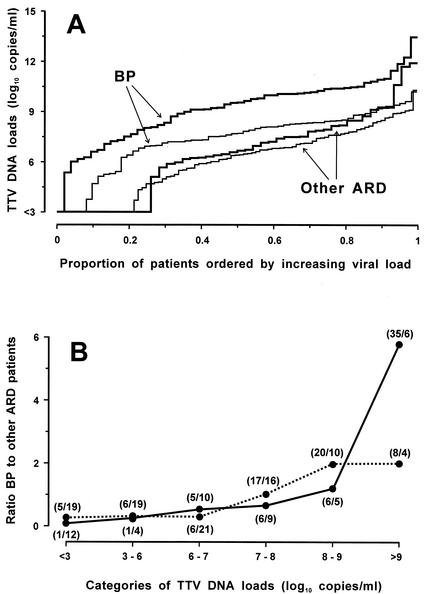
The above study had shown a tendency for children with BP to have especially high titers of TTV (data not shown). To further investigate this finding, we measured the loads of TTV in additional pediatric specimens sent to our laboratory for routine virological analysis. Thus, the eventual total number of specimens examined was 100 nasal swabs and 151 plasma samples from a total of 157 hospitalized children. A total of 54 nasal swabs and 62 plasma samples were from children with X-ray-confirmed BP. The others were from children with laryngitis (10 cases), bronchitis (18 cases), or bronchiolitis (65 cases), after checking that they showed no significant differences in TTV detection rates and load distributions, we treated them as a single category. As shown in Table 3 and Fig. 3A, TTV was detected more frequently and at higher titers in BP patients than in other ARD patients, regardless of whether the test was conducted on nasal or plasma samples. Furthermore, stratification by TTV load size showed that nasal swabs with TTV values of >9 log10 units per ml were almost sixfold more likely to originate from children with BP than were the ones with lower loads. Plasma load differences were much less informative about this finding (Fig. 3B). Also, nasal and plasma loads were again found to correlate (r = 0.87, corroborating what was observed in the pilot study. Of note, the mean ages of patients with BP or other ARD were not statistically different (12.8 ± 7.9 and 9.8 ± 6.8 months, respectively). No correlation was observed between the age of the children and TTV loads in nasal swabs (r = 0.33) or plasma (r = 0.25). However, the viremia rate was 63% in 26 subjects younger than 4 months and 95% in 42 subjects aged 12 months or more, thus comparing well with previous findings (26).
TABLE 3.
Detection and loads of TTV DNA in 100 nasal and 151 plasma samples, grouped by clinical diagnosis and CRV detection
TTV-positive samples only.
CRV include adenovirus group, cytomegalovirus, influenza A and B viruses, parainfluenza virus types 1 to 3, and RSV.
FIG. 3.
TTV loads in the 151 plasma and 100 nasal fluid specimens from children with ARD, subdivided by respiratory disease (BP versus other ARD). (A) Thick and thin lines represent nasal fluid and plasma samples, respectively. Most steps represent single patients. TTV loads in nasal fluid and plasma differed significantly between patients with BP and other ARD at P < 0.00001 and at P < 0.00001, respectively (Wilcoxon test). (B) Results stratified based on the TTV load in plasma (dotted line) and nasal fluid (continuous line). Each point represents the ratio between the number of patients with BP and the number of patients with other ARD. The numbers in parentheses are the number of patients with BP/the number of patients with other ARD.
Samples were also grouped according to whether the donors had been found positive or negative for CRV by routine virological analysis. The only significant difference detected was that the 14 CRV-positive BP patients had higher mean nasal TTV loads than did the 40 CRV-negative BP patients (Table 2), regardless of whether they harbored RSV (9 children) or other viruses (5 children).
Prevalence of group 4 TTV in patients with BP.
In the pilot study, the TTV groups were distributed uniformly regardless of clinical diagnosis, except for group 4 TTV which had been found only in four subjects with BP, raising the possibility that this group was especially closely associated with BP. To further investigate this aspect, we used the group 4-specific PCR assay to examine 42 of the 53 TTV-positive nasal swabs from children with BP and 31 of the 34 TTV-positive nasal swabs from children with ARD. We found that 9 (21%) and 1 (3%) of specimens, respectively, reacted positively (P = 0.03). Of the 10 patients with group 4-positive nasal swabs, 6 also yielded the same TTV group in plasma.
Longitudinal study of two children initially TTV positive in the respiratory tract and negative in plasma
In the cross-sectional study above, we had examined contemporaneous nasal and plasma samples from 94 patients. Of these patients, six had tested positive only for the nasal sample, thus indicating that they were experiencing a respiratory TTV infection in the absence of, or prior to, systemic TTV involvement. For two of these patients we could examine additional samples obtained at later times. Patient 3936 was 3 months old when first tested, and because he had obstructive BP with recurring clinical symptoms and was persistently negative for CRV, he was examined several times over 16 months. As shown in Table 4, the nasal cavity of this child was already TTV positive when he was first examined at 3.5 months of age, and it remained constantly positive with titers that increased steadily during the first 2.5 months of observation. On the other hand, the child's blood tested TTV negative three times before becoming positive at age 4.5 months; it subsequently remained positive with titers that also rose gradually but to a lower plateau than in the respective nasal samples. Nasal and plasma TTV sequences from this child were of group 1 and were identical at all times, but they could not be assigned with certainly to any currently known genotype (77 and 65% nucleotide homology to genotypes 6 and 4, respectively). For the second patient, a 3-month-old baby with laryngitis, we could examine only one additional plasma sample obtained 2 weeks after TTV had been detected in the nose but not in the plasma. At the second sampling, the plasma had converted from negative to positive (4.4 × 106 TTV DNA copies per ml). Thus, in both these patients, TTV infection of the respiratory tract had clearly preceded systemic infection.
TABLE 4.
Time course of detection of nasal and blood TTV loads in child 3936
| Age (mo.) | TTV DNA load (log10 copies/ml or μg of total DNA)
|
||||
|---|---|---|---|---|---|
| Nasal fluid | Nasal cells | Plasma | PBMC | ||
| 3 | Nega | ||||
| 3.5 | 6.25b
|
Neg | |||
| 4 | Neg | ||||
| 4.5 | 7.5 | 5.2 | 5.1 | 3.9 | |
| 5 | 7.3 | 7.1 | |||
| 6 | 10.8 | 9.4 | 8.1 | 8.7 | |
| 8 | 10.0 | 9.2 | 8.5 | 8.2 | |
| 9 | 9.4 | 7.8 | 7.6 | 7.9 | |
| 10 | 9.7 | 9.5 | 8.8 | 9.4 | |
| 12 | 11.4 | 9.5 | 9.0 | 6.9 | |
| 14 | 9.6 | 7.4 | 8.0 | 7.1 | |
| 16 | 10.4 | 8.1 | 8.0 | 7.4 | |
Neg, <1.0 × 103 log10 copies per ml.
TTV load measured in unseparated nasal fluid and cells.
DISCUSSION
The amazingly high prevalence of chronic TTV infections in healthy individuals worldwide represents a serious hurdle to the elucidation of their natural histories and clinical significances. Reported detection rates of viremia in the general population have varied considerably depending on the breadth and sensitivity of the diagnostic PCR assays used, but whenever these were adequate, the detection rates easily ranged over 80% (reviewed in reference 5). Additional obstacles are the extraordinary genetic heterogeneity of TTV (discussed in the introduction); the high frequency of mixed infections by multiple TTV genotypes (up to 90% in certain studies [29]), which presumably reflects a lack of cross-immunity; and the large variations in viral loads found in individual subjects (from 103 to 108 DNA copies per ml of adult plasma in our hands [27]).
The present investigation of TTV in juvenile patients with ARD consisted of two parts. The first was an in-depth study of 24 subjects in whom most of the above variables were taken into account, while the second enrolled a larger number of diseased children to focus on aspects that had appeared to merit a more extensive evaluation. Collectively, the results have confirmed the widespread diffusion and early acquisition of systemic TTV infection by showing that 84% of 151 children aged 1 to 24 months had the virus in their plasma. This prevalence is comparable to what was previously observed in healthy adults living in the same area as the study children (27) and in other geographical regions (25). According to available data, early TTV infection can be the result of transplacental transmission (11, 30) as well as of intra- or extrafamilial postnatal exposure to the virus (31).
The results have also shown that 87% of 100 children examined harbored TTV in the upper respiratory tract. Because nasal viral loads were often quite high, this indicates that at least in young children the respiratory tract may represent an important route of TTV excretion. Together with evidence discussed below, this suggests that respiratory transmission of TTV might be very significant and might make a greater contribution to the widespread diffusion of this virus than other proposed postnatal forms of transmission, including the fecal-oral route. In this regard, it is important to note that all specimens examined in the present study were collected at the time of hospital admission, thus excluding recent nosocomial acquisition of the observed TTV infections. Incidentally, the 24 children in the pilot study were also tested for TLMV; specimens from 23 were positive in plasma and specimens from 22 were positive in the respiratory tract (data not shown), thus suggesting that TTV and TLMV may share important epidemiological features.
Genetic characterization of TTV was limited to the children in the pilot study; nevertheless, it revealed a large heterogeneity, as shown by the detection of at least three groups and eight genotypes of the virus. That we detected no group 2 TTV sequences might reflect a relative infrequency of this group worldwide (12). It should also be noted that we made no attempt to further type, by sequence analysis or otherwise, the isolates that were not classified by the four group-specific PCR assays used; they may have belonged to the several genotypes of groups 4 and 5 described after such assays were designed (26) or to hitherto unrecognized genetic forms of TTV. As pointed out by Okamoto et al. (23), the extent of TTV diversity currently recognized is already unusually great for a DNA virus but is bound to increase substantially with the accumulation of sequence data worldwide. Importantly, detection of mixed populations of TTV belonging to more than one group (up to three groups) in a single subject was a frequent finding of this and previous studies (3, 9, 19, 28) and represents a further indication of the ubiquity and heterogeneity of TTV infection. Recent data have shown that adults can harbor different mixtures of TTV genotypes in different organs, suggestive of a preferential tropism of certain TTV strains for specific tissues (8, 23, 24). However, we found no evidence in the present study for the existence of genotypes exclusively tropic for the respiratory tract.
Too few cells could be retrieved from our nasal swabs for a reliable search of viral DNA intermediates or mRNA that might unequivocally demonstrate ongoing local TTV replication. However, overall data provided convincing evidence that at least part of the TTV present in the respiratory tract was produced locally. (i) As mentioned above, nasal TTV loads were often extremely high; (ii) in the children for whom a precise comparison of nasal fluid and plasma loads was possible, the former specimen constantly exhibited higher TTV copy numbers per ml than did the latter; (iii) thoroughly washed cells retrieved from nasal swabs were uniformly TTV positive and often contained more viral DNA copies per microgram of total DNA than did the corresponding PBMC; (iv) in some children, nasal and blood TTV were genetically discordant, as if they were partly or totally derived from independent infections (23); and, lastly but most importantly, (v) some children harbored TTV in the nose but not in plasma while the opposite situation was never observed when virus detection was performed by universal TM-PCR analysis. Coupled with findings revealing the presence of molecular markers of TTV replication in the lungs (4, 23, 32), these data leave little doubt that the respiratory tract is a site of continual TTV replication. Furthermore, examination of serial samples from two children demonstrated quite clearly that TTV infection of the respiratory tract can precede systemic infection, thus suggesting that the respiratory tract can also be a port of entry and a place of primary infection for TTV. This study was conducted with hospitalized children who had developed respiratory symptoms several days earlier. Examination of specimens collected earlier after clinical presentation should facilitate detection of primary respiratory TTV infections.
Ethical reasons did not permit the collection of suitable nasal samples from healthy children, thus preventing an evaluation of whether the underlying ARD could explain the high TTV prevalence rates and loads observed in the study patients. However, the data revealed an interesting correlation between TTV infection and clinical status. In fact, TTV prevalence and loads in both nasal swabs and plasma were significantly higher in patients with BP than in those with milder disease. The significance of such finding remains elusive; however, if TTV were directly involved in BP etiology, one would have expected that BP patients who tested negative for CRV would be more frequently TTV infected or would have higher TTV burdens than the CRV positive ones. Because, in contrast, the mean TTV loads were higher in CRV-positive BP patients, it seems more plausible that TTV infection may augment the severity of disease induced by other agents. Such a mechanism would be reminiscent of the pathogenic features of several related viruses of animals, including chicken anemia virus, porcine circovirus type 2, and beak and feather disease virus of parrots, which are known to augment the severity of concomitant bacterial and viral infections, presumably due to the ability to replicate in proliferating lymphocytes and to impair the antimicrobial defenses of the host (2, 13). Importantly, TTV has also been shown to replicate in stimulated PBMC (15). Also, because it is likely that TTV replicates solely or most effectively in the presence of active cell DNA synthesis (5), it is plausible that, as with other single-stranded DNA viruses, TTV replication in the respiratory tract is boosted by concomitant local infections by other agents, due to stimulation of resident lymphoid cells, influx of activated leukocytes, or ongoing regeneration of damaged tissue.
Another intriguing finding of the present study is that group 4 TTV was detected almost exclusively in children with BP, raising the possibility that this group is especially closely associated with severe ARD. That in several such cases group 4 TTV was detected in nasal swabs but not in plasma is indicative of an ongoing primary infection or superinfection in the respiratory tract and is therefore in line with this possibility. It should, however, be noted that our group 4-specific PCR assay had a restricted range of detection, since it was designed when the only group 4 sequences in GenBank were of genotype 21, while additional genotypes in this group have been identified by Peng et al. in a more recent investigation of Japanese and Chinese children (26). This, possibly in combination with geographical differences in the distribution of TTV genotypes, may explain the lower rate of group 4 TTV detection in the present study relative to what was observed by Peng et al. (26) and points directly to genotype 21 for a possible association with BP. Indeed, there are many examples of viral infections (papillomaviruses, adenoviruses, enteroviruses, etc.) that have different pathological consequences depending on the genotype of the virus involved. Further studies are clearly warranted.
Acknowledgments
This work was supported in part by grants from the Ministero dell'Istruzione, dell'Università e della Ricerca, Rome, Italy.
REFERENCES
- 1.Abe, K., T. Inami, K. Asano, C. Miyoshi, N. Masaki, S. Hayashi, K. Ishikawa, Y. Takebe, K. M. Win, A. R. El-Zayadi, K. H. Han, and D. Y. Zhang. 1999. TT virus infection is widespread in the general populations from different geographic regions. J. Clin. Microbiol. 37:2703-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adair, B. M. 2000. Immunopathogenesis of chicken anemia virus infection. Dev. Comp. Immunol. 24:247-255. [DOI] [PubMed] [Google Scholar]
- 3.Ball, J. K., R. Curran, S. Berridge, A. M. Grabowska, C. L. Jameson, B. J. Thomson, W. L. Irving, and P. M. Sharp. 1999. TT virus sequence heterogeneity in vivo: evidence for co-infection with multiple genetic types. J. Gen. Virol. 80:1759-1768. [DOI] [PubMed] [Google Scholar]
- 4.Bando, M., S. Ohno, K. Oshikawa, M. Takahashi, H. Okamoto, and Y. Sugiyama. 2001. Infection of TT virus in patients with idiopathic pulmonary fibrosis. Respir. Med. 95:935-942. [DOI] [PubMed] [Google Scholar]
- 5.Bendinelli, M., M. Pistello, F. Maggi, C. Fornai, G. Freer, and M. L. Vatteroni. 2001. Molecular properties, biology and clinical implications of TT virus, a recently identified widespread infectious agent of man. Clin. Microbiol. Rev. 14:98-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biagini, P., P. Gallian, H. Attoui, M. Touinssi, J. F. Cantaloube, P. de Micco, and X. De Lamballerie. 2001. Genetic analysis of full-length genomes and subgenomic sequences of TT virus-like mini virus human isolates. J. Gen. Virol. 82:379-383. [DOI] [PubMed] [Google Scholar]
- 7.Cleavinger, P. J., D. H. Persing, H. Li, S. B. Moore, M. R. Charlton, C. Sievers, T. M. Therneau, and N. N. Zein. 2000. Prevalence of TT virus infection in blood donors with elevated ALT in the absence of known hepatitis markers. Am. J. Gastroenterol. 95:772-776. [DOI] [PubMed] [Google Scholar]
- 8.Deng, X., M. Terunuma, R. Handema, M. Sakamoto, T. Kitamura, M. Ito, and Y. Akahane. 2000. Higher prevalence and viral load of TT virus in saliva than in the corresponding serum: another possible transmission route and replication site of TT virus. J. Med. Virol. 62:531-537. [PubMed] [Google Scholar]
- 9.Forns, X., P. Hegerich, A. Darnell, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. High prevalence of TT virus (TTV) infection in patients on maintenance hemodialysis: frequent mixed infections with different genotypes and lack of evidence of associated liver disease. J. Med. Virol. 59:313-317. [PubMed] [Google Scholar]
- 10.Gerner, P. 2000. TT virus infection in healthy children and in children with chronic hepatitis B or C. J. Pediatr. 136:573-575. [DOI] [PubMed] [Google Scholar]
- 11.Gerner, P., R. Oettinger, W. Gerner, J. Falbrede, and S. Wirth. 2000. Mother-to-infant transmission of TT virus: prevalence, extent and mechanism of vertical transmission. Pediatr. Infect. Dis. J. 19:1074-1077. [DOI] [PubMed] [Google Scholar]
- 12.Hallett, R. L., J. P. Clewley, F. Bobet, P. J. McKiernan, and C. G. Teo. 2000. Characterization of a highly divergent TT virus genome. J. Gen. Virol. 81:2273-2279. [DOI] [PubMed] [Google Scholar]
- 13.Krakowa, S., J. A. Ellis, F. McNeilly, S. Ringler, D. M. Rings, and G. Allan. 2001. Activation of the immune system is the pivotal event in the production of wasting disease in pigs infected with porcine circovirus-2 (PCV-2). Vet. Pathol. 38:31-42. [DOI] [PubMed] [Google Scholar]
- 14.Maggi, F., C. Fornai, M. L. Vatteroni, G. Siciliano, F. Menichetti, C. Tascini, S. Specter, M. Pistello, and M. Bendinelli. 2001. Low prevalence of TT virus in the cerebrospinal fluid of viremic patients with central nervous system disorders. J. Med. Virol. 65:418-422. [DOI] [PubMed] [Google Scholar]
- 15.Maggi, F., C. Fornai, L. Zaccaro, A. Morrica, M. L. Vatteroni, P. Isola, S. Marchi, A. Ricchiuti, M. Pistello, and M. Bendinelli. 2001. TT virus (TTV) loads associated with different peripheral blood cell types and evidence for TTV replication in activated mononuclear cells. J. Med. Virol. 64:1-6. [DOI] [PubMed] [Google Scholar]
- 16.Maggi, F., M. Pistello, M. L. Vatteroni, S. Presciuttini, S. Marchi, P. Isola, C. Fornai, S. Fagnani, E. Andreoli, G. Antonelli, and M. Bendinelli. 2001. Dynamics of persistent TT virus infection, as determined in patients treated with alpha interferon for concomitant hepatitis C virus infection. J. Virol. 75:11999-12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mushahwar, I. K. 2000. Recently discovered blood-borne viruses: are they hepatitis viruses or merely endosymbionts? J. Med. Virol. 62:399-404. [DOI] [PubMed] [Google Scholar]
- 18.Mushahwar, I. K., J. C. Erker, A. S. Muerhoff, T. P. Leary, J. N. Simons, L. G. Birkenmeyer, M. L. Chalmers, T. J. Pilot-Matias, and S. M. Dexai. 1999. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc. Natl. Acad. Sci. USA 96:3177-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niel, C., F. L. Saback, and E. Lampe. 2000. Coinfection with multiple TT virus strains belonging to different genotypes is a common event in healthy Brazilian adults. J. Clin. Microbiol. 38:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiguchi, S., M. Enomoto, S. Shiomi, M. Tanaka, K. Fukuda, A. Tamori, T. Tanaka, T. Takeda, S. Seki, Y. Yano, S. Otani, and T. Kuroki. 2000. TT virus infection in patients with chronic liver disease of unknown etiology. J. Med. Virol. 62:392-398. [DOI] [PubMed] [Google Scholar]
- 21.Nishizawa T., H. Okamoto, K. Konishi, H. Yoshizawa, Y. Miyakawa, and M. Mayumi. 1997. A novel DNA virus (TTV) associated with elevated transaminasi levels in posttransfusion hepatitis of unknown etiology. Biochem. Biophys. Res. Commun. 241:92-97. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto, H., and M. Mayumi. 2001. TT virus: virological and genomic characteristics and disease associations. J. Gastroenterol. 36:519-529. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto, H., T. Nishizawa, M. Takahashi, S. Asabe, F. Tsuda, and A. Yoshikawa. 2001. Heterogeneous distribution of TT virus of distinct genotypes in multiple tissues from infected humans. Virology 288:358-368. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto, H., M. Takahashi, N. Kato, M. Fukuda, A. Tawara, S. Fukuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 2000. Sequestration of TT virus of restricted genotypes in peripheral blood mononuclear cells. J. Virol. 74:10236-10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto, H., M. Takahashi, T. Nishizawa, M. Ukita, M. Fukuda, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1999. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology 259:428-438. [DOI] [PubMed] [Google Scholar]
- 26.Peng, Y. H., T. Nishizawa, M. Takahashi, T. Ishikawa, A. Yoshikawa, and H. Okamoto. 2002. Analysis of the entire genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Arch. Virol. 147:21-41. [DOI] [PubMed] [Google Scholar]
- 27.Pistello, M, A. Morrica, F. Maggi, M. L. Vatteroni, G. Freer, C. Fornai, F. Casula, S. Marchi, P. Ciccorossi, P. Rovero, and M. Bendinelli. 2001. TT virus levels in the plasma of infected individuals with different hepatic and extrahepatic pathologies. J. Med. Virol. 63:189-195. [PubMed] [Google Scholar]
- 28.Romeo, R., P. Hegerich, S. U. Emerson, M. Colombo, R. H. Purcell, and J. Bukh. 2000. High prevalence of TT virus (TTV) in naïve chimpanzees and in hepatitis C virus-infected humans: frequent mixed infections and identification of new TTV genotypes in chimpanzees. J. Gen. Virol. 81:1001-1007. [DOI] [PubMed] [Google Scholar]
- 29.Saback, F. L., S. A. Gomes, and C. Niel. 2002. High frequency of mixed TT virus infections in healthy adults and children detected by a simplified heteroduplex mobility assay. J. Virol. Methods 101:117-125. [DOI] [PubMed] [Google Scholar]
- 30.Schroter, M., S. Polywka, B. Zollner, P. Schafer, R. Laufs, and H. H. Feucht. 2000. Detection of TT virus DNA and GB virus type C/hepatitis G virus RNA in serum and breast milk: determination of mother to child transmission. J. Clin. Microbiol. 38:745-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugiyama, K., K. Goto, T. Ando, F. Mizutani, K. Terabe, Y. Kawabe, and Y. Wada. 1999. Route of TT virus infection in children. J. Med. Virol. 59:204-207. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, F., K. Chayama, A. Tsubota, N. Akuta, T. Someya, M. Kobayashi, Y. Suzuki, S. Saitoh, Y. Arase, K. Ikeda, and H. Kumada. 2001. Pathogenic significance and organic virus levels in patients infected with TT virus. Intervirology 44:291-297. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi, K., M. Hijikata, E. I. Samokhvalov, and S. Mishiro. 2000. Full or near full length nucleotide sequences of TT virus variants (types SANBAN and YONBAN) and the TT virus-like mini virus. Intervirology 43:119-123. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, K., Y. Iwasa, M. Hijikata, and S. Mishiro. 2000. Identification of a new human DNA virus (TTV-like minivirus, TMLV) intermediately related to TT virus and chicken anemia virus. Arch. Virol. 145:979-993. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka, Y., D. Primi, R. Y. H. Wang, T. Umemura, A. E. T. Yeo, M. Mizokami, H. J. Alter, and J. W. K. Shih. 2001. Genomic and molecular evolutionary analysis of a newly identified infectious agent (SEN virus) and its relationship to the TT virus family. J. Infect. Dis. 183:359-367. [DOI] [PubMed] [Google Scholar]
- 36.Tawara, A., Y. Akahane, M. Takahashi, T. Nishizawa, T. Ishikawa, and H. Okamoto. 2000. Transmission of human TT virus of genotype 1a to chimpanzees with fecal supernatant or serum from patients with acute TTV infection. Biochem. Biophys. Res. Commun. 278:470-476. [DOI] [PubMed] [Google Scholar]
- 37.Xia, X. 2000. Data analysis in molecular biology and evolution. Kluwer Academic Publishers, Boston, Mass.
- 38.Yokoyama, H., J. Yasuda, H. Okamoto, and Y. Iwakura. 2002. Pathological changes of renal epithelial cells in mice transgenic for the TT virus ORF1 gene. J. Gen. Virol. 83:141-150. [DOI] [PubMed] [Google Scholar]