Abstract
A novel strain of equine infectious anemia virus (EIAV) called vMA-1c that rapidly and specifically killed infected equine fibroblasts (ED cells) but not other infectible cell lines was established. This strain was generated from an avirulent, noncytopathic strain of EIAV, MA-1. Studies with this new cytolytic strain of virus have permitted us to define viral parameters associated with EIAV-induced cell killing and begin to explore the mechanism. vMA-1c infection resulted in induction of rapid cell death, enhanced fusogenic activity, and increased rates of spread in equine fibroblasts compared to other strains of EIAV. The highly cytolytic nature of vMA-1c suggested that this strain might be superinfecting equine fibroblasts. Receptor interference studies demonstrated that prior infection of equine fibroblasts with EIAV did not alter the ability of vMA-1c to infect and kill these cells. In similar studies in a canine fibroblast cell line, receptor interference did occur. vMA-1c infection of equine fibroblasts was also associated with large quantities of unintegrated viral DNA, a well-established hallmark of retroviral superinfection. Cloning of the vMA-1c genome identified nucleotide changes that would result in at least one amino acid change in all viral proteins. A chimeric infectious molecular clone containing the vMA-1c tat, S2, and env open reading frames recapitulated most of the characteristics of vMA-1c, including superinfection, fibroblast killing, and fusogenic activity. In summary, in vitro selection for a strain of EIAV that rapidly killed cells resulted in the generation of a virus that was able to superinfect these cells, presumably by the use of a novel mechanism of cell entry. This phenotype mapped to the 3′ half of the genome.
Induction of cell death by variant strains of retroviruses which are usually not cytotoxic has been recognized for more than 20 years (16, 45). However, the mechanisms that lie behind the cell death mediated by these variant strains are poorly understood. Several characteristics common to these retroviruses, in addition to their ability to kill the host cell, include the production of large amounts of unintegrated DNA (52) and the ability to superinfect cells due to the absence of receptor interference (11, 23, 34, 45). Whether these characteristics are directly responsible for the induction of cell death or simply serve as markers for viruses that also cause cell death is not clear. With each of these viruses, the virulence maps to the envelope, and these viruses either utilize novel receptors or bind to a different region on the same receptor as the less virulent strains (11, 23, 34). Subgroup B of avian leukosis virus, which rapidly induces cell death, may utilize an additional mechanism of cell killing. These viruses use a cellular receptor for entry that contains a death domain (4, 5). The death domain is activated during virus entry, and the caspase cascade is induced, resulting in apoptosis.
Equine infectious anemia virus (EIAV) is a lentivirus that in vivo primarily replicates in tissue macrophages (29, 36, 39). In tissue culture, in addition to infecting equine macrophages, EIAV strains persistently replicate in primary equine endothelial cells (24) and fibroblasts (17) as well as canine macrophage (13) and canine and feline fibroblast cell lines. While EIAV kills equine macrophage cultures during in vitro infection, other infected cells and cell lines are not killed but become chronically and productively infected with the virus. Thus, the same strain of EIAV (e.g., Th.1) can infect and kill macrophages while chronically infecting endothelial cells (W. J. Maury, unpublished data).
The mechanism of EIAV-induced cell killing is not known. A previous study exploring EIAV cytolysis demonstrated that a prototypic strain of tissue culture-derived virus can chronically infect fetal equine kidney cells while killing fetal donkey dermal cells over several weeks. In this study, cytopathology was associated with enhanced quantities of integrated proviral sequences and increased production of viral RNA (33). In addition to cell-specific differences that might be responsible for cytolytic activity, changes in the EIAV genome may also be responsible for induction of cell killing. Certainly, strains of EIAV that infect and kill macrophages differ from the tissue culture-derived isolates within env and long terminal repeat (LTR) sequences, and it is possible that these specific alterations may be required for cytolysis (30-32). For instance, changes within the EIAV SU protein may be responsible for differential interactions with the cellular receptor, resulting in the induction of cell death. To date, this possibility has not been explored.
In order to investigate the mechanism of EIAV cell killing, a cytolytic strain of EIAV called vMA-1c that specifically killed equine fibroblasts but not other infectible cell types was developed. With vMA-1c as a model for cytolytic EIAV strains, the interaction of this virus with cells was investigated. vMA-1c killing of equine fibroblasts correlated with absence of receptor interference, the production of increased quantities of virus-specific, unintegrated DNA, enhanced rates of virus spread within the infected culture, and increased fusogenic activity. These characteristics were consistent with vMA-1c superinfection of fibroblasts. Mapping studies indicated that sequences within the 3′ half of the genome were responsible for the ability of the strain to kill fibroblasts.
MATERIALS AND METHODS
Cells and viral strains.
Equine, feline, and canine cell lines were used to characterize the killing characteristics of vMA-1c. All cells used in this study were infectible with tissue culture strains of EIAV. All cells were maintained in high-glucose Dulbecco's modified Eagle's medium with 10% fetal calf serum, penicillin, and streptomycin. A highly transfectable feline fibroblast cell line, FEA, that is derived from fetal embryo cells was used for transfection of the molecular clones. In addition, Cf2Th, a canine fibroblastic cell line (ATCC CRL-1430) (28), and ED cells, an equine fibroblastic cell line derived from dermal cells (ATCC CCL57), were used.
Three different strains of EIAV were used in these studies. MA-1 is an avirulent, tissue culture-derived strain of EIAV that replicates well in fibroblasts and endothelial cells but poorly in primary equine macrophages (6, 24). vMA-1c, the strain studied in this paper, was derived by serial passage in ED cells from MA-1 as described in Results. pSP19-2 is an avirulent, infectious molecular clone of EIAV that replicates in most fibroblast cell lines as well as endothelial cells and macrophages (31).
Generation and titering of viral stocks.
Viral stocks of MA-1 and vMA-1c were generated in the equine fibroblast cell line ED. Supernatants were harvested from cells that were >95% positive for EIAV antigen as determined by immunostaining. Supernatants were aliquoted and frozen at −80°C until needed. Viral titers were determined by infection of ED cells with serial dilutions of stock, followed by fixation at 40 h postinfection and anti-EIAV immunostaining of cells as previously described (25). The number of EIAV antigen-positive cells within the infected cell monolayer was counted, and titers were determined. Viral stocks of pSP19-2 were generated by transfection of 2 μg of DNA into FEA cells with Gene Porter as the liposomal agent. Virus was permitted to spread through the culture, and supernatants were collected and passaged on ED cells, where supernatants from chronically infected cells were collected, used as viral stocks, and counted in ED cells as described above.
Detection of EIAV replication.
EIAV-infected cultures were identified and characterized for immunostaining of viral antigens as previously described (25). For immunostaining, polyclonal horse anti-EIAV antiserum (1:800) from a long-term-infected horse (WSU 2085) was used as the primary antiserum. This antiserum principally recognizes envelope (gp90 and gp46) and Gag proteins, as well as Gag precursor polyproteins. Incubation with primary antiserum was followed by several washes with phosphate-buffered saline and a 1:500 dilution of peroxidase-conjugated goat anti-horse immunoglobulin (Jackson Immunoresearch) as previously described (25). Peroxidase activity was detected with the substrate 3-amino-9-ethylcarbazole (Sigma).
Reverse transcription-PCR of vMA-1c-infected ED cells.
Initial sequence data from vMA-1c was generated by reverse transcription-PCR. mRNA was purified from 4-day vMA-1c-infected ED cells with the Fast Track poly(A) kit (Invitrogen). Negative-strand cDNA was synthesized from the mRNA with antisense oligonucleotides from EIAV as primers and avian myeloblastosis virus reverse transcriptase for 1 h at 42°C. Thirteen different pairs of sense and antisense EIAV primers were used to amplify 500 to 1,000 bp of DNA with 35 cycles (95°C for 30 s, 50°C for 30 s, and 72°C for 1.5 min). This approach produced a series of overlapping PCR products that were washed extensively with Amicon Microcon filters (100,000 molecular weight cutoff) and directly sequenced to determine the predominant RNA sequence present in the viral stock. The sequence data generated with this approach were compared to the sequences of the vMA-1c clones generated by long-template PCR to identify the 5′ and 3′ vMA-1c clones that best represented the predominant sequence present in the virus stock.
Cloning and sequencing of the vMA-1c genome with long-template PCR.
DNA from vMA-1c-infected ED cells at 4 days postinfection was isolated with the Wizard genomic purification kit (Promega) per the manufacturer's instructions. The DNA was amplified with the Expand long-template PCR system (Roche) with their buffer 3 system and 2.25 mM MgCl2. The primers that were used to create clones from the 5′ half of the genome were AatII5′ltr (GACGTCACTGTGGGGTTTTTATGA) and NcoIpolC′ (TACTTCCCATGGTGTCTGTCC), which produced a 4.9-kb fragment from the 5′ terminus of the LTR to the middle of the pol gene. The primers used to generate clones from the 3′ half of the genome were NcoIpol (GGGACAGCACCATGGGAAGTA) and 7886C′ (GTAGGATCTCGAACAGACAAAC), which produced a 3.3-kb fragment. The program for the amplification was 28 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 5 min. A single, subsequent cycle of amplification with Taq polymerase was performed to permit cloning of the fragment into a TA cloning vector. This amplification strategy divided the vMA-1c genome roughly in half at the naturally occurring NcoI site located within the pol gene. The amplified fragments were cloned into pGEM-T (Promega) and transformed into high-efficiency strain Escherichia coli JM109. Correctly sized clones were sequenced at the University of Iowa Sequencing Facility. All sequences were confirmed by sequencing both strands of DNA.
Isolation of unintegrated DNA, Southern analysis, and restriction polymorphism analysis.
Unintegrated, low-molecular-weight DNA was extracted from confluent T25 or T75 flask cultures. ED or Cf2Th cells were infected with either 5,000 or 8,000 infectious units of virus and harvested 40 h or 4 days postinfection. For chronic infections, EIAV-infected cultures were infected and maintained for several weeks until cultures were >98% seropositive for virus antigen. At that time, Hirt DNA extractions were performed. Cells that were dually infected with virus were harvested for Hirt supernatants either 40 h or 4 days following a secondary infection with 5,000 or 8,000 infectious units of virus.
Low-molecular-weight DNA was obtained as described by Hirt (14). Briefly, cells were gently lysed for 30 min at 37°C in 10 mM Tris (pH 8.0)-10 mM EDTA-1 μg of RNase per μl-1% sodium dodecyl sulfate. Pronase (500 μg/ml) and proteinase K (20 μg/ml) were added and incubated at 37°C for 2 h. NaCl was added to a final concentration of 1.1 M, the mixture was inverted gently, and the sample was stored at 4°C overnight. Genomic DNA and cellular debris were removed by centrifugation at 21,000 × g for 45 min at 4°C. Supernatant was removed with care to avoid any debris or viscous genomic DNA. The supernatant was extracted with equal volumes of phenol, 1:1 phenol-chloroform, and chloroform and precipitated with sodium acetate and 2.5 volumes of ethanol. The sample was stored overnight at −20°C, and DNA was pelleted at 21,000 × g for 30 min, washed in 80% ethanol, dried, and resuspended in sterile water for analysis.
Genomic DNA was isolated at 40 h postinfection with the Wizard genomic purification kit (Promega) per the manufacturer's instructions. Concentrations of DNA were determined spectrophotometrically.
For detection of unintegrated viral DNA, 5 μg of undigested DNA was run in each lane on a 0.9% Tris-acetate-EDTA-agarose gel. DNA was transferred to nitrocellulose, and a 32P-labeled full-length clone of EIAV (p29A) was used as a probe to identify EIAV-specific bands. Known quantities of a molecular clone of EIAV (p29A) that were digested with EcoRI to release the full-length, 8.3-kb viral sequence were run in parallel to permit the identification and quantification of linear DNA sequences.
For the restriction site polymorphism studies, a primer pair that amplified the transmembrane protein region of env that produced a 683-bp fragment from the Hirt DNA samples was used. The primers were 6743 (CAATTTGGCACAATCCATGA) and 7426C′ (CCATAGCCTGCTATGCGTCC). Thirty-eight cycles of amplification were performed as follows: 1 cycle of 95°C for 5 min, 50°C for 30 s, and 72°C for 1.5 min, followed by 36 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1.5 min, and followed by 1 cycle of 95°C for 30 s, 50°C for 30 s, and 72°C for 5 min. In the chronic MA-1 infection studies, amplified products were digested with MscI for 3 h, followed by the addition of more enzyme and continued digestion to maximize cutting. Two MscI sites are present within the parental MA-1 DNA, resulting in cleavage of the 683-bp fragment into 289-, 200-, and 194-bp fragments. The two smaller restriction fragments comigrated on the agarose gel and could not be resolved. The restriction sites were absent in the vMA-1c DNA. The fragments were separated on a 2.2% Tris-borate-EDTA (TBE) gel and visualized by ethidium bromide staining.
Restriction enzyme polymorphism studies with chronically infected pSP19-2 cells were performed by BstEII digestion of Hirt supernatant DNA, amplified with the primer set described above, producing a 683-bp fragment. pSP19-2 contains a BstEII site at position 1949 of the env gene, resulting in 439- and 244-bp bands. Neither the MA-1 nor the vMA-1c genome contained this site. Restriction digest analysis was performed as described above.
Neutralizing antibody studies.
Antiserum obtained from an MA-1-infected horse (7) was tested for neutralizing activity against vMA-1c. Serial dilutions of antiserum were incubated with vMA-1c for 30 min and added to ED cell monolayers for 40 h. Cells were fixed and immunostained for EIAV antigens as described above. Numbers of infectious hits were counted in each treatment, and the percent inhibition of infectivity was determined for each dilution of antibody. Antibody dilutions that were >98% effective at neutralizing infectivity were considered effective neutralization titers.
To test the ability of neutralizing antiserum to protect against vMA-1c cell killing, 50 or 5,000 infectious units of vMA-1 were incubated with or without antiserum for 30 min prior to addition to ED cells. Alternatively, antiserum was added to infections 2 h after the addition of virus to cells. Medium was changed, and fresh medium containing a 1:256 dilution of antiserum was maintained for 6 days. At the termination of the experiment, cells were washed with phosphate-buffered saline once and lysed. Cell viability was determined with an ATPLite-M assay (Packard Biosciences) per the manufacturer's instructions.
Generation of chimeric infectious molecular clones and production of virus.
The EIAV infectious molecular clone pSP19-2 was used as a backbone for the generation of chimeric infectious clones (31). A chimeric construct, p19/vEnv, that contained the second exon of tat, S2, and most of the env sequences from vMA-1c was generated. This construct was made by digestion of pSP19-2 and 7886-6/pGEM-T with NcoI (within pol) and BstBI (within the transmembrane protein), and the pieces were ligated and transformed into Stabl2 cells (Invitrogen) per the manufacturer's instructions to reduce the possibility of retroviral recombination and/or deletion. A chimeric construct, p19/vEnvltr, was generated by digestion of p19/vEnv and 7886-6/pGEM-T with BstBI and NotI. Appropriate fragments were separated on an agarose gel and cloned as described above.
Viral stocks were generated by transfection of molecular clone DNA via Gene Porter per the manufacturer's instructions into FEA cells. Virus was permitted to spread in the culture for several weeks. Supernatants were collected, clarified by centrifugation, added to ED cells, and maintained for several weeks. Spread of the chimeric viruses was monitored by immunostaining of cultures for EIAV antigens. Supernatants were collected for viral stocks once the cells were greater than 95% EIAV antigen positive.
Cell killing and receptor interference assays.
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) (42, 43) and ATPLite-M (Packard BioScience) assays were used to measure cell viability in the virally induced cell killing, receptor interference, and antibody neutralization assays. For these assays, ED or Cf2Th cells were infected with equal quantities of virus on the appropriate day postplating. At the termination of the experiment, cells were rinsed with phosphate-buffered saline. In the MTT assay, the cells were incubated with 500 μl of phosphate-buffered saline, 5% fetal calf serum, and 0.5 mg of MTT (Sigma) per ml for 3 to 4 h. Cells were rinsed again with phosphate-buffered saline, and the insoluble product was solubilized with acidic isopropanol. The optical density of the solution was read on a microtiter plate reader at 570 nm. ATPLite-M assays were performed per the manufacturer's instructions.
Receptor interference assays were performed on chronically infected ED or Cf2Th cells that were greater than 95% EIAV antigen positive. Equal quantities of infectious virus were added to the chronically infected cultures, and cell viability assays were performed 6 to 8 days postinfection.
RESULTS
Generation and initial characterization of vMA-1c.
A strain of EIAV that rapidly destroyed equine dermal fibroblast (ED) cell monolayers was established. This novel strain, called vMA-1c, was studied as a model of EIAV cell killing. vMA-1c was derived from MA-1, an avirulent, nonfusogenic, tissue culture-adapted strain of EIAV (6). vMA-1c was established from a stock of MA-1 that arose spontaneously and was identified as killing ED cells. To select a population of virus with high cytolytic activity, stocks of virus were serially passaged every 4 to 6 days onto a fresh monolayer of ED cells over a 2-month period. Then virus stocks of vMA-1c were biologically cloned in ED cell cultures by limiting dilution twice. During this cloning, the lowest concentration of virus as detected by reverse transcriptase activity and viral antigen immunostaining of cultures was also the lowest concentration to kill ED cultures.
This strain was found to readily infect and kill ED cells. By day 2 postinfection, syncytia were evident within ED cell cultures (Fig. 1). ED cultures were >95% EIAV antigen positive by 4 to 5 days postinfection, and the monolayer was destroyed within an additional 24 h. The rapid destruction of equine fibroblasts by vMA-1c was in contrast to infection with the parental strain of virus, MA-1, which did not fuse or kill ED cells by infection, and complete spread of which through the culture took several weeks (Fig. 1D).
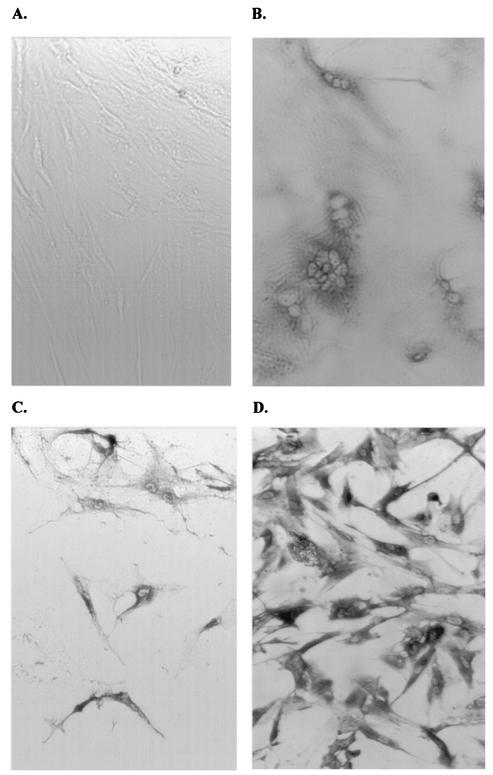
FIG. 1.
vMA-1c infection of equine dermal fibroblasts (ED cells). All cells were immunostained for EIAV antigens with horse anti-EIAV antiserum. (A) An uninfected ED cell culture. (B) vMA-1c-infected ED cell culture fixed and stained 2 days postinfection. Syncytia are prominent at this time. (C) vMA-1c-infected ED cell culture fixed and stained 5 days postinfection. (D) Immunostaining of ED cells chronically infected with parental MA-1 virus. Magnification, ×100.
With anti-EIAV immunostaining of infected cultures, the number of EIAV-infected ED cells was quantitated over time to determine if vMA-1c was spreading more rapidly than the parental virus (Fig. 2A). The numbers of parental MA-1- and vMA-1c-infected ED cells at early time points were similar (12 to 42 h); however, by 48 h postinfection, more cells were antigen positive in the vMA-1c infections, indicating enhanced rates of spread by vMA-1c. Additional experiments investigating virus spread at later times following infection indicated that vMA-1c infection in ED cells continued to spread more rapidly than MA-1 in ED cells until the experiments were terminated by vMA-1c-induced cell death (data not shown). Similar studies in a canine fibroblast cell line, Cf2Th, demonstrated that vMA-1c did not spread faster in this cell line and that vMA-1c spread was perhaps even delayed compared to that of MA-1 (Fig. 2B).
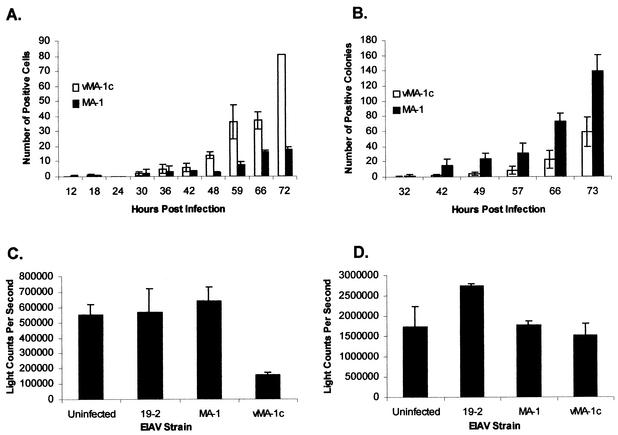
FIG. 2.
Characterization of vMA-1c infection. (A) vMA-1c spreads faster in ED cells than MA-1. Equivalent infectious units of vMA-1c or MA-1 were added to duplicate wells of ED cells (≈2 × 104/well), and infected wells were fixed at the times noted. Fixed cells were immunostained for EIAV antigens. (B) vMA-1c does not spread faster in Cf2Th cells than MA-1. Duplicate wells of Cf2Th cells (≈2 × 104/well) were infected with vMA-1c or MA-1, and wells were fixed at the times noted. Fixed cells were immunostained for EIAV antigens, and the numbers of infected cells/well were determined. (C) vMA-1c killing of ED cells. A total of 1,000 infectious units of vMA-1c, MA-1, or pSP19-2 were added to triplicate cultures (≈2 × 104/well) and maintained for 6 days. At the termination of the infection, cells were washed with phosphate-buffered saline and lysed. Detection of cellular ATP levels was determined with the ATPLite-M assay as a measure of cell viability. (D) Absence of vMA-1c cytolysis of Cf2Th cells. Assays were performed as described for C. Experiments were performed at least three times, and all treatments were performed in duplicate or triplicate. Shown are means of representative experiments ± standard deviation or range.
To characterize the killing properties of vMA-1c, equal numbers of cells were infected with 1,000 infectious units (multiplicity of infection of 0.05) of either vMA-1c, parental MA-1, or a stock of an unrelated infectious molecular clone of EIAV, pSP19-2. Cell viability of the infected monolayers was measured at 6 days postinfection (Fig. 2C and D). While the numbers of ED cells in the uninfected and MA-1- or pSP19-2-infected populations were equivalent at 6 days postinfection, as determined by an ATPLite-M assay (Packard BioScience), the number of cells in the vMA-1c infection was significantly lower (Fig. 2C). By visual inspection, most of the vMA-1c-infected ED monolayer had been destroyed (data not shown). In contrast, vMA-1c infection or the other EIAV infections of Cf2Th cells did not result in cell loss (Fig. 2D). Other EIAV-infectible cells such as equine endothelial cells and canine macrophage cell lines could also be productively infected with vMA-1c but were not rapidly killed by the infection (data not shown). These findings, along with the more rapid spread of vMA-1c in ED cells, suggested that a unique interaction between equine fibroblasts and vMA-1c was occurring.
Identification of genomic alternations in vMA-1c.
The LTR, gag, and env genes of vMA-1c were initially reverse transcription-PCR amplified, and the amplified fragments were directly sequenced to determine the predominant sequence present in the vMA-1c stock. In addition, proviral DNA was amplified and cloned from vMA-1c-infected ED cell DNA at 4 days postinfection with long-template PCR. Two clones from the 5′ half of the genome and two clones from the 3′ half of the genome were completely sequenced. In general, there was good agreement between the sequences obtained directly from the reverse transcription-PCR products and the provirus-generated clones, suggesting that there was limited sequence heterogeneity in the vMA-1c stock.
The cloned sequences were compared to the parental MA-1 sequence to identify changes (Fig. 3). Changes that would result in amino acid substitutions were present in all vMA-1c genes. The most notable changes were found in the S2 and env genes. S2 is a highly conserved gene located immediately upstream from and overlapping the start of env. The 65-amino-acid protein produced from this gene is not required for EIAV replication in tissue culture but enhances virulence in vivo (20, 21). A single-nucleotide insertion within the seventh codon of vMA-1c S2 resulted in a frameshift into env. Thus, it would be anticipated that vMA-1c does not produce an S2 protein and that the Env has two initiation codons: the methionine previously identified to be the start codon of Env, and the S2 methionine.
FIG. 3.
Nucleotide sequence of LTR and deduced amino acid sequences of vMA-1c open reading frames. Two long-template PCR-generated clones of the 5′ (5′ LTRPCR-2 and -3) and the 3′ (7886-1 and 7886-6) halves of the genome were sequenced and compared to parental MA-1 sequences. An NcoI site present in the integrase gene is the 3′ terminus for the 5′-half clones and the 5′ terminus for the 3′ clones. Dashes represent the identical amino acid (or nucleic acid within the LTR), and an asterisk represents a stop codon in the clones that is not present in MA-1. The number symbol represents the location of the NcoI site in the integrase gene. The amino acids, proline and tryptophan, which are located at the end of the 5′ clones and the beginning of the 3′ clones were duplicated during cloning and are shown in both sets of clones.
The greatest number of deduced amino acid changes were found in SU, with a total of 16 changes for clone 7886-1 and 17 amino acid changes for clone 7886-6. Some of these amino acid changes clustered into the V3, V4, and V6 regions of SU, previously identified as genetically hypervariable in vivo (19). Both 3′ clones contained premature stop codons in the transmembrane protein. The stop codon in clone 7886-1 would be predicted to produce a 156-amino-acid protein containing only the transmembrane protein ectodomain. In contrast, clone 7886-6 contained a stop codon at nucleotide 7328 of the proviral genome that would result in an abbreviated transmembrane protein of 228 amino acids with a full-length ectodomain and transmembrane domain, but a highly shortened cytoplasmic tail. From the initial reverse transcription-PCR sequencing studies, the predominant env sequence of vMA-1c was similar to that found in clone 7886-6, and, consequently, clone 7886-6 was used to generate the chimeric virus described below.
Absence of receptor interference during vMA-1c infection of ED cells.
Rapid spread within and destruction of the equine dermal monolayer by vMA-1c was reminiscent of previously reported retroviral infections where receptor interference was not detected, elevated levels of unintegrated DNA were present, and superinfection was occurring (10, 11, 45, 48, 52). Our initial receptor interference studies used cell death as a marker of vMA-1c infection. ED or Cf2Th cells were either uninfected or chronically infected with MA-1 or the unrelated strain of EIAV pSP19-2. Chronically infected cells were >98% EIAV antigen positive at the time of the studies. Cells were infected with 1,000 infectious units (multiplicity of infection = 0.05) of vMA-1c, MA-1, or pSP19-2, and cell viability was determined at day 6 or 8 postinfection (Fig. 4).
FIG. 4.
vMA-1c kills ED cells that are chronically infected with wild-type EIAV but not chronically infected Cf2Th cells. MA-1, vMA-1c, or pSP19-2 was incubated with (A) uninfected ED cells, (B) uninfected Cf2Th cells, (C) chronically pSP19-2-infected ED cells, (D) chronically pSP19-2-infected Cf2Th cells, (E) chronically MA-1-infected ED cells, or (F) chronically MA-1-infected Cf2Th cells. All chronically infected cultures were >98% EIAV antigen positive at the time of superinfection. At the termination of the infections, cells were lysed and cellular ATP levels were determined with ATPLite-M as a measure of cell viability. Experiments were performed at least three times, and all treatments were performed in triplicate in each experiment. Shown are means of a representative experiment ± standard deviation of the mean. The percentage of the uninfected-control value is shown above each bar.
As previously observed, vMA-1c infection of ED cells resulted in the loss of >90% of the monolayer (Fig. 4A); however, no decrease in cell viability was observed in the vMA-1c-infected Cf2Th cells (Fig. 4B). ED cells chronically infected with either MA-1 or pSP19-2 were also killed upon the addition of vMA-1c, although cell loss was not as pronounced as in the naïve cell population (Fig. 4C and 4E). In contrast, the addition of vMA-1c stocks to chronically MA-1-infected or pSP19-2-infected Cf2Th cultures had no deleterious effects on the monolayer (Fig. 4D and 4F). These findings suggested that vMA-1c was able to superinfect chronically infected ED cells and induce cell death.
Superinfection of ED cells by vMA-1c was supported by Southern blot detection of large quantities of EIAV-specific, unintegrated DNA present in vMA-1c-infected ED cells (Fig. 5A). Naïve ED cells or ED cells chronically infected with pSP19-2 were infected with either MA-1 or vMA-1c (multiplicity of infection = 0.004), and small, unintegrated DNA was extracted on day 4 postinfection. We then ran 5 μg of undigested Hirt DNA on a 0.9% agarose gel and transferred to nitrocellulose. The blot was probed with a 32P-labeled, full-length clone of EIAV. p29A, a molecular clone of EIAV, was digested with EcoRI, which linearizes the full-length sequence (6), and fourfold dilutions of digested p29A were run on the gel in parallel.
FIG. 5.
Detection of vMA-1c superinfection of ED cells. (A) Elevated levels of unintegrated DNA in vMA-1c-infected ED cells detected by Southern blotting. ED cells were acutely infected with virus, and Hirt supernatants were extracted at day 4 postinfection. In addition, chronically infected (>98% EIAV antigen positive) pSP19-2-infected ED cells were superinfected with either MA-1 or vMA-1c for 4 days prior to Hirt supernatant extraction. Southern blot analysis was performed on 5 μg of undigested low-molecular-weight DNA. DNA from a full-length clone of EIAV, p29A, was used as the probe. A total of 2.4 ng (not shown), 7.2 ng, and 28.2 ng of EcoRI-digested p29a that is 8.3 kb in length (6) was run in parallel to identify full-length linear viral sequences.
Unintegrated viral sequences could not be detected in the lanes containing either MA-1-infected or chronically pSP19-2-infected cells. Two bands were evident in the lane containing vMA-1c-infected ED DNA, an 8.3-kb band that comigrated with the full-length EIAV genome and a larger band of approximately 16 kb. The larger band may represent nicked circular DNA, as has been reported in Southern blots of other superinfecting retroviruses (27). An 8.3-kb band was also evident in the lane containing Hirt DNA from ED cells that were chronically infected with pSP19-2 and secondarily infected with vMA-1c. These findings indicated that vMA-1c infections of ED cells resulted in the production of large quantities of unintegrated linear viral DNA even when cells had previously been infected with other strains of EIAV.
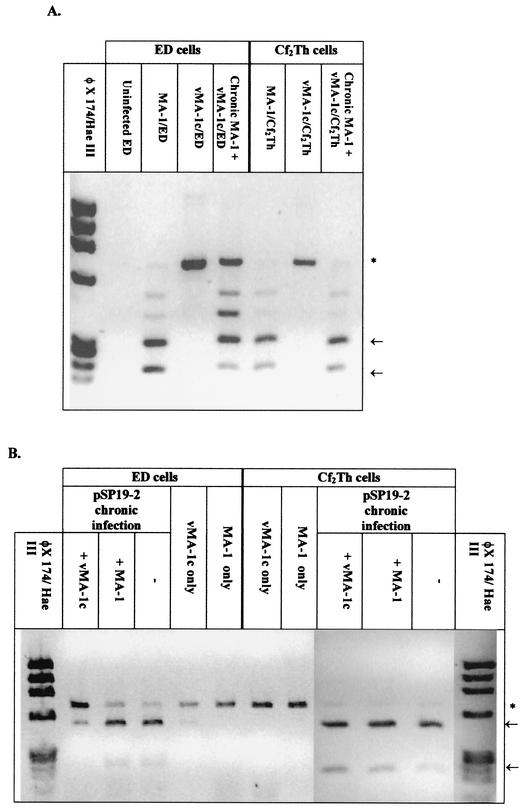
While these studies indicated that vMA-1c could superinfect ED cells producing elevated levels of virus-specific, unintegrated DNA, these studies did not directly address whether vMA-1c superinfection of Cf2Th cells was occurring. To test the specificity of vMA-1c superinfection of ED cells, restriction enzyme site polymorphisms in the envelope gene that distinguished between the vMA-1c, pSP19-2, and MA-1 genomes were identified. Two MscI sites are located at nucleotides 1947 and 2145 of the MA-1 env gene that are not present in vMA-1c. A 683-bp fragment was amplified from unintegrated DNA from EIAV-infected ED and Cf2Th cells and digested with MscI.
Digestion of MA-1 DNA resulted in three fragments of 286, 200, and 194 bp (Fig. 6A). The smaller two bands could not be distinguished within the gel. In contrast, MscI digestion of DNA from vMA-1c-infected cells gave a single band of 683 bp. MscI restriction analysis of DNA amplified from MA-1-chronically infected ED cells that were secondarily infected with vMA-1c resulted in the presence of both digested and undigested fragments, indicating that vMA-1c was able to enter and reverse transcribe in ED cells that had previously been infected with MA-1. In contrast, the presence of vMA-1c DNA was not evident upon vMA-1c infection of chronically MA-1-infected Cf2Th cells. While the data shown in Fig. 5 and 6 are from unintegrated DNA from cells acutely infected for 4 days, similar results were found in both unintegrated and genomic DNA studies at 40 h postinfection, a time point prior to virus spread (data not shown). These results indicate that detection of vMA-1c DNA in chronically MA-1-infected ED cells but not chronically MA-1-infected Cf2Th cells was due to the ability of vMA-1c to superinfect ED cells and not due to the more rapid spread of vMA-1c.
FIG. 6.
Restriction site polymorphism analysis of PCR-amplified DNA from Hirt supernatants demonstrated that vMA-1c is able to superinfect ED cells but not Cf2Th cells. Naïve or chronically infected ED cells were infected with MA-1 or vMA-1c for 4 days. Low-molecular-weight (Hirt) DNA was isolated from infected cells, and DNA was amplified with EIAV env primers 6743 and 7426C′. (A) vMA-1c is able to superinfect MA-1-infected ED cells but not MA-1-infected Cf2Th cells. env DNA was amplified from MA-1-infected cells and contained two MscI sites producing 286-, 200-, and 194-bp bands upon MscI digestion. The sites were not present in vMA-1c, resulting in a 683-bp band. MscI did not cutthe MA-1-amplified DNA to completion, presumably due to restriction site polymorphisms present in the MA-1 stock, and partially digested bands are evident in the gel. An asterisk indicates the 683-bp band, and the arrows identify the completely digested fragments. (B) vMA-1c is able to superinfect pSP19-2-infected ED cells but not pSP19-2-infected Cf2Th cells. The parental MA-1 virus was unable to superinfect either of the chronically pSP19-2-infected cell populations. Low-molecular-weight DNA was PCR amplified with primers 6743 and 7426C′ from chronically pSP19-2-infected ED and Cf2Th cells superinfected with vMA-1c or MA-1. Restriction site polymorphism analysis was then performed with BstEII digestion. pSP19-2 contains a BstEII site, producing 439- and 244-bp bands upon digestion; the site was not present in vMA-1c or MA-1, resulting in a 683-bp band. An asterisk indicates the 683-bp band, and the arrows identify the completely digested fragments.
The Southern blot results of Hirt supernatant DNA suggested that only vMA-1c could superinfect ED cells. However, to verify these findings, restriction enzyme polymorphism studies were also performed with cells chronically infected with pSP19-2 and subsequently infected with either MA-1 or vMA-1c. Within the 683-bp fragment of env that was amplified in these restriction enzyme polymorphism studies, pSP19-2 contains a BstEII site at position 1949 of the env gene that is not present in either the MA-1 or vMA-1c genome. PCR amplification of DNA from chronically pSP19-2-infected ED or Cf2Th cells followed by BstEII digestion demonstrated the presence of two bands of 439 and 244 bp (Fig. 6B).
Identical to the findings with chronically MA-1-infected ED cells, vMA-1c was able to superinfect pSP19-2-infected ED cells, as demonstrated by the presence of the 683-bp fragment in BstEII-digested DNA from the dually infected ED cells. In contrast, DNA from ED cells chronically infected with pSP19-2 and secondarily infected with MA-1 did not contain increased amounts of the 683-bp fragment over that present in the pSP19-2-infected culture, indicating that MA-1 was unable to superinfect ED cells. Neither MA-1 nor vMA-1c was able to superinfect pSP19-2-infected Cf2Th cell cultures. In total, these studies demonstrate that vMA-1c can specifically superinfect ED cells but not Cf2Th cells and that ED superinfection was a unique characteristic of vMA-1c.
Recent studies with feline leukemia virus have demonstrated that superinfection of certain strains of feline leukemia virus result from SU protein binding to an endogenous Env protein secreted by T cells, which in turn binds to the cellular receptor Pit-1 (2). In a similar manner, it was possible that vMA-1c was using a soluble protein synthesized by equine fibroblasts to mediate superinfection. To test this possibility, supernatants from uninfected ED cells were filtered and added to either Cf2Th cells or equine endothelial cultures in the presence or absence of vMA-1c, and cultures were monitored for cell death. No cell death was observed in either cell type following vMA-1c infection, suggesting that a soluble factor from the equine fibroblasts was not mediating vMA-1c-induced cell killing (data not shown).
Antibodies that neutralize vMA-1c infection block cell killing.
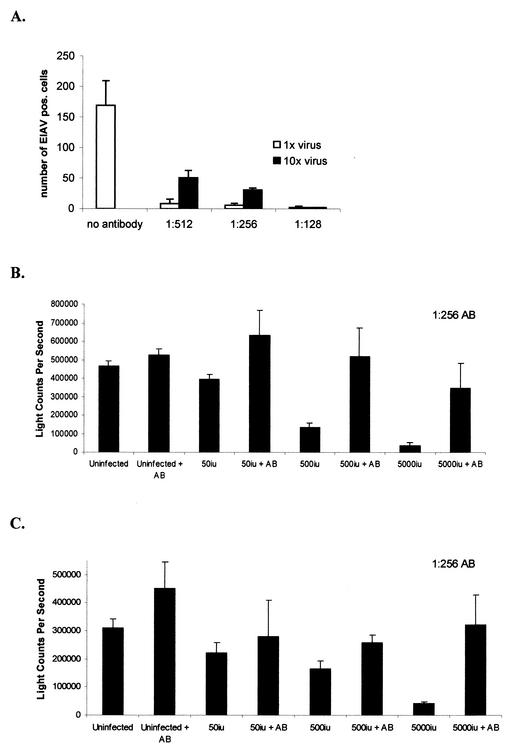
Equine antiserum directed against MA-1 infection was found to neutralize vMA-1c infection (Fig. 7A). At a dilution of 1:256, the antiserum preincubated with vMA-1c for 30 min was able to neutralize >98% of the infectious virus, as determined by cell immunostaining of viral antigens at 40 h postinfection. Thus, we were able to test if the addition of neutralizing antiserum could prevent vMA-1c-induced ED cell death. As expected, incubation of antiserum with the viral inoculum 30 min prior to infection effectively blocked cell killing by vMA-1c at 6 days postinfection (Fig. 7B). Antiserum also protected cells from death when added 2 h postinfection at multiplicities of infection ranging from 0.0025 (50 infectious units) to 0.25 (5,000 infectious units) (Fig. 7C). These findings indicated that vMA-1c-induced cell killing of the monolayer required ongoing infection that extracellular neutralizing antibodies prevented.
FIG. 7.
Neutralizing antibodies block vMA-1c-induced ED killing. (A) Serial dilutions of horse anti-EIAV antibodies (AB) (6) directed against the MA-1 strain of virus were incubated with vMA-1c for 30 min prior to addition to ED cells. A multiplicity of infection of either 0.0035 (1×) or 0.035 (10×) was used to determine virus neutralization. Cells were fixed and immunostained for virus antigens at 40 h postinfection. (B) Either 0, 500, or 5,000 infectious units of vMA-1c were incubated for 30 min with a 1:256 dilution of neutralizing antiserum and then added to 2 × 104 ED cells. At 7 days postinfection, cells were washed and lysed, and cellular ATP levels were determined by ATPLite-M as a measure of cell viability. (C) Either 50, 500, or 5,000 infectious units of vMA-1c were incubated with ED cells (2 × 104) for 2 h. Medium was removed, and medium containing a 1:256 dilution of horse anti-EIAV was added for the remainder of the study. At 7 days postinfection, cells were washed and lysed, and cellular ATP levels were determined by ATPLite-M as a measure of cell viability. Experiments were performed at least three times, and infections were performed in quadruplicate in each experiment. Shown are means of a representative experiment ± standard deviation of the mean.
Cell killing maps to the 3′ half of the vMA-1c genome.
Two chimeric infectious molecular clones, p19/vEnvltr and p19/vEnv, that contained either the entire 3′ half of the vMA-1c genome (from the NcoI site through the 3′ LTR) or just the coding regions of the 3′ half of vMA-1c (from the NcoI site to the BstBI site), respectively, were created (Fig. 8A). The clones in parallel with pSP19-2 were transfected into FEA cells, and over a period of several weeks, the culture became 1 to 10% EIAV antigen positive. In a manner similar to vMA-1c and other EIAV strains, neither of the chimeric clones induced cytopathology in FEA cells.
FIG. 8.
Chimeric EIAV infectious clones containing vMA-1c tat, S2, and env sequences resulted in increased syncytium formation within the ED cell culture, decreased cell numbers, and EIAV-infected ED cell superinfection. (A) Schematic diagrams of the chimeric clones p19/vEnvltr and p19/vEnv. (B) A photomicrograph of p19/vEnv fusogenic activity. Magnification, ×200. Syncytia were readily apparent in cultures infected with the chimeric virus 4 days postinfection. (C) Cell death resulted from p19/vEnv infection of ED cells. ED cells were infected at 24-h intervals with either vMA-1c or p19/vEnv. MTT assays were performed at the termination of the experiment as a measure of cell viability. Cell killing by p19/vEnv lagged behind vMA-1c killing by approximately 2 days. (D) Chronic EIAV infection does not protect ED cells from p19/vEnv-induced killing. ED cells or ED cells that were chronically infected with either MA-1 or pSP19-2 were infected with vMA-1c or p19/vEnv. At 8 days postinfection, cell viability was determined by ATPLite-M assay. The percentage of the uninfected-control value is shown above each bar.
Supernatants from transfected cells were collected, centrifuged twice to eliminate cellular debris, and added to ED cells. The two clones had similar replication and cell killing characteristics in ED cells, so subsequent studies focused on p19/vEnv only. p19/vEnv-infected ED cultures contained notable syncytia, similar to those identified in vMA-1c-infected ED cultures (Fig. 8B), and caused destruction of the monolayer with a slightly delayed time course compared to vMA-1c (Fig. 8C). The ability of p19/vEnv to superinfect and kill chronically EIAV-infected ED monolayers was tested. As Fig. 8D demonstrates, p19/vEnv was able to kill chronically MA-1-infected or pSP19-2-infected ED cultures in a manner similar to vMA-1c, although neither vMA-1c nor p19/vEnv was as effective in killing the chronically infected cultures as at killing naïve cells. These findings indicated that the coding sequences present within the 3′ half of the genome are responsible for the superinfectivity, fusogenic activity, and cell killing caused by vMA-1c.
DISCUSSION
In this study, we have developed and characterized a strain of EIAV, vMA-1c, that rapidly and specifically killed equine fibroblasts. While vMA-1c infected other cells used for propagation of EIAV, it was specifically cytolytic for equine fibroblasts. vMA-1c cell killing correlated with rapid spread of the virus within the equine fibroblast culture, an absence of receptor interference, and the production of large quantities of unintegrated viral DNA. Mapping studies with a chimeric virus generated from pSP19-2 and vMA-1c sequences indicated that genes within the 3′ half of vMA-1c were responsible for the absence of viral interference as well as the fusogenic and cell killing properties of vMA-1c. Whether all three of these characteristics of vMA-1c map to the same region of the 3′ end of genome has not been determined.
In general, high levels of fusogenic activity in retroviruses have not been associated with a superinfection phenotype. This suggests that vMA-1c induction of syncytia and superinfection may map to different regions of the vMA-1c genome even though both characteristics were only observed in equine fibroblasts and mapped to the 3′ end of the genome. Enhanced fusogenic activity of retroviruses has been previously mapped to several regions within Env (15, 18, 53). Amino acid changes in the proline-rich hinge region of murine leukemia virus SU, the transmembrane protein fusion peptide, the transmembrane protein membrane-spanning domain, and the transmembrane protein cytoplasmic tail have been reported to alter the fusogenic properties of retroviruses (15, 18, 26, 37, 40, 44, 46, 47, 50, 51, 53). The vMA-1c transmembrane protein contains a truncated cytoplasmic tail, and we predict that the shortened cytoplasmic tail of the transmembrane protein may be responsible for the highly fusogenic properties of vMA-1c.
The shortened transmembrane protein cytoplasmic tail may also be responsible for the enhanced rate of spread of the virus. Sauter et al. have demonstrated that in simian immunodeficiency virus, tyrosine at position 723 within the cytoplasmic tail is involved in recycling of transmembrane protein from the surface of the cells (38). The authors demonstrated enhanced infection kinetics when the transmembrane protein was truncated. Furthermore, the cytoplasmic tail of lentiviral transmembrane proteins has been shown to be associated with several cellular proteins involved in trafficking (3, 12, 49). Transmembrane protein truncations in tissue culture-derived EIAV stocks have been described in an earlier study, but enhanced fusogenic activity and rates of spread of this strain were not noted (35).
The inability of prototypic strains of EIAV to interfere with vMA-1c entry into ED cells suggested that vMA-1c was entering equine fibroblasts via a novel interaction not used by other strains of EIAV. This mechanism of entry could involve the utilization a different cellular receptor or an altered interaction of the viral SU with the normal cellular receptor (1, 10). Interestingly, vMA-1c infection of other cell types that were not killed by the infection (e.g., Cf2Th cells) appeared to be mediated through the normal cellular receptor, because prior infection of these cells prevented vMA-1c infection, as detected by viral genome restriction enzyme site polymorphism analysis. vMA-1c also was not fusogenic in these other cell types.
Correlation of superinfection with enhanced in vivo virulence and cell cytolysis has been described for a number of different retroviruses, including feline leukemia virus (2, 10, 34), avian leukosis virus (1, 4, 5, 45, 48), and a mink cell focus-forming murine leukemia virus (23, 52). Unlike our novel strain of EIAV, many of these retroviruses do not cause destruction of the infected monolayer but do cause decreased rates of increasing cell numbers (48, 52). Recently, Yoshimura et al. proposed that superinfection may induce apoptosis by the production of large amounts of unintegrated DNA as a consequence of superinfection (52). Further studies with vMA-1c will be needed to determine if the virus is inducing apoptosis and, if so, the mechanism mediating cell death.
The association of elevated levels of unintegrated viral DNA and cell killing has been noted for years (16, 48); however, the mechanism by which unintegrated retroviral sequences cause cell death has remained unclear. Recently, Li et al. demonstrated that cell lines deficient in enzymes of the nonhomologous DNA end-joining repair system did not produce two LTR circles and were more susceptible to retrovirus-induced cell death (22). The authors propose that linear retroviral sequences may mimic irradiation-induced double-stranded DNA breaks, a phenomenon known to induce apoptosis. This finding suggests that superinfection that results in the rapid buildup of linear DNA may kill cells through the presence of uncontrolled quantities of linear DNA, triggering apoptosis. Alternatively, the buildup of preintegration complexes within the cell may lead to numerous double-stranded breaks of the genome that may induce cell death (8, 9).
The rate of spread and the cytolytic activity of vMA-1c in ED cells are reminiscent of EIAV infection of primary equine macrophages. All strains of EIAV that replicate in macrophages also kill macrophages but not necessarily other infectible cell types. This suggests the intriguing possibility that EIAV superinfection may be occurring in macrophages. Consistent with this possibility, Steagall et al. have demonstrated that large quantities of unintegrated DNA as well as some integrated proviral sequences are found in pSP19-2-infected macrophage cultures at 48 and 72 h postinfection (41). In contrast, most of the pSP19-2 DNA was integrated in a chronically infected culture of the feline fibroblast cell line FEA (41). Receptor interference studies in primary macrophages have not been performed but would provide insight into the mechanism of EIAV entry into macrophages. Such interference studies are currently not possible due to the rapidity with which all macrophage-tropic strains of virus kill these cells. The future identification and characterization of the EIAV cellular receptor should allow more detailed studies on the mechanism of viral entry into macrophages.
Acknowledgments
We thank Susan Carpenter, C. Martin Stoltzfus, and Paul Cassella for helpful comments on the manuscript.
This study was supported by a grant from the National Cancer Institute (CA72063) to W.M. and by a University of Iowa Comprehensive Cancer Center Collaborative grant.
REFERENCES
- 1.Adkins, H. B., S. C. Blacklow, and J. A. Young. 2001. Two functionally distinct forms of a retroviral receptor explain the nonreciprocal receptor interference among subgroups B, D, and E avian leukosis viruses. J. Virol. 75:3520-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 3.Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brojatsch, J., J. Naughton, H. B. Adkins, and J. A. Young. 2000. TVB receptors for cytopathic and noncytopathic subgroups of avian leukosis viruses are functional death receptors. J. Virol. 74:11490-11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845-855. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter, S., and B. Chesebro. 1989. Change in host cell tropism associated with in vitro replication of equine infectious anemia virus. J. Virol. 63:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter, S., L. H. Evans, M. Sevoian, and B. Chesebro. 1987. Role of the host immune response in selection of equine infectious anemia virus variants. J. Virol. 61:3783-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel, R., R. A. Katz, G. Merkel, J. C. Hittle, T. J. Yen, and A. M. Skalka. 2001. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol. Cell. Biol. 21:1164-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel, R., R. A. Katz, and A. M. Skalka. 1999. A role for DNA-PK in retroviral DNA integration. Science 284:644-647. [DOI] [PubMed] [Google Scholar]
- 10.Donahue, P. R., S. L. Quackenbush, M. V. Gallo, C. M. deNoronha, J. Overbaugh, E. A. Hoover, and J. I. Mullins. 1991. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus feline leukemia virus-FAIDS. J. Virol. 65:4461-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorner, A. J., and J. M. Coffin. 1986. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell 45:365-374. [DOI] [PubMed] [Google Scholar]
- 12.Evans, D. T., K. C. Tillman, and R. C. Desrosiers. 2002. Envelope glycoprotein cytoplasmic domains from diverse lentiviruses interact with the prenylated Rab acceptor. J. Virol. 76:327-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hines, R., and W. Maury. 2001. DH82 cells: a macrophage cell line for the replication and study of equine infectious anemia virus. J. Virol. Methods 95:47-56. [DOI] [PubMed] [Google Scholar]
- 14.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 15.Januszeski, M. M., P. M. Cannon, D. Chen, Y. Rozenberg, and W. F. Anderson. 1997. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol. 71:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keshet, E., and H. M. Temin. 1979. Cell killing by spleen necrosis virus is correlated with a transient accumulation of spleen necrosis virus DNA. J. Virol. 31:376-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klevjer-Anderson, P., W. P. Cheevers, and T. B. Crawford. 1979. Characterization of the infection of equine fibroblasts by equine infectious anemia virus. Arch. Virol. 60:279-289. [DOI] [PubMed] [Google Scholar]
- 18.Lavillette, D., M. Maurice, C. Roche, S. J. Russell, M. Sitbon, and F. L. Cosset. 1998. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J. Virol. 72:9955-9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leroux, C., C. J. Issel, and R. C. Montelaro. 1997. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J. Virol. 71:9627-9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, F., C. Leroux, J. K. Craigo, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2000. The S2 gene of equine infectious anemia virus is a highly conserved determinant of viral replication and virulence properties in experimentally infected ponies. J. Virol. 74:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, F., B. A. Puffer, and R. C. Montelaro. 1998. The S2 gene of equine infectious anemia virus is dispensable for viral replication in vitro. J. Virol. 72:8344-8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the nonhomologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marin, M., C. S. Tailor, A. Nouri, S. L. Kozak, and D. Kabat. 1999. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J. Virol. 73:9362-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maury, W., J. L. Oaks, and S. Bradley. 1998. Equine endothelial cells support productive infection of equine infectious anemia virus. J. Virol. 72:9291-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maury, W. J., S. Carpenter, K. Graves, and B. Chesebro. 1994. Cellular and viral specificity of equine infectious anemia virus. Tat transactivation. Virology 200:632-642. [DOI] [PubMed] [Google Scholar]
- 26.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, Jr., F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 66:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullins, J. I., J. W. Casey, M. O. Nicolson, and N. Davidson. 1980. Sequence organization of feline leukemia virus DNA in infected cells. Nucleic Acids Res. 8:3287-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson-Rees, W. A., R. B. Owens, P. Arnstein, and A. J. Kniazeff. 1976. Source, alterations, characteristics and use of a new dog cell line (Cf2Th). In Vitro 12:665-669. [DOI] [PubMed] [Google Scholar]
- 29.Oaks, J. L., T. C. McGuire, C. Ulibarri, and T. B. Crawford. 1998. Equine infectious anemia virus is found in tissue macrophages during subclinical infection. J. Virol. 72:7263-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payne, S. L., X. M. Qi, H. Shao, A. Dwyer, and F. J. Fuller. 1998. Disease induction by virus derived from molecular clones of equine infectious anemia virus. J. Virol. 72:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Payne, S. L., J. Rausch, K. Rushlow, R. C. Montelaro, C. Issel, M. Flaherty, S. Perry, D. Sellon, and F. Fuller. 1994. Characterization of infectious molecular clones of equine infectious anaemia virus. J. Gen. Virol. 75:425-429. [DOI] [PubMed] [Google Scholar]
- 32.Perry, S. T., M. T. Flaherty, M. J. Kelley, D. L. Clabough, S. R. Tronick, L. Coggins, L. Whetter, C. R. Lengel, and F. Fuller. 1992. The surface envelope protein gene region of equine infectious anemia virus is not an important determinant of tropism in vitro. J. Virol. 66:4085-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasty, S., B. R. Dhruva, R. L. Schiltz, D. S. Shih, C. J. Issel, and R. C. Montelaro. 1990. Proviral DNA integration and transcriptional patterns of equine infectious anemia virus during persistent and cytopathic infections. J. Virol. 64:86-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinhart, T. A., A. K. Ghosh, E. A. Hoover, and J. I. Mullins. 1993. Distinct superinfection interference properties yet similar receptor utilization by cytopathic and noncytopathic feline leukemia viruses. J. Virol. 67:5153-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice, N. R., L. E. Henderson, R. C. Sowder, T. D. Copeland, S. Oroszlan, and J. F. Edwards. 1990. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J. Virol. 64:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice, N. R., A. S. Lequarre, J. W. Casey, S. Lahn, R. M. Stephens, and J. Edwards. 1989. Viral DNA in horses infected with equine infectious anemia virus. J. Virol. 63:5194-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritter, G. D., Jr., M. J. Mulligan, S. L. Lydy, and R. W. Compans. 1993. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology 197:255-264. [DOI] [PubMed] [Google Scholar]
- 38.Sauter, M. M., A. Pelchen-Matthews, R. Bron, M. Marsh, C. C. LaBranche, P. J. Vance, J. Romano, B. S. Haggarty, T. K. Hart, W. M. Lee, and J. A. Hoxie. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J. Cell Biol. 132:795-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sellon, D. C., S. T. Perry, L. Coggins, and F. J. Fuller. 1992. Wild-type equine infectious anemia virus replicates in vivo predominantly in tissue macrophages, not in peripheral blood monocytes. J. Virol. 66:5906-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spies, C. P., and R. W. Compans. 1994. Effects of cytoplasmic domain length on cell surface expression and syncytium-forming capacity of the simian immunodeficiency virus envelope glycoprotein. Virology 203:8-19. [DOI] [PubMed] [Google Scholar]
- 41.Steagall, W. K., M. D. Robek, S. T. Perry, F. J. Fuller, and S. L. Payne. 1995. Incorporation of uracil into viral DNA correlates with reduced replication of EIAV in macrophages. Virology 210:302-313. [DOI] [PubMed] [Google Scholar]
- 42.Tada, H., O. Shiho, K. Kuroshima, M. Koyama, and K. Tsukamoto. 1986. An improved colorimetric assay for interleukin 2. J. Immunol. Methods 93:157-165. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, S., T. Abe, J. Gotoh, and Y. Fukuuchi. 2002. Substrate-dependence of reduction of MTT: a tetrazolium dye differs in cultured astroglia and neurons. Neurochem. Int. 40:441-448. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, G. M., and D. A. Sanders. 1999. The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol. Biol. Cell 10:2803-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Temin, H. M. 1988. Mechanisms of cell killing/cytopathic effects by nonhuman retroviruses. Rev. Infect. Dis. 10:399-405. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, A., K. D. Gray, and M. J. Roth. 1997. Analysis of mutations within the cytoplasmic domain of the Moloney murine leukemia virus transmembrane protein. Virology 227:305-313. [DOI] [PubMed] [Google Scholar]
- 47.Weimin Wu, B., P. M. Cannon, E. M. Gordon, F. L. Hall, and W. F. Anderson. 1998. Characterization of the proline-rich region of murine leukemia virus envelope protein. J. Virol. 72:5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weller, S. K., A. E. Joy, and H. M. Temin. 1980. Correlation between cell killing and massive second-round superinfection by members of some subgroups of avian leukosis virus. J. Virol. 33:494-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Honing, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adapter. J. Virol. 75:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, C., C. P. Spies, and R. W. Compans. 1995. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl. Acad. Sci. USA 92:9871-9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao, Q., and R. W. Compans. 1995. Differences in the role of the cytoplasmic domain of human parainfluenza virus fusion proteins. J. Virol. 69:7045-7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshimura, F. K., T. Wang, and S. Nanua. 2001. Mink cell focus-forming murine leukemia virus killing of mink cells involves apoptosis and superinfection. J. Virol. 75:6007-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, N. L., P. M. Cannon, D. Chen, and W. F. Anderson. 1998. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J. Virol. 72:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]