Abstract
We developed a new molecular method of typing Streptococcus suis based on polymerase chain reaction (PCR) amplification of a large fragment of rRNA genes, including a part of the 16S and 23S genes and the 16S–23S intergenic spacer region (ISR), followed by restriction fragment length polymorphism (RFLP) analysis with RsaI or MboII endonuclease. The 16S–23S ISRs of 5 S. suis isolates were sequenced and compared. Size and sequence polymorphisms were observed between the S735 reference strain and the 4 wild-type strains. The genetic relationships between 138 independent S. suis strains belonging to various serotypes, isolated from swine or human cases, were determined. The discriminatory power of the method was > 0.95, the threshold value for interpreting typing results with confidence (0.954 with RsaI and 0.984 with RsaI plus MboII). The in vitro reproducibility was 100%. The strains isolated from humans were less genetically diverse than the strains isolated from pigs. For the first time, 2 molecular patterns (R6, M9) were significantly associated with S. suis serotype 2 strains. This genetic tool could be valuable in distinguishing individual isolates of S. suis during epidemiologic investigations.
Résumé
Une nouvelle méthode de typage moléculaire de Streptococcus suis a été développée. Elle est basée sur l’amplification génique des régions intergéniques présentes entre les gènes codant pour l’ARNr 16S et 23S de S. suis (ISR) et sur une microrestriction du produit amplifié à l’aide des enzymes de restriction RsaI et MboII. Les régions intergéniques présentes entre les gènes codant pour l’ARN 16S et 23S de cinq isolats ont été séquencés et comparés. Des polymorphismes de taille et de séquence ont été observés entre la souche de référence S735 et les quatre souches sauvages. Les relations phylogénétiques entre 138 souches de S. suis ont été étudiées. Elles n’étaient pas reliées entre elles, étaient isolées chez l’homme ou chez le porc et appartenaient à différents sérotypes. La technique s’est révélée discriminante (indice de discrimination supérieur à 0,95 : valeur limite pour interpréter les résultats de typage avec confiance) et reproductible (D = 0,954 avec RsaI et D = 0,984 avec RsaI et MboII). Les profils génétiques obtenus à partir des souches humaines sont apparus plus homogènes que ceux des souches isolées chez le porc. Pour la première fois, deux profils (R6, M9) ont été significativement associés aux souches appartenant au sérotype 2. Cette méthode pourrait donc être fort utile pour identifier S. suis, lors de suivis épidémiologiques de l’infection en élevage.
(Traduit par les auteurs)
Introduction
Streptococcus suis is a worldwide cause of a variety of porcine infections. One of the most important agents of swine meningitis, it also causes meningoencephalitis, septicemia, arthritis, endocarditis, pericarditis, polyserositis, and sudden death of weaning piglets as well as growing pigs (1). This pathogen is also responsible for meningitis, septicemia, arthritis, and endocarditis in humans, usually people who have worked with raw pork or had contact with pigs (2,3). More recently, several cases of human S. suis infection acquired from wild boars have been reported (4–6).
Streptococcus suis can be classified into types according to capsular polysaccharide antigens. To date, 35 serotypes have been described: 1, 2, 1/2, and 3 to 34. Serotype 2 has always been considered the most virulent and is the most prevalent type isolated from diseased pigs in most countries where the swine industry is important (7–9). However, S. suis serotype 2 strains have been isolated from healthy pigs, and S. suis strains of other serotypes can also cause disease in pigs (1,10). Moreover, the mere presence of the putative virulence factors (such as muramidase-released protein, extracellular protein factor, and hemolysin) does not necessarily define a strain as virulent (8,11).
Several molecular typing schemes have been developed to determine the relatedness of strains of S. suis associated with infection, including multilocus enzyme electrophoresis (12), restriction endonuclease analysis with HaeIII (13), ribotyping (14,15), repetitive extragenic palindromic (REP) or enterobacterial repetitive intergenic consensus (ERIC) polymerase chain reaction (PCR) (16,17), arbitrarily primed PCR (18,19), and pulsed-field gel electrophoresis (PFGE) (20–22). Among these methods, PFGE has been shown to be highly discriminatory and reproducible (21). However, it is laborious and time-consuming. The recently introduced multilocus sequence typing scheme (23), a nucleotide-sequence-based method with a high discriminatory power and a high degree of reproducibility, is difficult to apply for routine diagnostic testing. Characterization of the 16S–23S ribosomal (r) DNA intergenic spacer region (ISR) has also been used to compare bacterial strains (24–27). Schlegel and colleagues (25) reported that ISR analysis is a powerful method for identifying species within the genus Streptococcus, including S. suis. However, this method, based on PCR amplification and restriction of a fragment of rRNA genes, including the 16S and 23S genes, and the ISR, was not used to differentiate S. suis strains. On the other hand, Hassan and associates (27) showed size and sequence variations of the ISR but with a limited number of S. suis strains.
The objective of our study was to combine these 2 last techniques to develop a new method of differentiating S. suis strains in an easily and rapidly reproducible and discriminatory manner. This new molecular typing method would be based on PCR amplification of a larger fragment of rRNA genes, including a part of the 16S and 23S genes, and the 16S–23S rDNA ISR, followed by restriction fragment length polymorphism (RFLP) analysis with RsaI and MboII endonucleases. We then analyzed the genetic relationships between 138 S. suis strains of various serotypes, isolated from swine or human cases.
Materials and methods
Bacterial strains
We studied 138 epidemiologically unrelated strains of S. suis: 86 strains isolated from pigs with meningitis, septicemia, or arthritis; 22 strains isolated from clinically healthy pigs, the specimens being from the nasal cavity or palatine tonsil; 29 strains isolated from humans; and reference strain S735 serotype 2, isolated from a diseased pig by one of us (M.G.). Capsular typing revealed serotype 1 (1 strain), serotype 2 (98 strains, 29 from humans and 69 from swine, 1 of the 69 being the reference strain), serotype 1/2 (7 strains), serotype 3 (6 strains), serotype 4 (1 strain), serotype 5 (1 strain), serotype 7 (12 strains), and serotype 9 (10 strains), as previously reported (28); 2 strains were autoagglutinable and thus not typable. The specificity of the ISR-RFLP method was evaluated with 31 strains belonging to 25 other bacterial species related to S. suis or isolated from pigs (Table I).
Table I.
Bacterial strains used to test the specificity of a new method of differentiating Streptococcus suis
| Species | Strains | n |
|---|---|---|
| Streptococcus agalactiae | ATCC 13813 | 1 |
| S. acidominimus | NCDO 2025 | 1 |
| S. alactolyticus | ATCC 43077 | 1 |
| S. anginosus | ATCC 33397 | 1 |
| S. bovis | ATCC 33317 | 1 |
| S. constellatus | ATCC 27823 | 1 |
| S. difficilis | ATCC 51487 | 1 |
| S. gordonii | ATCC 10558 | 1 |
| S. hyointestinalis | CCUG 27888 | 1 |
| S. intestinalis | ATCC 43492 | 1 |
| S. pneumoniae | ATCC 33400 | 1 |
| S. porcinus | ATCC 43138 and field strain | 2 |
| S. pyogenes | ATCC 12344 | 1 |
| Escherichia coli | Field strains | 2 |
| Campylobacter jejuni | Field strain | 1 |
| Campylobacter coli | Field strain | 1 |
| Mycoplasma hyopneumoniae | ATCC 25934 and field strain | 2 |
| Mycoplasma hyosynoviae | ATCC 25591 and field strain | 2 |
| Mycoplasma hyorhinis | ATCC 17981 and field strain | 2 |
| Mycoplasma flocculare | ATCC 27399 | 1 |
| Actinobacillus pleuropneumoniaea | ATCC 27088 and field strain | 2 |
| Actinobacillus lignieresii | ATCC 49236 | 1 |
| Actinobacillus rossi | ATCC 27072 | 1 |
| Pasteurella multocida | Field strain | 1 |
| Staphylococcus aureus | Field strain | 1 |
ATCC — American Type Culture Collection, Rockville, Maryland, USA; NCDO — National Collection of Dairy Organisms, Shinfield, Reading, England; CCUG — Culture Collection, University of Göteborg, Göteborg, Sweden
Serotype 1
Sixteen S. suis strains isolated from pigs at 5 farms were used to evaluate the reproducibility and stability of the ISR-RFLP patterns after 1 to 32 passages on artificial media.
New molecular typing method
Genomic DNA from each strain was prepared as described by Kellog and Kwok (29). Briefly, 20 colonies were suspended in a mixture of 250 μL of 10 mM Tris HCl (pH 8.3), 100 mM KCl, 2.5 mM MgCl2, 1% (v/v) Tween 20 (Sigma-Aldrich Chimie, Saint Quentin Fallavier, France), 1% (v/v) Triton X-100 (Sigma-Aldrich Chimie), 0.01% (v/v) Nonidet P-40 (Sigma-Aldrich Chimie), and 120 μg/mL proteinase K (Sigma-Aldrich Chimie). The suspensions were incubated for 1 h at 60°C and then for 10 min at 95°C to inactivate the proteinase K; after cooling to room temperature, they were kept at −20°C. For each S. suis strain used to verify the stability of patterns, 2 independent extractions of DNA were performed.
The template DNA was then amplified with the forward primer 16S-489(f) (5′-TTCTCACTTGACGGTATCTTAC-3′), complementary to the 3′ end of the 16S rRNA gene (AF009477) (30), and the reverse primer 23S-206(r) (5′-GGTACCTTAGATGTTTCAGTTC-3′), complementary to the 5′ end of the 23S rRNA gene (31). The length of the PCR product was approximately 1714 base pairs (bp). The PCR mixture contained PCR buffer (67 mM Tris-HCl, 16 mM [NH4]2SO4, 0.01% Tween 20, and 2.5 mM MgCl2 [pH 8.8]), 1.5 mM of deoxyribonucleoside triphosphate (Eurobio, Les Ulis, France), 400 nM of each primer, 2 units of Taq DNA polymerase (Eurobio), and 5 μL of the DNA template in a total volume of 50 μL. The DNA template was replaced by double-distilled water for the negative control of the PCR step. Amplification was performed in a GeneAmp PCR system 9600 (Applied Biosystems, Courtaboeuf, France). The reaction procedure consisted of 40 cycles of amplification at 94°C for 30 s, 60°C for 30 s, and 72°C for 5 min, and then final extension at 72°C for 5 min.
Sequencing of the ISR
The 16S–23S rDNA ISR of 5 strains (field strains 298, 332, 347, 353, and reference strain S735) was sequenced. Strains 298 and 332 (serotype 2) had been isolated from cases of septicemia; strain 347 (serotype 2) and strain 353 (serotype 1/2) had been isolated from palatine tonsils of clinically healthy pigs. Characterization by PFGE with the use of SmaI endonuclease (21) showed that strains 298, 332, and 353 had the same pattern, whereas strain 347 and reference strain S735 had different patterns.
Nucleotide sequences were determined by cycle sequencing based on the dideoxynucleoside termination chain method described by Sanger (32). The PCR products were purified by means of the QIAquick PCR Purification Kit (Qiagen, Courtaboeuf, France), quantified spectrophotometrically, and sequenced by means of the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit and 373A sequencer (Applied Biosystems), according to the manufacturers’ instructions. Two internal primers were designed to sequence totally the 1.7-kb ISR product: Rib-s2 (5′-TTTCTCTTCGGAGCATCGGTGA-3′) and Rib-as2 (5′-GACGTACAGGTTTCCATTTCC-3′).
Analysis of RFLP
To characterize the genetic variations in the ISR of the 138 S. suis strains, the ISR products were digested singly with RsaI, MboII, AvaII, Bsp1286I, AccII, BcnI, and Eco0109I (Amersham, Les Ulis, France; Roche Diagnostics, Meylan, France). The reactions were performed with 15 μL of ISR products as described by the manufacturers.
The ISR products digested with MboII, AvaII, Bsp1286I, AccII, BcnI, or Eco0109I were separated in a 2% standard agarose gel in TBE buffer (90 mM Tris, 90 mM borate, 2.5 mM EDTA [pH 8]) for 2 h at a constant voltage of 125 V. The ISR products digested with RsaI were separated in a 2.5% low-melting-point agarose gel in TBE buffer for 2.5 h at a constant voltage of 125 V. The patterns were detected by ultraviolet transillumination after ethidium bromide staining. A 50-bp “ladder” was used as a molecular size standard (Pfizer, Paris, France).
The patterns were digitized and analyzed with use of the Biogene package (Vilber-Lourmat, Marne la Vallée, France) as previously described (21). The unweighted pair group method with arithmetic mean was used with a confidence interval of 8%. The numerical index of discrimination described by Hunter and Gaston (33), calculated with RsaI or MboII alone or in combination, was used to rate the discriminatory power of the ISR-RFLP method.
The relationships between the patterns of strains isolated from diseased subjects versus clinically healthy pigs, including serotype and origin (pig or human), were analyzed with the Fisher exact test (n ≤ 5) or the chi-squared test (n > 5) of independence in 2 × 2 tables by means of the Systat 9.0 program for Windows. Differences were considered statistically significant when the P-value was less than 0.05.
Results
Specificity of the ISR-RFLP method
When the microorganisms listed in Table I were used as DNA templates, no amplification product was observed (data not shown).
Characterization of the 16S–23S ISR of 5 S. suis strains
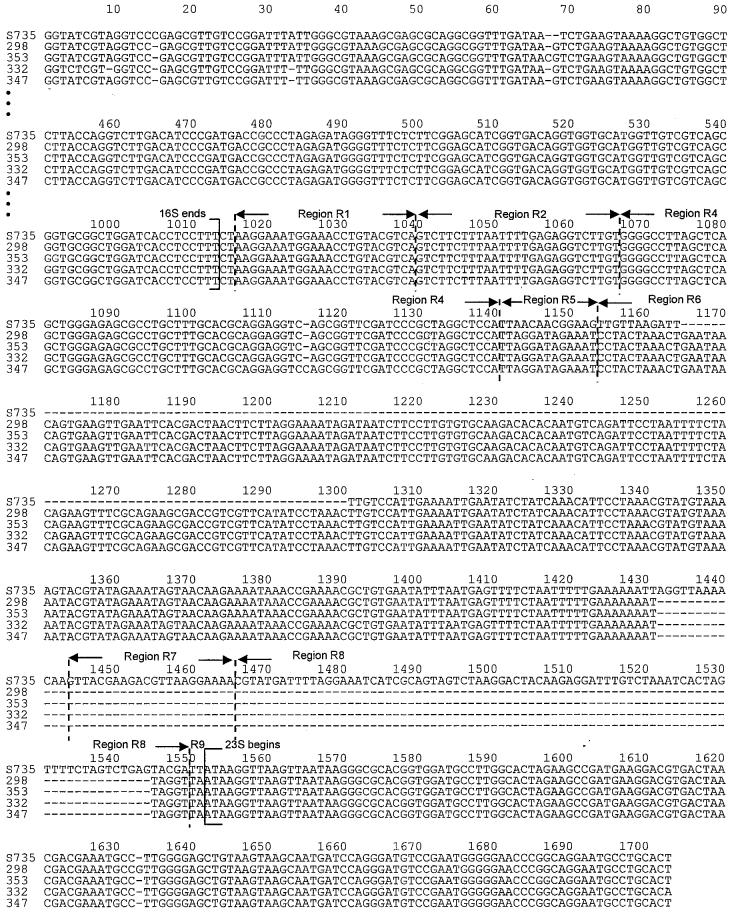
Sequences spanning the 3′ end of the 16S gene, the 16S–23S rISR, and the 5′ end of the 23S gene of the 5 S. suis strains studied were deposited in GenBank under the accession numbers AY585196, AY585197, AY585198, AY585199, and AY585200. The sequencing allowed us to determine the exact size of each ISR: for strains 298, 332, and 353, 424 bp; for strain 347, 425 bp; and for strain S735, 401 bp. The multiple-alignment result of the 5 nucleotide sequences is shown in Figure 1. The 5 sequences were 84% identical; the sequences of strains 298, 332, 347, and 353 were 98% identical.
Figure 1.
Multiple-sequence alignment of 5 strains of Streptococcus suis. The nucleotide sequence numbers are given from a consensus alignment. Dashes indicates spacers between adjacent nucleotides introduced for maximum alignment. Arrows indicate the 3′ end of the 16S ribosomal (r) DNA gene (position 1013), the 5′ end of the 23S rDNA gene (1552), and the 8 regions of the 16S–23S intergenic spacer region (ISR) (1014 to 1551): R1 (1016 to 1039), R2 (1040 to 1066), R4 (1067 to 1140; identical to the transfer RNAAla gene), R5 (1141 to 1153), R6 (1154 to 1443), R7 (1444 to 1465), R8 (1466 to 1549), and R9 (1550 to 1551), as defined by Chanter and coworkers (24).
The ISR had distinct subregions with an ordered relationship. As previously described (24), these regions were defined after multiple alignment with the ISR sequence of S. equi (AF489598). The number of regions varied from 7 to 8. Region R1 (positions 1016 to 1039 of the multiple alignment), following the end of the 16S rRNA gene, and region 2 (positions 1040 to 1066) were present and identical in all 5 strains. Region R3 was not found among the 5 isolates. Region R4 (positions 1067 to 1140) was present in all 5 strains and was 100% identical to the sequence for the transfer (t) RNA gene for alanine found in several other streptococcal ISRs and 97% identical to the DNA sequence of Staphylococcus aureus coding for tRNAAla (NC_002758). The size of R4 was 73 bp for strains S735, 298, 332, and 353 and 74 bp for strain 347. The 13-bp region R5 (positions 1141 to 1153) was present in all 5 strains, was 54% homologous for all the strains, and was identical in strains 298, 332, 347, and 353. Region R6 (positions 1154 to 1443) was the longest (154 to 278 bp) and was identical in all the strains except S735, which showed a deletion in positions 1165 to 1300 and an insertion in positions 1432 to 1443. The 22-bp region R7 (positions 1444 to 1465) was present only in strain S735. Region R8 (positions 1466 to 1549) was present in all the strains but was only 5 bp in strains 298, 332, 347, and 353, whereas it was 84 bp in strain S735. Region R9 (positions 1550 to 1551) was present and identical in all 5 strains.
The ISR sequences of the 5 strains did not share significant homology with the sequences of any proteins in GenBank.
Choice of endonucleases for RFLP analysis
The positions of the RsaI, MboII, AvaII, Bsp1286I, AccII, BcnI, and Eco0109I restriction sites were determined in the 16S–23S ISR of the same 5 strains (Table II). Only endonucleases with 4 or 5 restriction sites in the ISR (RsaI, AccII, BcnI, Eco0109I, and MboII) were selected for the RFLP analysis. To further select endonucleases that would give a good distribution of restriction length fragments, the ISR-RFLP method was initially performed on 10 epidemiologically unrelated S. suis strains. All patterns obtained with AccII, BcnI, and Eco0109I digestion were composed of 1 to 4 fragments, whereas RsaI and MboII digestion resulted in fragments of a large range of sizes and was therefore chosen for typing the 138 S. suis strains.
Table II.
Number of restriction sites of the endonucleases considered for restriction fragment length polymorphism (RFLP) analysis observed in the 16S–23S intergenic spacer region (ISR) of 5 strains of S. suis
| Strain; number of sites
|
||||||
|---|---|---|---|---|---|---|
| Endonuclease | Recognition sequence 5′–3′ | S735 | 298 | 332 | 347 | 353 |
| RsaI | GT↓AC | 8 | 5 | 5 | 5 | 5 |
| MboII | GAAGA(N)8↓ | 4 | 4 | 4 | 4 | 4 |
| AvaII | G↓G(A,T)CC | 2 | 2 | 2 | 3 | 2 |
| Bsp1286I | G(A,G,T)GC(A,C,T)↓ C | 3 | 2 | 2 | 2 | 2 |
| AccII | CG↓CG | 4 | 4 | 3 | 4 | 4 |
| BcnI | CC(G,C)↓GG | 4 | 4 | 4 | 4 | 4 |
| Eco0109I | (A,G)G↓GNCC(C,T) | 4 | 3 | 3 | 3 | 3 |
Stability of ISR-RFLP patterns
Under the conditions used, the in vitro stability of the ISR-RFLP patterns, estimated with strains isolated from different farms after 1 to 32 passages on artificial media, was 100% (results not shown). Moreover, this molecular typing method was reproducible: a similar pattern was shown for each strain after the 2 independent DNA extractions.
Genetic diversity of S. suis as defined by ISR-RFLP analysis
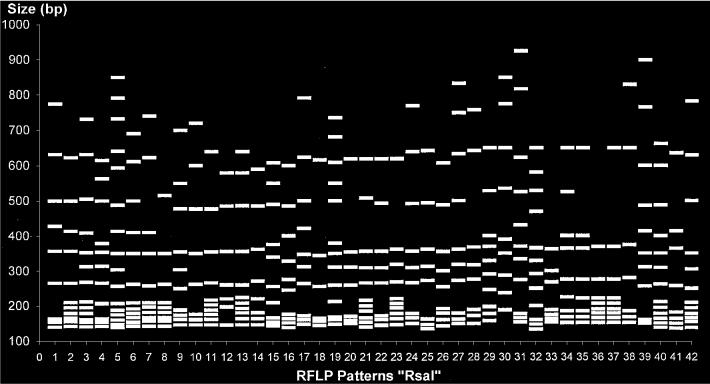
The RsaI ISR-RFLP patterns were composed of 6 to 21 fragments of 134 to 925 bp; 42 patterns were identified among the epidemiologically unrelated 138 S. suis strains analyzed. Figure 2 schematically represents all the patterns, and Figure 3 presents the deduced relationships among the strains. The 138 strains diverged by up to 33% (67% homology). At 72% homology, 5 groups (A to E) were identified. At 75% homology, group A was divided into subgroups a and b. Some fragments were present in multiple patterns; there were 3 potential markers: a 700-bp fragment, a 500-bp fragment, and 6 fragments (sextuplet) of 140, 155, 165, 175, 195, and 224 bp (± 8%) (Figure 2).
Figure 2.
Schematic representation of the 42 patterns obtained by restriction fragment length polymorphism (RFLP) after restriction of the 16S–23S ISR of 138 S. suis strains with the endonuclease RsaI. bp — base pairs.
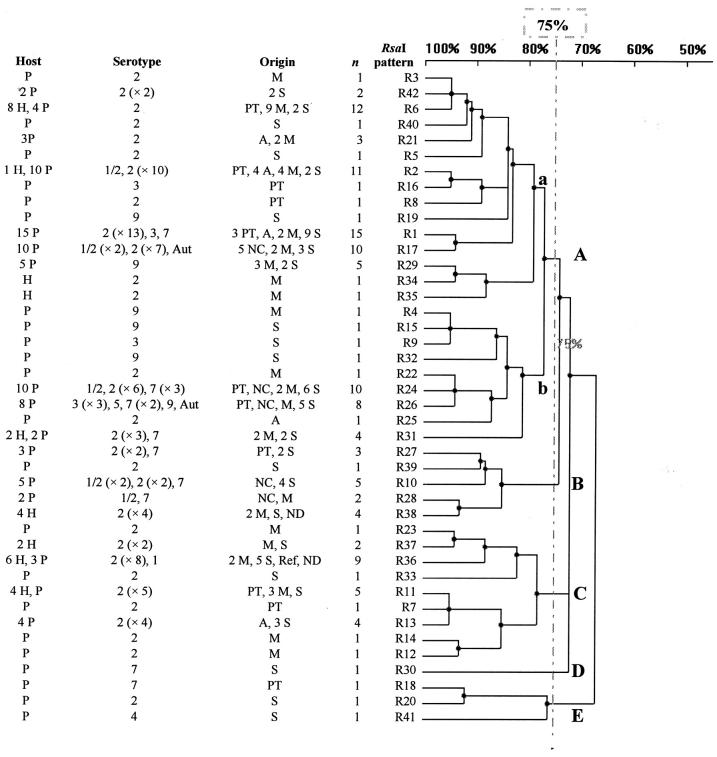
Figure 3.
Genetic relationships among the 138 S. suis strains, estimated by clustering analysis of RFLP patterns obtained after restriction of the 16S–23S ISR with RsaI. The human or swine origin, serotype, and tissue origin of the strains, number of strains for each pattern, and level of homology (%) among the strains are reported in the dendrogram. P — pig; H — human; Aut — autoagglutinable; M — meningitis; S — septicemia; PT — palatine tonsil; A — arthritis; NC — nasal cavity; ND — not done; Ref — reference strain.
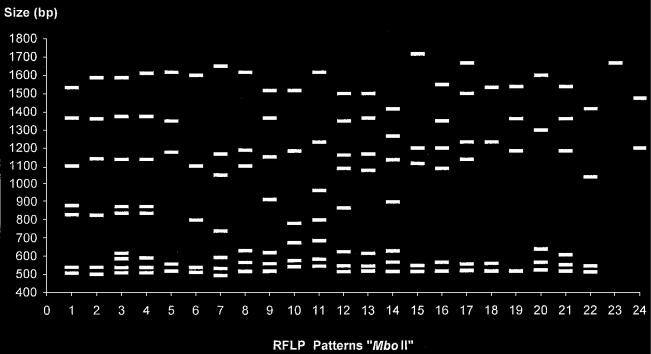
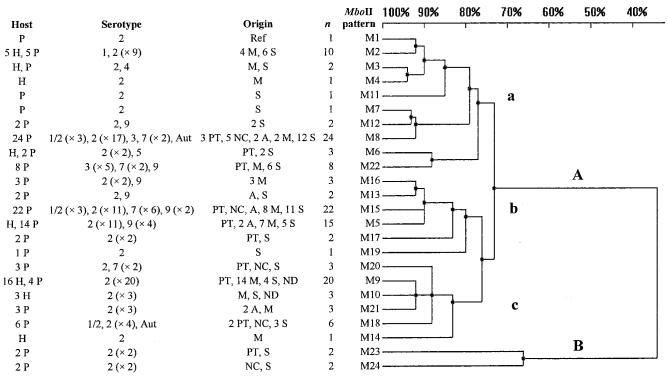
The MboII ISR-RFLP patterns were composed of 1 to 9 fragments of 493 to 1717 bp; 24 patterns were identified among the epidemiologically unrelated 138 S. suis strains analyzed. Figure 4 schematically represents all the patterns, and Figure 5 presents the deduced relationships among the strains. The 138 strains diverged by up to 66% (34% homology). At 34% homology, 2 groups (A and B) were identified. At 72% homology, group A was divided into subgroups a, b, and c.
Figure 4.
Schematic representation of the 24 RFLP patterns obtained after restriction of the 16S–23S ISR of the 138 S. suis strains with the endonuclease MboII.
Figure 5.
Genetic relationships among the 138 S. suis strains, estimated by clustering analysis of RFLP patterns obtained after restriction of the 16S–23S ISR with MboII.
Table III presents the ISR-RFLP characterization of the 138 strains.
Table III.
Characterization of the 138 S. suis strains by the ISR-RFLP method with the use of RsaI and MboIIa
| Strain | Serotype | Origin | Host | RsaI pattern | MboII pattern | Strain | Serotype | Origin | Host | RsaI pattern | MboII pattern |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 2 | S | P | R2 | M23 | 144 | 2 | A | P | R21 | M5 |
| 9 | 2 | S | P | R1 | M8 | 145 | 2 | M | P | R21 | M5 |
| 12 | 2 | M | P | R3 | M9 | 147 | 2 | M | P | R21 | M5 |
| 13 | 9 | M | P | R4 | M5 | 148 | 2 | M | P | R22 | M15 |
| 18 | 2 | S | P | R1 | M18 | 149 | 2 | M | P | R23 | M5 |
| 20 | 2 | M | P | R1 | M8 | 152 | 2 | NC | P | R24 | M15 |
| 24 | 2 | S | P | R2 | M12 | 165 | 2 | M | P | R14 | M16 |
| 25 | 2 | S | P | R1 | M18 | 167 | 2 | A | P | R25 | M15 |
| 27 | 2 | S | P | R5 | M19 | 171 | 2 | S | P | R1 | M8 |
| 29 | 2 | M | P | R2 | M21 | 174 | Aut | NC | P | R26 | M18 |
| 30 | 2 | M | P | R2 | M8 | 176 | 7 | PT | P | R27 | M8 |
| 33 | 2 | M | P | R2 | M2 | 181 | 7 | NC | P | R28 | M20 |
| 36 | 2 | S | P | R1 | M8 | 191 | 9 | S | P | R29 | M5 |
| 39 | 2 | S | P | R1 | M8 | 193 | 9 | S | P | R29 | M15 |
| 40 | 2 | A | P | R2 | M21 | 194 | 9 | M | P | R29 | M15 |
| 41 | 2 | A | P | R2 | M13 | 195 | 9 | M | P | R29 | M16 |
| 42 | 2 | A | P | R2 | M21 | 196 | 7 | M | P | R24 | M15 |
| 43 | 2 | A | P | R1 | M8 | 198 | 7 | M | P | R1 | M15 |
| 44 | 2 | A | P | R2 | M8 | 199 | 7 | M | P | R24 | M15 |
| 54 | 2 | M | H | R6 | M5 | 201 | 9 | M | P | R29 | M5 |
| 56 | 1/2 | PT | P | R2 | M18 | 202 | 7 | S | P | R24 | M15 |
| 59 | 2 | PT | P | R1 | M17 | 203 | 7 | S | P | R26 | M22 |
| 62 | 2 | PT | P | R1 | M15 | 204 | 7 | S | P | R30 | M15 |
| 63 | 2 | PT | P | R7 | M23 | 205 | 7 | S | P | R31 | M15 |
| 64 | 2 | PT | P | R6 | M5 | 206 | 7 | S | P | R26 | M22 |
| 65 | 2 | PT | P | R8 | M18 | 208 | 9 | M | P | R26 | M22 |
| 69 | 3 | PT | P | R1 | M8 | 209 | 3 | S | P | R26 | M22 |
| 71 | 2 | S | P | R1 | M24 | 210 | 3 | S | P | R26 | M22 |
| 78 | 2 | S | P | R1 | M8 | 211 | 3 | S | P | R26 | M22 |
| 79 | 3 | S | P | R9 | M22 | 212 | 9 | S | P | R32 | M5 |
| 80 | 7 | NC | P | R10 | M8 | 215 | 1/2 | M | P | R28 | M15 |
| 81 | 1/2 | S | P | R10 | M15 | 261 | 2 | M | H | R6 | M9 |
| 86 | 2 | S | P | R11 | M17 | 262 | 2 | M | H | R6 | M9 |
| 89 | 2 | A | P | R13 | M5 | 263 | 2 | M | H | R6 | M9 |
| 90 | 1/2 | S | P | R10 | M15 | 264 | 2 | M | H | R6 | M9 |
| 91 | 2 | S | P | R10 | M20 | 265 | 2 | M | H | R6 | M9 |
| 93 | 2 | M | P | R12 | M5 | 266 | 2 | M | H | R6 | M9 |
| 100 | 1/2 | S | P | R17 | M8 | 267 | 2 | M | H | R6 | M9 |
| 102 | 9 | S | P | R19 | M12 | 268 | 2 | M | H | R34 | M9 |
| 103 | 2 | S | P | R20 | M15 | 269 | 2 | M | H | R11 | M9 |
| 104 | 9 | S | P | R15 | M13 | 270 | 2 | M | H | R35 | M9 |
| 105 | 2 | S | P | R1 | M8 | 271 | 2 | M | H | R11 | M9 |
| 106 | 3 | PT | P | R16 | M22 | 272 | 2 | ND | H | R36 | M9 |
| 109 | 1/2 | NC | P | R17 | M8 | 273 | 2 | S | H | R36 | M2 |
| 114 | 7 | PT | P | R18 | M20 | 274 | 2 | M | H | R11 | M2 |
| 117 | 2 | S | P | R17 | M8 | 275 | 2 | M | H | R37 | M9 |
| 118 | 2 | S | P | R17 | M8 | 276 | 2 | M | H | R36 | M9 |
| 127 | 2 | NC | P | R17 | M8 | 277 | 2 | S | H | R37 | M2 |
| 130 | 2 | NC | P | R17 | M8 | 278 | 2 | M | H | R38 | M10 |
| 131 | Aut | NC | P | R17 | M8 | 279 | 2 | ND | H | R38 | M10 |
| 132 | 2 | NC | P | R17 | M24 | 280 | 2 | S | H | R38 | M10 |
| 135 | 2 | M | P | R17 | M15 | 281 | 2 | S | H | R36 | M9 |
| 138 | 2 | M | P | R17 | M15 | 282 | 2 | PT | H | R11 | M9 |
| 298 | 2 | S | P | R24 | M8 | 365 | 5 | PT | P | R26 | M6 |
| 301 | 2 | S | P | R33 | M6 | 366 | 2 | S | P | R27 | M15 |
| 302 | 2 | S | P | R36 | M2 | 372 | 2 | S | P | R27 | M18 |
| 304 | 2 | M | H | R36 | M2 | 374 | 2 | S | P | R42 | M5 |
| 306 | 2 | S | H | R36 | M6 | 376 | 2 | S | P | R42 | M5 |
| 316 | 1 | S | P | R36 | M2 | 380 | 2 | S | P | R39 | M15 |
| 321 | 2 | M | H | R31 | M3 | 390 | 2 | S | P | R13 | M5 |
| 322 | 2 | M | H | R31 | M4 | 394 | 2 | S | P | R24 | M8 |
| 327 | 2 | S | P | R6 | M9 | 395 | 2 | S | P | R13 | M9 |
| 328 | 2 | S | P | R6 | M9 | 398 | 2 | S | P | R24 | M8 |
| 333 | 2 | M | H | R38 | M2 | 400 | 2 | S | P | R24 | M15 |
| 335 | 4 | S | P | R41 | M3 | 405 | 2 | S | P | R40 | M2 |
| 337 | 2 | S | P | R24 | M7 | 407 | 2 | S | P | R13 | M2 |
| 343 | 2 | S | P | R10 | M11 | 735 | 2 | Ref | P | R36 | M1 |
| 347 | 2 | S | P | R31 | M15 | 166’ | 2 | M | P | R6 | M16 |
| 353 | 1/2 | PT | P | R24 | M8 | 303 | 2 | M | H | R2 | M14 |
P — pig; H — human; Aut — autoagglutinable; M — meningitis; S — septicemia; PT — palatine tonsil; A — arthritis; NC — nasal cavity; ND — not done; Ref — reference strain
Distribution of ISR-RFLP patterns and groups in relation to S. suis tissue source
Among the 113 strains isolated from diseased pigs and humans, 38 RsaI patterns and 23 MboII patterns were identified. After restriction with RsaI, 78 of the 113 strains were observed to be in group A, and after restriction with MboII, 83 of the 113 were observed to be in subgroups a and b. No significant association was observed between the tissue from which the strains were isolated and RsaI or MboII group.
Among the 22 strains isolated from the nasal cavity or palatine tonsil of healthy pigs, 14 RsaI patterns and 11 MboII patterns were identified. In the 2 dendrograms, these strains were not clustered in a specific group. However, patterns R17 and M8 were significantly associated with the strains isolated from the healthy pigs (P = 0.011 and 0.028, respectively).
The 3 potential markers (700-bp fragment, 500-bp fragment, and sextuplet) observed after restriction with RsaI were not significantly associated with the tissue source of the strains.
Distribution of ISR-RFLP patterns and groups in relation to S. suis serotype
Among the 98 strains of serotype 2, 30 RsaI patterns and 23 MboII patterns were identified. The dendrogram obtained after restriction with RsaI (Figure 3) showed that 53 of the 98 strains were in subgroup a and 24 in group C; the relationships between these strains and groupings were significant (P = 0.021 and 0.002, respectively). The dendrogram obtained after restriction with MboII (Figure 5) showed that 32 of the 98 strains were in subgroup c, and the relationships between these strains and subgroup c were significant (P = 0.006). The patterns R6 and M9 were significantly associated with S. suis serotype 2 strains (P = 0.019 and 0.002, respectively).
The S. suis capsular types 1, 1/2, 3, 4, 5, 7, and 9 were clustered in subgroup b (P < 0.001) in the dendrogram obtained after restriction with RsaI. Significant associations were observed between these strains and patterns R26 (P < 0.001), R29 (P = 0.002), M15 (P = 0.018), and M22 (P < 0.001).
The sextuplet observed in the RsaI patterns was detected in 54 of the 98 S. suis serotype 2 strains and in 2 of the 38 S. suis strains of serotypes 1, 1/2, 3, 4, 5, 7, and 9 (P < 0.001). The other potential markers were not significantly associated with serotype.
Distribution of ISR-RFLP patterns and groups in relation to S. suis host species
The S. suis strains isolated from humans were genetically less diverse than those isolated from pigs. After restriction with RsaI, 9 patterns were identified among the 29 strains isolated from humans, whereas 38 patterns were identified among the 109 strains isolated from pigs. Of the 29 strains isolated from humans, 12 (41%) were in group C, whereas of the 109 strains isolated from pigs, 96 (88%) were distributed among the other groups (P < 0.001). The patterns R6, R36, R11, and R38 were significantly associated with strains isolated from humans (P < 0.001, P = 0.003, P = 0.007, and P = 0.002, respectively), because they were identified in 22 (76%) of the 29 strains. The 700-bp fragment was detected in 8 (26%) of the 29 strains isolated from humans and in 5 (4%) of the 109 strains isolated from pigs (P < 0.001). The 500-bp fragment was observed in 13 (45%) of the 29 strains isolated from humans and in 88 (81%) of the 109 strains isolated from pigs (P < 0.001). The sextuplet was not significantly associated with the host species.
After restriction with MboII, 8 patterns were identified in the 29 strains isolated from humans, whereas 21 patterns were identified in the 109 strains isolated from pigs. Of the 29 strains isolated from humans, 20 (69%) were in subgroup c (P < 0.001) and 24 (83%) had the patterns M10, M2, and M9 (P < 0.001, P = 0.034, and P < 0.001, respectively).
The new method as an epidemiologic tool
An epidemiologic tool would be valuable in investigating cross-colonization or possible epidemic invasive S. suis infections. Therefore, the discriminatory power of the ISR-RFLP method was evaluated for 3 classes of isolates: all S. suis strains; strains isolated from cases of meningitis, septicemia, or arthritis; and serotype 2 strains. Table IV shows that the index of discrimination was greater than 0.95 with RsaI alone and with RsaI and MboII in combination for the 3 populations.
Table IV.
Numerical index of discrimination of the ISR-RFLP method, calculated from data for 1 or 2 endonucleases, in typing the total population and subsets of S. suis isolates
| Endonuclease; index
|
|||
|---|---|---|---|
| Population | RsaI | MboII | RsaI + MboII |
| Total (n = 138) | 0.954 | 0.904 | 0.984 |
| Strains from disease cases (n = 113) | 0.955 | 0.900 | 0.982 |
| Serotype 2 strains (n = 98) | 0.954 | 0.897 | 0.980 |
Discussion
In our study, the ISR-RFLP method and the sequencing of 5 S. suis strains (the reference strain S735 and 4 field strains) showed a size polymorphism of the ISR: from 401 to 425 bp. Gel electrophoresis of the amplified ISR from these 5 strains revealed only 1 band, and the sequence data showed a quality compatible with the presence of a single template. This suggests that the copies of this operon in the chromosome of S. suis are homogeneous. In some bacteria, there are different versions of the operon in the same cell, so that there is more than 1 PCR product when DNA is amplified from primers in the 16S and 23S genes (34). In our study, the structure of the spacers appeared to be dominated by 3 variable regions: R6, R7, and R8. We noticed some differences between our results and those of Hassan and associates (27); for example, in the S735 strain, the ISR size (401 bp in our study versus 406 bp in the study of Hassan and associates), the size of regions R5 (13 bp versus 17 bp), R6 (154 bp versus 125 bp), R8 (84 bp versus 95 bp), and R9 (2 bp versus 45 bp), and the fact that region R7 (22 bp in our study) was not described by Hassan and associates. According to Chanter and coworkers (24), the occurrence of strains with very distinct spacer regions might be caused by DNA recombination. Moreover, the S735 strain is 40 y old and was subcultured many times. In our study, among the 4 field strains (serotype 2 or 1/2) isolated from disease cases or healthy pigs, no significant size or sequence polymorphism of the ISR was observed. The serotype 2 strain isolated from palatine tonsil of 1 healthy pig (strain 347), which had a PFGE pattern distinct from the patterns of the 3 other field strains, had an ISR of 425 bp (versus 424 bp in the other field strains) and a region R4, encoding the tRNA for alanine, of 74 bp (versus 73 bp). Therefore, molecular typing of S. suis strains on the basis of characterization of a fragment of rRNA genes, including a part of the 16S and 23S genes and the 16S–23S rDNA ISR, appears to be a potential method to characterize S. suis serotype 2.
Sequencing of the ISR cannot be used routinely with a large number of strains; thus, a PCR-RFLP analysis of the spacer 16S–23S rDNA was developed to characterize S. suis strains. This approach had previously been used to differentiate species of streptococci (25), including S. suis, but not to characterize S. suis strains. Under the conditions described in this paper, this technique was discriminatory with RsaI and with the combination of RsaI and MboII. The ISR-RFLP method is also reproducible: a similar pattern was shown for each strain after the 2 independent DNA extractions and for some strains after 1 to 32 passages on artificial media. The location of the ISR, flanked by the highly conserved 16S rRNA and 23S rRNA genes, allows a specific amplification of the spacer region of S. suis and good reproducibility.
Using the ISR-RFLP method with RsaI and MboII to analyze a collection of 138 independent S. suis strains (86 isolated from diseased pigs, 22 from healthy pigs, and 29 from humans, most of whom had meningitis, as well as a reference strain from a pig), we confirmed that strains isolated from humans are genetically more homogeneous than strains isolated from pigs (21). Most strains isolated from humans are clustered in group C (RsaI), subgroup c (MboII), and have the RsaI patterns R6, R36, R11, and R38 and the MboII patterns M10, M2, and M9. A 700-bp fragment is present more frequently in these strains than in strains isolated from pigs. As observed by PFGE (21) and in this study, some S. suis strains isolated from humans and pigs show the same pattern.
We found that the S. suis serotype 2 strains were significantly associated with the same clusters (subgroup a and group C with RsaI and subgroup c with MboII) and with 2 molecular patterns (R6 and M9). A marker of serotype 2 was described, a fragment sextuplet of 140 to 224 bp, observed after restriction with RsaI. This marker seems to be a good indicator of strain virulence, because among the 98 strains of serotype 2, 88% were isolated from cases of septicemia, meningitis, or arthritis; serotype 2 has been considered the most virulent and prevalent type isolated from diseased pigs (7–9). Two patterns were significantly associated with strains isolated from healthy pigs (R17 and M8). However, the S. suis strains corresponding to these patterns cannot be considered potentially avirulent, because these patterns were also observed in strains isolated from cases of septicemia, meningitis, or arthritis. Moreover, it would be necessary to confirm this hypothesis by analyzing a new collection of S. suis strains isolated from healthy pigs and to test them in a standardized virulence model.
In conclusion, this genetic tool could be valuable in distinguishing individual isolates of S. suis to establish the origin of the infection in a herd and to monitor the kinetics of an outbreak.
Acknowledgments
The authors thank Claire de Boisséson and Véronique Béven, of Agence Française de Sécurité Sanitaire des Aliments, Ploufragan, for skilled technical assistance.
References
- Higgins R, Gottschalk M. Streptococcal diseases. In: Straw BE, D’Allaire S, Mengeling WL, Taylor DJ, eds. Diseases of Swine. 8th ed. Ames, Iowa: Iowa State University Press, 1999:563–578.
- Matsuo H, Sakamoto S. Purulent meningitis caused by Streptococcus suis in a pig breeder. Kansenshogaku Zasshi. 2003;77:340–342. doi: 10.11150/kansenshogakuzasshi1970.77.340. [DOI] [PubMed] [Google Scholar]
- Strangmann E, Froleke H, Kohse KP. Septic shock caused by Streptococcus suis: case report and investigation of a risk group. Int J Hyg Environ Health. 2002;205:385–392. doi: 10.1078/1438-4639-00165. [DOI] [PubMed] [Google Scholar]
- Bensaid T, Bonnefoi-Kyriacou B, Dupel-Pottier C, Bellon O, Lagier E, Chardon H. Streptococcus suis meningitis after wild boar hunting. Presse Med. 2003;32:1077–1078. [PubMed] [Google Scholar]
- Pedroli S, Kobisch M, Beauchet O, Chaussinand JP, Lucht F. Streptococcus suis bacteremia. Presse Med. 2003;32:599–601. [PubMed] [Google Scholar]
- Rosenkranz M, Elsner HA, Sturenburg HJ, Weiller C, Rother J, Sobottka I. Streptococcus suis meningitis and septicemia contracted from a wild boar in Germany. J Neurol. 2003;250:869–870. doi: 10.1007/s00415-003-1103-3. [DOI] [PubMed] [Google Scholar]
- Berthelot-Hérault F, Morvan H, Kéribin AM, Gottschalk M, Kobisch M. Production of muramidase released protein (MRP), extracellular factor (EF) and haemolysin by field isolates of Streptococcus suis capsular type 2, 1/2, 9, 7 and 3 isolated from swine in France. Vet Res. 2000;31:473–479. doi: 10.1051/vetres:2000133. [DOI] [PubMed] [Google Scholar]
- Segura M, Gottschalk M. Extracellular virulence factors of streptococci associated with animal diseases. Front Biosci. 2004;9:1157–1188. doi: 10.2741/1287. [DOI] [PubMed] [Google Scholar]
- Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. 1997;21:381–407. doi: 10.1023/a:1005870317757. [DOI] [PubMed] [Google Scholar]
- Cloutier G, D’Allaire S, Martinez G, Surprenant C, Lacouture S, Gottschalk M. Epidemiology of Streptococcus suis serotype 5 infection in a pig herd with and without clinical disease. Vet Microbiol. 2003;97:135–151. doi: 10.1016/j.vetmic.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Gottschalk M, Segura M. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol. 2000;76:259–272. doi: 10.1016/s0378-1135(00)00250-9. [DOI] [PubMed] [Google Scholar]
- Mwaniki CG, Robertson ID, Trott DJ, Atyeo RF, Lee BJ, Hampson DJ. Clonal analysis and virulence of Australian isolates of Streptococcus suis type 2. Epidemiol Infect. 1994;113:321–334. doi: 10.1017/s095026880005175x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amass SF, SanMiguel P, Clark LK. Demonstration of vertical transmission of Streptococcus suis in swine by genomic finger-printing. J Clin Microbiol. 1997;35:1595–1596. doi: 10.1128/jcm.35.6.1595-1596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okwumabua O, Staats J, Chengappa MM. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping) J Clin Microbiol. 1995;33:968–972. doi: 10.1128/jcm.33.4.968-972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarradas C, Luque I, de Andres D, et al. Epidemiological relationship of human and swine Streptococcus suis isolates. J Vet Med B Infect Dis Vet Public Health. 2001;48:347–355. doi: 10.1046/j.1439-0450.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- Oliveira S, Batista L, Torremorell M, Pijoan C. Experimental colonization of piglets and gilts with systemic strains of Haemophilus parasuis and Streptococcus suis to prevent disease. Can J Vet Res. 2001;65:161–167. [PMC free article] [PubMed] [Google Scholar]
- Torremorell M, Pijoan C. Prolonged persistence of an epidemic Streptococcus suis strain in a closed pig population. Vet Rec. 1998;143:394–395. doi: 10.1136/vr.143.14.394. [DOI] [PubMed] [Google Scholar]
- Chatellier S, Gottschalk M, Higgins R, Brousseau R, Harel J. Relatedness of Streptococcus suis serotype 2 isolates from different geographic origins as evaluated by molecular fingerprinting and phenotyping. J Clin Microbiol. 1999;37:362–366. doi: 10.1128/jcm.37.2.362-366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Harel J, Lacouture S, Gottschalk M. Genetic diversity of Streptococcus suis serotypes 2 and 1/2 isolates recovered from carrier pigs in closed herds. Can J Vet Res. 2002;66:240–248. [PMC free article] [PubMed] [Google Scholar]
- Allgaier A, Goethe R, Wisselink HJ, Smith HE, Valentin-Weigand P. Relatedness of Streptococcus suis isolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J Clin Microbiol. 2001;39:445–453. doi: 10.1128/JCM.39.2.445-453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot-Hérault F, Marois C, Gottschalk M, Kobisch M. Genetic diversity of Streptococcus suis strains isolated from pigs and humans as revealed by pulsed-field gel electrophoresis. J Clin Microbiol. 2002;40:615–619. doi: 10.1128/JCM.40.2.615-619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela AI, Goyache J, Tarradas C, et al. Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J Clin Microbiol. 2003;41:2498–2502. doi: 10.1128/JCM.41.6.2498-2502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Leigh JA, Heath PJ, et al. Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulence clones and potential capsular serotype exchange. J Clin Microbiol. 2002;40:3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanter N, Collin N, Holmes N, Binns M, Mumford J. Characterization of the Lancefield group C streptococcus 16S–23S RNA gene intergenic spacer and its potential for identification and sub-specific typing. Epidemiol Infect. 1997;118:125–135. doi: 10.1017/s0950268896007285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel L, Grimont F, Grimont PA, Bouvet A. Identification of major streptococcal species by rrn-amplified ribosomal DNA restriction analysis. J Clin Microbiol. 2003;41:657–666. doi: 10.1128/JCM.41.2.657-666.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaver YJ, Nagpal ML, Rudner R, Nakamura LK, Fox KF, Fox A. Restriction fragment length polymorphism of rRNA operons for discrimination and intergenic spacer sequences for cataloging of Bacillus subtilis sub-groups. J Microbiol Methods. 2002;50:215–223. doi: 10.1016/s0167-7012(02)00036-2. [DOI] [PubMed] [Google Scholar]
- Hassan AA, Khan IU, Abdulmawjood A, Lammler C. Inter- and intraspecies variations of the 16S–23S rDNA intergenic spacer region of various streptococcal species. Syst Appl Microbiol. 2003;26:97–103. doi: 10.1078/072320203322337371. [DOI] [PubMed] [Google Scholar]
- Gottschalk M, Higgins R, Jacques M, Mittal KR, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27:2633–2636. doi: 10.1128/jcm.27.12.2633-2636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellog DE, Kwok S. Detection of human immunodeficiency virus. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR Protocols: Guide to Methods and Applications. San Diego, California: Academic Press, 1990:339–343.
- Chatellier S, Harel J, Zhang Y, et al. Phylogenetic diversity of Streptococcus suis strains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int J Syst Bacteriol. 1998;48:581–589. doi: 10.1099/00207713-48-2-581. [DOI] [PubMed] [Google Scholar]
- Kostman JR, Alden MB, Mair M, Edlind TD, LiPuma JJ, Stull TL. A universal approach to bacterial molecular epidemiology by polymerase chain reaction ribotyping. J Infect Dis. 1995;171:204–208. doi: 10.1093/infdis/171.1.204. [DOI] [PubMed] [Google Scholar]
- Sanger F. Determination of nucleotide sequences in DNA. Science. 1981;214:1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürtler V, Rao Y, Pearson SR, Bates SM, Mayall BC. DNA sequence heterogeneity in the 3 copies of the long 16S–23S rDNA spacer of Enterococcus faecalis isolates. Microbiology. 1999;145:1785–1796. doi: 10.1099/13500872-145-7-1785. [DOI] [PubMed] [Google Scholar]