Abstract
We have previously shown that Sindbis virus RNA polymerase requires an N-terminal aromatic amino acid or histidine for wild-type or pseudo-wild-type function; mutant viruses with a nonaromatic amino acid at the N terminus of the polymerase, but which are otherwise wild type, are unable to produce progeny viruses and will not form a plaque at any temperature tested. We now show that such mutant polymerases can function to produce progeny virus sufficient to form plaques at both 30 and 40°C upon addition of AU, AUA, or AUU to the 5′ terminus of the genomic RNA or upon substitution of A for U as the third nucleotide of the genome. These results are consistent with the hypothesis that (i) 3′-UA-5′ is required at the 3′ terminus of the minus-strand RNA for initiation of plus-strand genomic RNA synthesis; (ii) in the wild-type virus this sequence is present in a secondary structure that can be opened by the wild-type polymerase but not by the mutant polymerase; (iii) the addition of AU, AUA, or AUU to the 5′ end of the genomic RNA provides unpaired 3′-UA-5′ at the 3′ end of the minus strand that can be utilized by the mutant polymerase, and similarly, the effect of the U3A mutation is to destabilize the secondary structure, freeing 3′-terminal UA; and (iv) the N terminus of nsP4 may directly interact with the 3′ terminus of the minus-strand RNA for the initiation of the plus-strand genomic RNA synthesis. This hypothesis is discussed in light of our present results as well as of previous studies of alphavirus RNAs, including defective interfering RNAs.
Sindbis virus (SINV) is the type species of the genus Alphavirus in the family Togaviridae (39) and has been well characterized at the molecular level (35). The alphaviruses belong to a larger assemblage of viruses referred to as the alphavirus-like superfamily, which includes many plus-stranded plant RNA viruses in addition to the alphaviruses (1, 9, 10). Among plant virus counterparts, SINV shares a particular similarity with Tobacco mosaic virus (TMV) in genome organization and gene expression, although the structures of the two virions are unrelated. There are three conserved domains in the replicase proteins of the two viruses, namely, a methyltransferase (capping enzyme) domain, a helicase domain, and an RNA polymerase domain, the last of which is located downstream of a leaky termination codon in TMV as well as in most alphaviruses. Structural protein genes are carried in the 3′-terminal region and translated from subgenomic RNAs (35). An important difference between the RNA replicase complexes of SINV and TMV, however, is that SINV has a protease activity that produces a number of intermediate cleavage products as well as four final polypeptides, which serves to regulate the virus life cycle (12, 16, 30, 31, 33). In contrast, TMV does not have a protease activity, and two large multifunctional proteins of 126 and 183 kDa function in RNA replication (8, 11, 14, 17, 38).
The N-terminal amino acid of nsP4, the viral RNA polymerase that is formed by cleavage of a polyprotein precursor in alphaviruses, is of particular importance for the stability of nsP4 and for the activities of the RNA replicase. Cleavage to release nsP4 is absolutely required for RNA synthesis (31), and the N terminus of nsP4 is invariably Tyr in all alphaviruses sequenced (35). Since Tyr is a destabilizing amino acid in the N-end rule pathway (37), free nsP4 is rapidly degraded in lysates of rabbit reticulocytes (4) as well as in infected cells (18). Mutants of SINV that contain N-terminal Phe, Trp, or His in place of Tyr are pseudo-wild type in cultured cells, but virus with N-terminal Met is attenuated and viruses with other amino acids at the N terminus are not viable (32). Previously, we identified suppressor mutations at three different sites in the nonstructural proteins that would allow nsP4 bearing N-terminal Ala, Leu, or Arg to function at 30°C (29). One of these sites was in nsP1 (T349K), and the other two were in nsP4 itself (Q191L and E315G,V,K). Pseudorevertants containing these suppressors were temperature sensitive, however, and did not form plaques at 40°C. Since stocks of pseudorevertant viruses obtained by passing the mutant viruses contained virus that plaqued at both 30 and 40°C as well as virus that plaqued only at 30°C, it was obvious that there existed other suppressor mutations that allowed the mutant RNA polymerase to function at 40°C. We have now characterized the 5′-terminal regions of pseudorevertant viruses, which could not be analyzed by the mapping methodology previously used. We found that addition of AU, AUA, or AUU to the 5′ end of the genomic RNA or the mutation U3→A suppressed the lethality of nonaromatic amino acids at the N terminus of nsP4, allowing the formation of small plaques at both 30 and 40°C.
MATERIALS AND METHODS
Cells and virus.
Secondary chicken embryo fibroblast monolayer cells cultured in Eagle's minimum essential medium supplemented with 3% fetal bovine serum were used throughout. Stocks of revertant viruses were produced by extended growth at 30°C following transfection of mutant RNA. Clones of individual viruses were isolated directly from the 30°C stock, or after passage of rescued virus at 40°C, by the method of limiting end point dilution using 96-well plates with incubation at 30°C.
RNA sequencing.
Virus was propagated in chicken cells at 30°C and purified by sucrose gradient centrifugation. RNA was extracted from purified virus, and the 5′-terminal region was directly sequenced by the dideoxy method, using a minus-sense primer annealing to nucleotides 140 to 156. After the primer extension reaction, the template RNA-cDNA duplex was denatured by heating in formamide at 95°C for 1 min and treated with RNase A and alkali to hydrolyze the template RNA, and the cDNA product was precipitated with ethanol. The 3′ terminus of any cDNA product with a free 3′ hydroxyl group was extended with terminal deoxynucleotidyl transferase (Bethesda Research Laboratories), using a mixture of four deoxyribonucleotides (1 mM each) (3). This serves to remove the cross bands that result from termination at the 5′ ends of the templates and that obscure the last few nucleotides of the sequence.
Changes at the 5′ ends of viral RNAs.
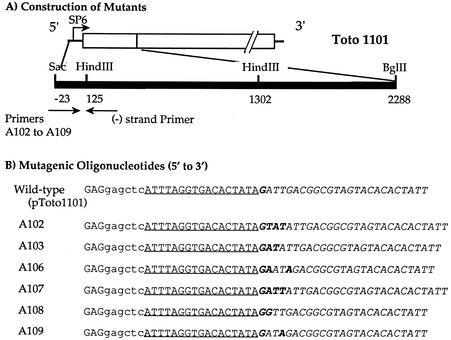
Site-directed mutations that have specific 5′-terminal nucleotide extensions in the genomic RNA, or that have U3→A, were created in the background of pToto1101 (27), a SINV full-length cDNA clone from which infectious RNA transcripts can be produced in vitro. With pToto1101 DNA as a template, the 5′-terminal region was amplified by PCR, using one of the mutagenic primers shown in Fig. 1 and a minus-sense primer annealing to a sequence downstream of the HindIII site at position 125. The PCR product was digested with SacI and HindIII, and the mutant fragment was purified by electrophoresis in a low-melting-point agarose gel. The purified fragment was cloned into an intermediate vector called pSCV12 (30), and the nucleotide sequence between the SacI and HindIII sites was determined to ensure that no unwanted changes had been introduced into the sequence. The SacI-BglII insert from pSCV12 was cloned into SacI- and BglII-digested pToto1101 or a derivative having the codon for Ala, Met, Leu, or Arg in place of the wild-type Tyr codon at the 5′ end of the nsP4 gene (32).
FIG. 1.
Construction of SINV genomes with altered 5′ residues. (A) Diagram of the 5′ region of the genome of SINV with restriction sites used for the construction of mutants indicated. The bases of the arrows indicate the locations of the two primers used for PCR, a 5′ mutagenic primer and a 3′ universal primer. The PCR products were digested with SacI and HindIII and transferred via a shuttle vector into the full-length SINV cDNA clone Toto1101. (B) Oligonucleotides (5′ to 3′) used to obtain constructs with different 5′ ends. The SacI site is shown in lowercase letters, the SP6 promoter is underlined, and the 5′-terminal sequence of SINV is shown in italics. Modified nucleotides introduced by the primers are shown in boldface.
RESULTS
5′-terminal sequencing of mutant RNAs.
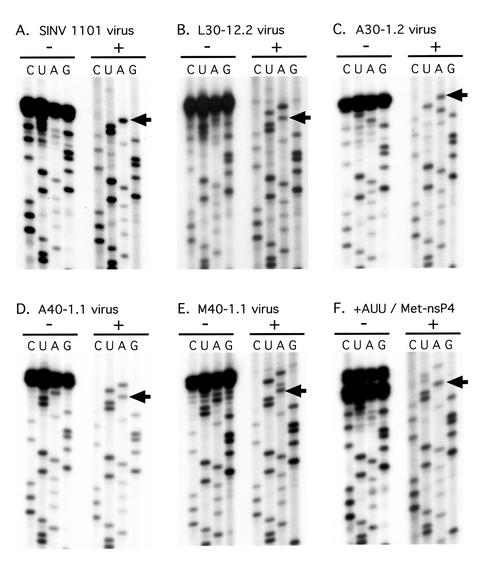
We previously described mutants that contained different amino acids at the N terminus of nsP4 (32). Stocks of pseudorevertants of such mutants have been prepared by extended incubation at 30°C of cells transfected with mutant RNA, and a number of suppressor mutations in such stocks have been mapped (29). We have now isolated cloned viruses from several such stocks by limiting dilution and determined the sequences of the 5′ termini of the genomic RNAs of 13 pseudorevertants. Representative sequencing gels are shown in Fig. 2. By using the chase procedure first described by DeBorde et al. (3), in which terminal deoxynucleotidyl transferase treatment is used to resolve the terminal nucleotides, it was possible to determine the sequence at the exact 5′ end, as illustrated in Fig. 2. The results from the 13 pseudorevertants, grouped into different categories, are summarized in Table 1. The revertants are named according to the amino acid at the N terminus of nsP4 and the method used to isolate the revertant. Thus, for example, L30-22.2 means that the N-terminal residue of nsP4 is Leu, the revertant stock was prepared by growth at 30°C, this is the 22nd independent stock of revertants obtained with this mutant, and this is the second virus clone obtained from the stock by limiting dilution. Note that the 40°C stocks were obtained by passing revertants that originally arose at 30°C at 40°C (29). For all revertants, the identity of the amino acid at the N terminus of nsP4 was determined by sequencing reverse transcription-PCR products from the region of the RNA encoding this residue.
FIG. 2.
The 5′-terminal nucleotide sequence determined from genomic RNA. RNA was purified from virions and sequenced by the dideoxy method. For each virus, two sets of sequencing reactions are shown. In the set on the left (−), the reaction products were not further treated. In the set on the right (+), the reaction products were further treated with terminal deoxynucleotidyl transferase. (A) Wild-type virus. (B) L30-12.2 virus. (C) A30-1.2 virus. (D) A40-1.1 virus. (E) M40-1.1 virus. (F) Virus rescued after transfection with transcripts containing Met-nsP4 and a GAUU extension at the 5′ terminus. The arrows indicate the A residue corresponding to the 5′-terminal A of the wild-type virus.
TABLE 1.
Revertant virus clones and their plaque phenotypes and genotypes
| Virus clonea | Plaquesb at 30/40°C | N terminus of nsP4 | 5′ terminusc |
|---|---|---|---|
| Wild type | L/L | Tyr | ·AUUGAC |
| A30-1.1 | S/none | Ala | ·------ |
| A30-1.2 | S/Min | Ala | ·--A--- |
| A30-1.3 | S/none | Ala | ·------ |
| L30-12.1 | S/none | Leu | ·------ |
| L30-12.2 | S/none | Leu | ·AU------ |
| L30-22.1 | S/Min | Leu | ·--A--- |
| L30-22.2 | S/S | Leu | ·--A--- |
| R30-13.1 | S/none | Arg | ·------ |
| R30-13.2 | S/none | Arg | ·------ |
| A40-1.1 | S/S | Ala | ·AU------ |
| A40-1.2 | S/S | Ala | ·AU------ |
| M40-1.1 | S/S | Met | ·AUA------ |
| M40-1.2 | S/S | Met | ·AU------ |
Clones are designated with the amino acid at the nsP4 N-terminus, temperature at which virus stock was grown, revertant stock number, and individual clone number.
L, large plaques; S, small plaques; Min, minute plaques.
·, a cap structure; −, same nucleotide as in the wild type. The 5′-terminal six nucleotides are shown.
The 5′-terminal sequence of the genome of the parental HR strain of SINV used in this study was confirmed to be Cap-AUUGAC (Fig. 2), as previously described (34). Five revertant viruses (A30-1.1, A30-1.3, L30-12.1, R30-13.1, and R30-13.2) were found to have the parental sequence at the 5′ end. These five revertants were previously shown to have a suppressor at nsP4-E315 (E315G, except L31-12.1, which had the suppressor E315K) (29), and all are temperature sensitive, forming plaques at 30°C but not at 40°C.
The remaining eight revertants have changes at the 5′ end of the genome. Four have an extra AU at the 5′ end (results for two of these, L30-12.2 and A40-1.1, are shown in Fig. 2), one has an extra AUA (M40-1.1, illustrated in Fig. 2), and three have the change U3→A (the result for one of these, A30-1.2, is shown in Fig. 2). These eight revertants fall into three different classes. A30-1.2, L30-22.1, L30-22.2, A40-1.1, and A40-1.2 form plaques at 40°C as well as at 30°C but appear to be attenuated in some way, as do all of the revertants studied, in that the plaques formed are small or minute in comparison to the large plaques formed by the parental virus. However, we have previously found that plaque size does not necessarily correlate with virus growth (24), and this appears to be the case here. Although small plaque size suggests that the revertants grow poorly in comparison to the wild type, in fact all of the revertants produce high yields of virus, i.e., >109 PFU/ml after 48 h at 30°C when grown in chicken cells. Other possibilities for the smaller plaque sizes are that they result because the mutant viruses spread more slowly during plaque assay due to a delay in the replication of the mutants or that cells infected by the mutants have longer survival times such that they do not score as dead cells during the plaque assay.
As shown below, the changes at the 5′ end are responsible for suppressing the otherwise lethal substitution of Ala or Leu for the N-terminal Tyr of nsP4. Notice that in the case of the Ala revertants, both the addition of AU at the 5′ end and the U3A mutation occurred and both changes give rise to virus that is able to plaque at both 30 and 40°C. However, the AU addition may be more effective in suppressing the Ala mutation because it was selected by passage at 40°C. Further, the revertant carrying this change makes larger plaques (classified as small) than does the revertant carrying the change at nucleotide 3, which makes minute plaques.
The phenotypes of the two Met revertants tested, M40-1.1 and M40-1.2, are similar to those of the Ala and Leu revertants that are able to plaque at 40°C, but there is an important distinction. Met-nsP4 is viable, and virus with this mutation forms small plaques at 30°C and minute plaques at 40°C. The pseudorevertants with the extra nucleotides at the 5′ terminus differ from the parental Met mutant in that the plaques formed at 40°C are larger, being small rather than minute. Passage of the Met mutant at 40°C must have resulted in the selection of a virus that replicates more efficiently at this temperature, since it was selected during passage, and this virus forms somewhat larger plaques at 40°C. Note that although both Met variants were obtained by cloning the same virus stock (M40-1), the two variants have different changes at the 5′ end. They may represent independently arising variants, or one variant may have given rise to the other by insertion or deletion of a single nucleotide. Similarly, revertants L30-12.1 and -12.2 differ even though they were derived from the same stock, as do revertants L30-22.1 and -22.2. Differences in sequence between the two L30-12 variants were seen, as was the case for the two M40-1 variants, but as described below, there are probably additional differences between the two L30-12 variants that are present in regions of the genome that were not sequenced. In the case of the two L30-22 variants, no differences were found, and there must be one or more differences in the unsequenced regions that result in a difference in plaque size.
The result with L30-12.2 is puzzling. The addition of AU at the 5′ end would appear to be responsible for suppressing the lethality of Leu-nsP4, given the results with the other mutants. However, this pseudorevertant does not plaque at 40°C. One possibility is that this suppressor is not effective for the Leu mutation at 40°C, even though it does suppress Ala-nsP4 at this temperature and improves the efficiency of replication of Met-nsP4 virus at 40°C. This seems unlikely, because U3→A suppresses Leu-nsP4 at 40°C and, as noted above, addition of AU appears to be more effective than the U3A mutation. A second possibility is that there is another mutation in the revertant that renders it temperature sensitive. We have found that two of the three suppressors of nonaromatic amino acids at the N terminus at nsP4 that were previously identified render the virus temperature sensitive (29). Although L30-12.2 has the wild-type Glu-315 in nsP4 (29), it is possible that another suppressor that rendered the virus temperature sensitive was first selected and the AU-containing virus arose later because of better growth at 30°C.
Reconstruction of mutants with 5′-terminal modifications.
To determine if the changes observed at the 5′ end of the RNA were responsible for suppressing the lethality of the changes in the N-terminal residue of nsP4, we constructed mutants that had changes at the 5′ end of the RNA combined with different residues at the N terminus of nsP4. This procedure eliminates any other changes that might have arisen during passage of the mutant that contributed to the rescued phenotype. The full-length clone of the SINV HR strain called pToto1101 was used as the starting point (27). RNA was transcribed in vitro from the wild-type or mutant constructs and transfected into chicken cells, and the ability of the transfected RNA to directly cause a plaque at 30 or 40°C was tested by overlaying the transfected cells with agarose. Virus was also rescued from transfected cells incubated in liquid medium at 30°C and was tested for plaque formation at the two temperatures, and the 5′ end sequence of the genome of rescued virus was determined as before. The results are summarized in Table 2.
TABLE 2.
Effect of adding or modifying nucleotides at the 5′ terminus of the genomic RNA on SINV mutants with altered amino acids at the N terminus of nsP4
| Virus | 5′ terminus of Transcript RNAa | N-terminal amino acid of nsP4 | Plaque sizeb after transfection at 30/40° | 5′ terminus of RNA from rescued virus | Plaque size after infection at 30/40°C |
|---|---|---|---|---|---|
| Wild type | ·GAUUGAC | Tyr | L/L | ·AUUGAC | L/L |
| Mutants | ·GUAU------ | Tyr | L/L | ·------ | L/L |
| ·GUAU------ | Ala | S/Min | ·AU------ | S/S | |
| ·GAU------ | Ala | S/S | ·AU------ | S/S | |
| ·GA--A--- | Ala | S/none | ·--A--- | S/Min | |
| ·GAUU------ | Met | S/S | ·AUU------ | S/S | |
| ·G--A--- | Leu | S/Min | ·--A--- | S/Min | |
| ·G--A--- | Arg | S/Min | ·--A--- | S/Min | |
| ·GG----- | Leu | None/none | |||
| ·GG----- | Arg | None/none |
·, cap structure; −, same nucleotide as in the wild type.
L, large plaques; S, small plaques; Min, minute plaques.
This parental cDNA clone has an SP6 RNA polymerase promoter positioned immediately upstream of the SINV RNA sequence, but with an extra G at the 5′ end that is required for efficient initiation by SP6 polymerase. In the wild-type transcript, this G is removed during replication of the RNA to give viral genomes that are identical to those of wild-type virus (Table 2). Similarly, when the transcript with Tyr-nsP4 contained an extra GUAU at the 5′ end, these four nucleotides were also removed to give a genome with the authentic starting sequence. However, when GUAU was added to transcripts with Ala-nsP4, only the GU was removed and the rescued virus has an extra AU at the 5′ end of its RNA (Table 2). Similarly, when GAU was added to Ala-nsP4 transcripts, only the G was removed, to give virus with an extra AU at the 5′ end of the RNA. In either case, the rescued virus formed small plaques at both 30 and 40°C (Table 2) and behaved as did the pseudorevertants rescued by passage of the Ala-nsP4 virus (Table 1). Thus, it is clear that addition of 5′-terminal AU is sufficient to suppress the lethality of the Ala substitution in nsP4.
We also tested the ability of the U3A mutation to suppress Ala-nsP4, Leu-nsP4, and Arg-nsP4. In all cases, the extra G was removed and virus that retained the mutation at the third nucleotide was rescued (Table 2). Thus, this mutation is sufficient to suppress the lethality of Ala, Leu, or Arg at the N terminus of nsP4.
Two transcripts that have a change of A1 to G were tested. No virus could be rescued, illustrating the requirement for a 5′-terminal AU (Table 2).
Finally, we also tested the effect of adding 5′-terminal GAUU to Met-nsP4 RNA. As described above, Met-nsP4 is viable, but addition of the extra nucleotides increases the efficiency of replication, especially at 40°C. The rescued virus, which was grown at 30°C, consisted of two populations, as shown in Fig. 2 by the presence of two terminal cross bands. In one population, only the G was removed to give virus containing an extra AUU sequence at the 5′ end of the RNA (Fig. 2 and Table 2). The second population had the entire GAUU removed so that it began with the parental sequence, as is obvious in Fig. 2 by the increased strength of the signal at this position. The phenotype of the rescued virus was similar to that of Met-nsP4 virus passaged at 40°C, which was found to have the extra sequence AUA (Table 1).
DISCUSSION
Requirement for AU at the start of alphavirus RNA.
Our results make it clear that AU is required as the start of alphavirus plus-strand RNA. AU is the start of all alphavirus genomic and subgenomic RNAs that have been sequenced (reviewed in reference 35). However, the third and following nucleotides are variable (see, for example, reference 25), and a change at nucleotide 3 can suppress mutations at the N terminus of nsP4 (Table 2). RNAs with extra 5′ nucleotides are always trimmed so that the viral genome begins with AU. This may be the AU of the parental virus, or an extra AU may be retained if it is required to suppress nsP4 mutations.
Studies of defective interfering (DI) RNAs of alphaviruses support the conclusion that the starting AU is important for viral RNA replication. In all cases in which the sequence of the 5′ end of DI RNA has been obtained, the starting nucleotides are AU. Where the starting sequence is that of the parental virus, this is to be expected. However, other start sequences have often been found. Pettersson (26) found that DI RNAs of Semliki Forest virus (SFV) had heterogeneous 5′ termini that began cap-(AU)nCAUG, where n = 5 to 9. Since the SFV genome begins cap-AUG, this suggests that five to nine extra AUs have been added to the 5′ end of the genome, reminiscent of our finding of one extra AU added to suppress nsP4 mutants. DI RNAs of SINV have been found to start with the parental RNA sequence, with the sequence of the subgenomic RNA, or with a sequence derived from a cellular tRNA (21, 36). The most interesting 5′ end for the present discussion is that of a sequence derived from a cellular tRNAAsp when the DI RNA arises in chicken cells. The sequence at the 5′ end of the DI RNA lacks several nucleotides at the 5′ end of tRNAAsp, and, significantly, the starting nucleotides are AU in the DI RNA. The sequenced tRNAs from chicken as well as from a number of mammals possess G in the position of the starting A in the DI RNA (19, 20, 28) (GenBank accession no. AF037471). Thus, acquisition of this 5′-terminal sequence from the cellular tRNA appears to require a change from G to A as well as deletion of about 10 nucleotides, so that the DI RNA begins with AU.
Why is the extra AU required to suppress nsP4 mutants?
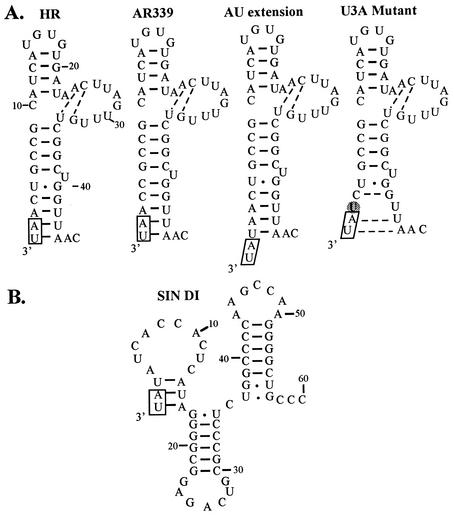
Comparative sequencing of the 5′ ends of several alphavirus RNAs led us to propose that the 5′-terminal 44 nucleotides of SINV, and the corresponding sequences of other alphaviruses, are present in a stem-loop structure (25). A complementary stem-loop structure could also form at the 3′ end of the minus-strand RNA replication intermediate produced in infected cells. Mutagenesis studies provided support for the existence of this structure in the infected cell and for its importance in the replication of RNA (6, 23). The template for plus-strand RNA synthesis is believed to be a single-stranded minus-strand RNA, and we hypothesized that one function of the stem-loop structure was to serve as a promoter at the 3′ end of the minus strand for the initiation of genomic plus-strand RNA. The 3′ end structure of minus-strand RNA is shown in Fig. 3 for two strains of SINV, the parental AR339 virus and the HR strain derived from it. Wengler et al. (40) have shown that the 3′ end of the minus strand of SFV, and presumably all alphaviruses, has a nontemplated G added to the 3′ end, which is not shown in the Fig. 3. The enzyme that adds this G could possibly add other nucleotides as well and result in the extra AU or AUA at the 5′ ends of mutant RNAs. The enzyme responsible is not known but might be the viral RNA polymerase nsP4. Poliovirus polymerase 3D and bovine viral diarrhea virus NS5B polymerase have been shown to possess terminal nucleotidyl transferase activity (22, 41), and various cellular DNA polymerases also possess such activity (2). The 3′-terminal G is not copied into genomic RNA, or if it is copied, is quickly removed, similar to our finding that extra nucleotides added to the 5′ end disappear upon replication of wild-type RNA.
FIG. 3.
Putative secondary structure at the 3′ end of the minus strand. The 3′ sequences of the viral genomes of two strains of SINV and of several mutants (A), as well as that from a DI RNA (B), are shown.
In this model, the 3′-terminal AU is present in a stem structure. To initiate RNA synthesis, we postulate that the replicase complex binds to the 44-nucleotide secondary structure and then opens the 3′-terminal stem. This stem is more robust in the AR339 sequence than in the HR sequence because of the extra GC pair, and AR339 grows to higher titer in cultured cells than does HR. However, the HR mutant A5G, which has the AR339 terminal structure, is attenuated relative to HR (23). Thus, during selection of HR, a change in the replicase complex that required the weakening of the 3′-terminal stem for efficient replication may have occurred, and this change in the replicase also attenuated HR relative to AR339. One possibility is a change in the helicase domain of nsP2, L438P, that appears to have arisen upon passage to produce the HR strain (J. Corver and J. H. Strauss, unpublished observations). In any event, the effect appears to be similar to the present findings with the nsP4 mutants.
The sequences of the 3′ structures with an extra AU and with the U3A mutation are also shown in Fig. 3. The extra AU is unpaired. The effect of the U3A mutation is to eliminate a base pair such that the 3′ stem is not stable, and the effect of this mutation is also to give rise to unpaired AU at the 3′ end. We propose that mutant replicase containing nsP4 with a nonaromatic residue at the N terminus is unable to open the parental stem structure to initiate RNA synthesis, and the unpaired AU allows the mutant replicase to initiate synthesis. In the case of Tyr-nsP4 constructs, the extra AU is quickly deleted (Table 2).
The 3′ terminus derived from chicken tRNAAsp is also shown in Fig. 3. This structure presumably serves the same function as the viral structure. The tRNA structure also terminates in AU that is base paired into a stem. As noted above, a change in the tRNA sequence from G to A appears to have been required to produce this terminal AU.
Although this model, in which an unpaired AU is required for initiation of plus-strand RNA synthesis, is consistent with the mutational data, the regulation of plus- and minus-strand RNA synthesis during alphavirus RNA replication is in actuality quite complex. We have shown previously that mutants with a nonaromatic residue as the N terminus of nsP4 are defective in minus-strand RNA synthesis as well as in genomic RNA synthesis. Because defects in the synthesis of minus-strand templates must result in reduced synthesis of plus strands, it is difficult to disentangle the relative effects of mutations on synthesis of the two RNAs. To further complicate the picture, Frolov et al. (6) have shown in an elegant series of experiments that elements in the 5′ end of SINV genomic RNA encompassing the 44-nucleotide stem-loop structure referred to above serve as promoters for both plus- and minus-strand RNA synthesis. They were able to only partially separate the elements of the plus and minus promoters. Deletion of A5 (genomic RNA sense) gave rise to a construct that could be used by the wild-type replicase to synthesize minus-strand RNA but not to synthesize plus-strand RNA, whereas other changes affected both plus- and minus-strand RNA synthesis. Given that the structures of the wild-type RNA, of the wild-type subgenomic RNA, and of a modified tRNA can all serve as promoters for RNA synthesis, as described above, the identity of the elements required to promote RNA synthesis will be difficult to establish. It is noteworthy that the results of Frolov et al. (6) show that the viral replicase must interact simultaneously with both ends of the viral RNA to initiate RNA replication. It was shown many years ago that the RNA of SINV forms H-bonded circles that are stable under physiological conditions and that the formation of these circles does not require protein (5, 13). Thus, the necessity for interactions of the replicase with both ends of the RNA for RNA replication furnishes a rationale for the cyclization of SINV RNA. For the present discussion, however, the finding that the replicase must interact with both the 5′ structure and the 3′ structure and the fact that mutants with “nonpermitted” residues at the N terminus are defective in minus-strand RNA synthesis (29) mean that it will be difficult to test directly the relative importance of the terminal AU in plus- and minus-strand synthesis.
Importance of the N-terminal Tyr of nsP4.
Our results on the mutagenesis of the N-terminal residue of nsP4 (references 29 and 32 and the present results) clearly show that N-terminal Tyr is required for efficient viral replication, consistent with the absolute requirement for cleavage to release nsP4 from the polyprotein precursor before RNA synthesis is possible (31). The polymerizing functions of the mutant nsP4 polymerases appear to remain intact, however, because, as shown here, changes in the promoter region of the RNA give rise to variants that yield in excess of 109 PFU/ml at 30°C. Thus, we hypothesize that the mutant polymerases are defective in the initiation of RNA synthesis through interaction with a promoter. If so, the N-terminal Tyr could function by direct interaction with the promoter or, perhaps more likely, could interact with other viral or cellular proteins to form a complex that interacts with the promoter. The existence of a suppressor in nsP1 suggests that one function of the Tyr is to interact with nsP1 (29).
Although replacement of the N-terminal Tyr with a nonaromatic residue is lethal in that the resulting virus will not form a plaque, the RNA is quasi-infectious as defined by Gmyl et al. (7). The viral replicase with the mutant nsP4 cannot replicate the RNA efficiently under the conditions of plaque assay because no plaques are formed. However, RNA replication must occur at some low level at 30°C, because variants arise after extended incubation that are then able to produce sufficient progeny virus at 30°C, and in some revertants at 40°C as well, to form a plaque. Such variants could not arise if no RNA replication was taking place.
The various routes by which revertants can arise are summarized in Fig. 4. If a single nucleotide substitution in the mutant codon can give rise to a permitted N-terminal residue, namely, Tyr, Phe, Trp, or His, then this is the dominant revertant found (32). Second-site pseudorevertants with suppressor mutations can also arise, and these are the only revertants found if more than one nucleotide substitution is required to change to a permitted residue. We have identified suppressors in nsP1 (T349K) and in nsP4 (Q191L and E315G,V,K) (29). We have now shown that the mutation U3A will also suppress nonpermitted residues at the N terminus of nsP4, as will addition of AU, AUA, or AUU to the 5′ end of the genomic RNA.
FIG. 4.
Four types of reversion mutations that suppress the nsP4 N-terminal mutants. At the 5′ terminus, the first three nucleotides are shown adjacent to the cap structure. The remainder of the genome is shown schematically, with translated regions indicated by the boxes. The Tyr residue at the start of nsP4 is indicated. For the mutant constructs, the modified residues at the N terminus of nsP4 are indicated by X. Four different types of changes in these mutant constructs that have been found to result in the production of infectious virus are diagrammed.
It is interesting that the N-terminal amino acid of nsP4 plays such an important role in RNA replication of SINV whereas the equivalent region of TMV RNA polymerase is present within the large multifunctional 183-kDa protein. The similarities in gene organization and in the sequences of the nonstructural proteins in these viruses show that their RNA replication machinery descended from a common ancestor, but the acquisition of a protease by the alphaviruses has allowed them to regulate their life cycle in a precise way. Proteolytic cleavages have been shown to change the ability of the alphavirus replicase to recognize the promoters for plus- and minus-strand synthesis (16, 31), thereby allowing minus-strand synthesis only early after infection and possibly resulting in the downregulation of virus replication in mosquitoes that follows infection and allows the insect to survive and subsequently spread the virus (15). This regulation may result from specific roles played by the N termini released upon protease cleavage, from conformational changes that follow cleavage, or from both. We suggest that the precise regulation of RNA synthesis evolved by alphaviruses may be important for their persistence as arboviruses in which they alternate between invertebrate and vertebrate hosts.
Acknowledgments
This work was supported by grant AI 10793 from the NIH.
REFERENCES
- 1.Ahlquist, P., E. G. Strauss, C. M. Rice, J. H. Strauss, J. Haseloff, and D. Zimmern. 1985. Sindbis virus proteins nsP1 and nsP2 contain homology to nonstructural proteins from several RNA plant viruses. J. Virol. 53:536-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark, J. M. 1988. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 16:9677-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeBorde, D. C., C. W. Naeve, M. L. Herlocher, and H. F. Maassab. 1986. Resolution of a common RNA sequencing ambiguity by terminal deoxynucleotidyl transferase. Anal. Biochem. 157:275-282. [DOI] [PubMed] [Google Scholar]
- 4.de Groot, R. J., T. Rümenapf, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1991. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. USA 88:8967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frey, T. K., D. L. Gard, and J. H. Strauss. 1979. Biophysical studies on circle formation by Sindbis virus 49S RNA. J. Mol. Biol. 132:1-18. [DOI] [PubMed] [Google Scholar]
- 6.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gmyl, A. P., E. V. Pilipenko, S. V. Maslova, G. A. Belov, and V. I. Agol. 1993. Functional and genetic plasticities of the poliovirus genome: quasi-infectious RNAs modified in the 5′-untranslated region yield a variety of pseudorevertants. J. Virol. 67:6309-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goelet, P., G. P. Lomonossoff, P. J. G. Butler, M. E. Akam, M. J. Gait, and J. Karn. 1982. Nucleotide sequence of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. USA 79:5818-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldbach, R. 1987. Genome similarities between plant and animal RNA viruses. Microbiol. Sci. 4:197-201. [PubMed] [Google Scholar]
- 10.Goldbach, R., O. Le Gall, and J. Wellink. 1991. Alpha-like viruses in plants. Semin. Virol. 2:19-25. [Google Scholar]
- 11.Goregaoker, S. P., D. J. Lewandowski, and J. N. Culver. 2001. Identification and functional analysis of an interaction between of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology 282:320-328. [DOI] [PubMed] [Google Scholar]
- 12.Hardy, W. R., and J. H. Strauss. 1989. Processing the nonstructural polyproteins of Sindbis virus: nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J. Virol. 63:4653-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, M. T., H. J. Kung, and N. Davidson. 1973. An electron microscope study of Sindbis virus RNA. Cold Spring Harbor Symp. Quant. Biol. 38:943-950. [DOI] [PubMed] [Google Scholar]
- 14.Ishikawa, M., T. Meshi, F. Motoyoshi, N. Takamatsu, and Y. Okada. 1986. In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res. 14:8291-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karpf, A. R., E. Lenches, E. G. Strauss, J. H. Strauss, and D. T. Brown. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 71:7119-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus-strand and plus-strand RNA-synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewandowski, D. J., and W. O. Dawson. 2000. Functions of the 126- and 183-kDa proteins of tobacco mosaic virus. Virology 271:90-98. [DOI] [PubMed] [Google Scholar]
- 18.Li, G., and C. M. Rice. 1989. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J. Virol. 63:1326-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Looney, J. E., and J. D. Harding. 1983. Structure and evolution of a mouse tRNA gene cluster encoding tRNAAsp, tRNAGly, and tRNAGlu and an unlinked, solitary gene encoding tRNAAsp. Nucleic Acids Res. 11:8761-8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mezquita, J., B. Lopez-Ibor, M. Pau, and C. Mezquita. 1993. Intron and intronless transcription of the chicken polyubiquitin gene UbII. FEBS Lett. 319:244-248. [DOI] [PubMed] [Google Scholar]
- 21.Monroe, S. S., and S. Schlesinger. 1983. RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5′ ends. Proc. Natl. Acad. Sci. USA 80:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neufeld, K. L., J. M. Galarza, O. C. Richards, D. F. Summers, and E. Ehrenfeld. 1994. Identification of terminal adenylyl transferase activity of the poliovirus polymerase 3Dpol. J. Virol. 68:5811-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niesters, H. G. M., and J. H. Strauss. 1990. Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA. J. Virol. 64:4162-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niesters, H. G. M., and J. H. Strauss. 1990. Mutagenesis of the conserved 51 nucleotide region of Sindbis virus. J. Virol. 64:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ou, J.-H., E. G. Strauss, and J. H. Strauss. 1983. The 5′-terminal sequences of the genomic RNAs of several alphaviruses. J. Mol. Biol. 168:1-15. [DOI] [PubMed] [Google Scholar]
- 26.Pettersson, R. F. 1981. 5′-Terminal nucleotide sequence of Semliki Forest virus 18S defective-interfering RNA is heterogeneous and different from the genomic 42S RNA. Proc. Natl. Acad. Sci. USA 78:115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibuya, K., S. Noguchi, S. Nishimura, and T. Sekiya. 1981. Mammalian tRNA genes: nucleotide sequence of rat genes for tRNAAsp, tRNAGly, and tRNAGlu. J. Biochem. 97:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirako, Y., E. G. Strauss, and J. H. Strauss. 2000. Suppressor mutations that allow Sindbis virus RNA polymerase to function with nonaromatic amino acids at the N-terminus: evidence for interaction between nsP1 and nsP4 in minus-strand RNA synthesis. Virology 276:148-160. [DOI] [PubMed] [Google Scholar]
- 30.Shirako, Y., and J. H. Strauss. 1990. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology 177:54-64. [DOI] [PubMed] [Google Scholar]
- 31.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirako, Y., and J. H. Strauss. 1998. Requirement for an aromatic amino acid or histidine at the N terminus of Sindbis virus RNA polymerase. J. Virol. 72:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss, E. G., R. J. de Groot, R. Levinson, and J. H. Strauss. 1992. Identification of the active site residues in the nsP2 proteinase of Sindbis virus. Virology 191:932-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 35.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsiang, M., S. S. Monroe, and S. Schlesinger. 1985. Studies of defective interfering RNAs of Sindbis virus with and without tRNAAsp sequences at their 5′ termini. J. Virol. 54:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varshavsky, A. 1992. The N-end rule. Cell 69:725-735. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe, T., A. Honda, A. Iwata, S. Ueda, T. Hibi, and A. Ishihama. 1999. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J. Virol. 73:2633-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weaver, S. C., L. Dalgarno, T. K. Frey, H. V. Huang, R. M. Kinney, C. M. Rice, J. T. Roehrig, R. E. Shope, and E. G. Strauss. 2000. Family Togaviridae, p. 879-889. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 40.Wengler, G., G. Wengler, and H. J. Gross. 1979. Replicative form of Semliki Forest virus RNA contains an unpaired guanosine. Nature (London) 282:754-756. [DOI] [PubMed] [Google Scholar]
- 41.Zhong, W., L. L. Gutshall, and A. M. Del Vecchio. 1998. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J. Virol. 72:9365-9369. [DOI] [PMC free article] [PubMed] [Google Scholar]