Abstract
The equine herpesvirus 1 (EHV-1) immediate-early (IE) and EICP0 proteins are potent trans-activators of EHV-1 promoters; however, in transient-transfection assays, the IE protein inhibits the trans-activation function of the EICP0 protein. Assays with IE mutant proteins revealed that its DNA-binding domain, TFIIB-binding domain, and nuclear localization signal may be important for the antagonism between the IE and EICP0 proteins. In vitro interaction assays with the purified IE and EICP0 proteins indicated that these proteins interact directly. At late times postinfection, the IE and EICP0 proteins colocalized in the nuclei of infected equine cells. Transient-transfection assays showed that the EICP0 protein trans-activated EHV-1 promoters harboring only a minimal promoter region (TATA box and cap site), suggesting that the EICP0 protein trans-activates EHV-1 promoters by interactions with general transcription factor(s). In vitro interaction assays revealed that the EICP0 protein interacted directly with the basal transcription factors TFIIB and TBP and that the EICP0 protein (amino acids [aa] 143 to 278) mediated the interaction with aa 125 to 174 of TFIIB. Our unpublished data showed that the IE protein interacts with the same domain (aa 125 to 174) of TFIIB and with TBP. Taken together, these results suggested that interaction of the EICP0 protein with the IE protein, TFIIB, and TBP may mediate the antagonism between the IE and EICP0 proteins.
The immediate-early (IE) gene of equine herpesvirus 1 (EHV-1) is essential for replication (16), lies within each of the two inverted repeats, and encodes a 1,487-amino-acid (aa) polypeptide (21). The IE protein trans-activates EHV-1 and heterologous viral promoters and trans-represses its own expression (56, 57). Residues 422 to 597 of the IE protein are sufficient for its sequence-specific DNA binding to the consensus binding sequence 5′-ATCGT-3′ that overlaps the transcription initiation site of the IE promoter and to sequences in the E and L promoters that contain a degenerate version of this cognate cis element (36). A potent transcriptional activation domain lies within the first 89 aa residues of the IE protein (58), and aa 963 to 970 are necessary for nuclear localization of truncated IE polypeptides (57). The IE protein binds to the transcription initiation site of the glycoprotein K (gK) promoter sequences, thereby repressing transcription of this true late gene (35). The EICP0 protein is able to release the γ2 L gK promoter from repression mediated by the IE protein (35). EHV-1 EICP22 (ICP22 homolog) (27), EICP27 (ICP27 homolog) (64), and EICP0 (2) are regulated as E genes, in contrast to the case for herpes simplex virus type 1 (HSV-1), in which the homologs of these three regulatory genes are members of the IE gene family (29, 42). The EICP22 protein physically interacts with the IE protein (11, 12) and increases the in vitro DNA-binding activity of the IE protein for sequences in the IE, E, and L promoters (37).
The EICP0 gene of the KyA virus encodes a protein of 419 aa that is a powerful, promiscuous trans-activator that activates all classes of EHV-1 promoters (1, 2). The EHV-1 EICP0 protein and its homologs in HSV-1 (10, 15), VZV (45, 59), BHV-1 (62), and PRV-1 (7, 61) contain a cysteine-rich zinc RING finger (C3HC4 type) near the N terminus, a motif that is found in numerous viral and cellular proteins (54) and is important for its trans-activating functions (6, 13, 45). Trans-activation assays with EICP0 mutants d8-46 and d19-30 revealed that the RING finger (aa 8 to 46) is essential for activation of the E and L (γ1 and γ2) promoters (1). The EICP0 protein is phosphorylated during infection, and deletion of the serine-rich region (aa 210 to 217), a potential site for phosphorylation, reduced by >70% the ability of the EICP0 protein to activate the γ2 L class of promoters (1). The EICP0 protein is highly antagonistic with the IE protein and thus differs from HSV-1 ICP0, which functions synergistically with ICP4 to activate expression of HSV-1 E and L promoters (6, 15).
Little is known about the mechanisms of promoter targeting during trans-activation by the EICP0 protein. Transcription of viral genes during a productive infection is mediated by the interaction between viral activator proteins and various components of the cellular transcriptional machinery (8, 17, 20, 31, 43). These interactions are important, at least in part, for facilitating transcription by increasing the assembly of a preinitiation complex (PIC) (8, 9, 20, 38, 40, 55). The PIC contains multiple components of the cellular transcriptional machinery, including RNA polymerase II (Pol II) and general transcription factors (GTFs), and can be formed on promoter templates in vitro from the individual assembly of the GTFs and Pol II (3, 25). Among the GTFs, TFIID and TFIIB function in an early stage of PIC assembly by acting as a scaffold for the assembly of the remaining transcriptional machinery. The recognition of promoters is most frequently mediated by TFIID through the binding of the TATA box-binding protein (TBP) subunit to TATA box elements and/or recognition of non-TATA box cis elements by TBP-associated factors (3, 24, 25). TFIIB interacts with TBP and Pol II and is responsible for the recruitment of Pol II to the complex (23). The complex of TBP-TFIIB-Pol II represents the minimal requirement for PIC formation but is not sufficient for activation of gene expression. Thus, processes other than factor recruitment are potentially influenced by trans-activators. TFIID and TFIIB have been implicated to be direct targets for viral trans-activators. The HSV-1 ICP4 forms a tripartite complex with TFIIB and TBP (55) and promotes PIC formation by enhancing the binding of TFIID to the TATA box element (20). Our previous results have shown that the interaction of the EHV-1 IE protein with TFIIB is necessary for its full trans-activation function and that the IE-TFIIB interaction may be part of the mechanism by which the IE protein activates transcription (30). The large T antigen of simian virus 40 (SV40) (31), the E1A protein of adenovirus (17), and the EBNA2 and ZEBRA proteins of Epstein-Barr virus (8, 43) are other examples of viral trans-activators that directly interact with TFIID and/or TFIIB.
We present here our findings that the EHV-1 EICP0 protein interacts directly with TFIIB and TBP, and this interaction may contribute to its transcriptional activation of EHV-1 genes. The EICP0 protein (aa 144 to 287) binds to the domain within TFIIB at aa 125 to 174. The IE protein also binds to the same domain within TFIIB (R. A. Albrecht, H. K. Jang, S. K. Kim, and D. J. O'Callaghan, unpublished data). The studies presented in here suggest that interaction of the EICP0 protein with the IE protein, TFIIB, and TBP may mediate the antagonism between the IE and EICP0 proteins.
MATERIALS AND METHODS
Virus and cell culture.
The equine transitional cell carcinoma (ETCC) and mouse fibroblasts L-M cells were maintained at 37°C in complete Eagle minimal essential medium supplemented with 100 U of penicillin/ml, 100 μg of streptomycin/ml, nonessential amino acids, and 5% fetal bovine serum. The Kentucky A (KyA) strain of EHV-1 was propagated in suspension cultures of L-M cells as previously described (49, 50).
Plasmids.
Plasmids were constructed and maintained in Escherichia coli HB101 by standard methods (53). Plasmids pGST-IE(1-1487), pGST-IE(407-539), pGST-IE(422-575), and pGST-IE(898-1487), which express IE aa 1 to 1487, 407 to 539, 422 to 575, and 898 to 1487 as glutathione S-transferase (GST) fusion proteins, respectively, have been described previously (36). Plasmids pGST-IE(1-960), pGST-IE(1-424), and pGST-IE(1-289), pGST-IE(1-88), pGST-IE(179-424), pGST-IE(539-910), and pGST-IE(323-1487) have been described previously (30).
(i) Mammalian IE and EICP0 expression plasmids.
To generate pSVIEΔ89/242 (pSVIEΔSRT2), plasmid pSVIE (56) was first digested with HindIII and BamHI, and the resultant HindIII-BamHI fragment was cloned into the HindIII and BamHI sites of pUC19 and designated pUIESRT2. pUIESRT2 was digested with NaeI and self-ligated with T4 DNA ligase to generate plasmid pUIEΔSRT2. The HindIII and BamHI fragment of pUIEΔSRT2 was cloned into the HindIII and BamHI sites of the pSVIE, creating pSVIEΔ89/242 (pSVIEΔSRT2). To generate pSVIE(243-1029), plasmid pUIEΔSRT2 was first digested with NcoI and NaeI, filled with Klenow fragment, and self-ligated to generate the pUIEΔSRT2-K. The HindIII-BamHI fragment of pUIEΔSRT2-K was cloned into the HindIII and BamHI sites of the pSVIE to generate pSVIE(243-1487). The HindIII-BamHI fragment of pSVIE(1-1029) (57) was cloned into the HindIII and BamHI sites of the pSVIE(243-1487), resulting in pSVIE(243-1029). To generate pSVIE(539-1029), plasmid pSVIE(1-1029) was digested with NcoI and self-ligated. To generate pSVIEΔ644/824, the XhoI-BamHI fragment of pSVIE was cloned into the XhoI and BamHI sites of pGEX-4T-1 (Pharmacia) and designated pGEX-IE. Plasmid pGEX-IE was digested with EagI and self-ligated to generate the pGEX-IEΔ644/824. The XhoI-BamHI fragment of pGEX-IEΔ644/824 was cloned into the XhoI and BamHI sites of pSVIE, yielding pSVIEΔ644/824. Plasmids pSVIE(323-1487) (58), pSVIE(323-1487)AAD (58), and pSVIE(1-951) (57) have been described previously. To generate plasmid pTri-EICP0-His, which expresses His-tagged EICP0 protein, the 280-bp C-terminal DNA fragment of the EICP0 gene from pSVICP0K (2) was amplified by PCR with the primers EICP0#F1 (5′-GTTTTAGTCGACCGAAGCTCTG-3′) and EICP0#R1 (5′-GCAAGCTTTTGGTTTCTCCGGTATCTTTGGCAG-3′), which contained SalI and HindIII sites, respectively, cloned back into the SalI and HindIII sites of the pSVICP0K; this plasmid was designated pSVEICP0-1. The 1.3-kb NcoI-HindIII fragment of pSVEICP0-1 was cloned into the NcoI and HindIII sites of the pTriEx-1 (Novagen), resulting in pTri-EICP0-His.
(ii) In vitro transcription and translation plasmids.
The IVTT EICP0 mutants (Fig. 7A) were prepared with the SV40 mammalian expression vector pSVSPORT1 (Gibco-BRL), which also contains the Sp6 promoter. To generate pSVEICP0(106-419), the 1.15-kb NcoI-EcoRI fragment of pSVICP0K (2), which expresses the full-length EICP0 protein, was cloned back into the NcoI (first ATG codon) and EcoRI sites of pSVICP0K. To generate pSVEICP0Δ203/277, pSVICP0K was digested with NaeI, an SstII linker (5′-TCCCCGCGGGGA-3′) was inserted, and the plasmid was designated pSVEICP0-S. pSVEICP0-S was digested with SstII and self-ligated to generate pSVEICP0Δ203/277. To generate pSVEICP0-SK1, the 0.95-kb NcoI-EcoRI fragment of pSVEICP0Δ203/277 was cloned into the NcoI and HindIII sites of pSVEICP0(106-419). To generate pSVEICP0(1-323), the pSVICP0K was digested with SalI and self-ligated. To generate pSVEICP0(1-278), the pSVICP0K was digested with NaeI and XbaI, filled with Klenow fragment, and self-ligated. To generate pSVEICP0(1-202), pUC19 was digested with SmaI, an SstII linker (5′-TCCCCGCGGGGA-3′) was inserted, and the plasmid was designated pUC19SII. The 0.66-kb EcoRI and SacII fragment of pSVICP0K was cloned into the EcoRI and SacII sites of pUC19SII to generate pUEICP0(1-202). The 0.68-kb EcoRI-HindIII fragment of pUEICP0(1-202) was cloned into the EcoRI and HindIII sites of pSVSPORT1, resulting in pSVEICP0(1-202). To generate pSVEICP0(1-164), pSVICP0K was digested with SstI and then self-ligated. To generate pSVEICP0(1-143), pSVICP0K was digested with NheI, filled with Klenow fragment, and self-ligated. To generate pSVEICP0(106-323), pSVEICP0(106-419) was digested with SalI and self-ligated. To generate plasmid pSVEICP0Δ144/314, the 1.5-kb EcoRI-HindIII fragment of pSVEICP0 was cloned into the EcoRI and HindIII sites of pUC19 and designated pUEICP0. The 0.32-kb C-terminal DNA fragment of the EICP0 gene from pSVICP0K was amplified by PCR with the primers EICP0#F5 (5′-GGTCTAGAAGCCATGGACAGCGATGGGCCTGTTGCGGTTG-3′) and EICP0#R1 (5′-GCAAGCTTTTGGTTTCTCCGGTATCTTTGGCAG-3′), which contained XbaI and HindIII sites, respectively, cloned into the NheI and HindIII sites of the pUEICP0 and designated pUEICP0Δ144/314. The 0.75-kb EcoRI-HindIII fragment of pUEICP0Δ144/314 was cloned into the EcoRI and HindIII sites of the pSVSPORT1, resulting in the pSVEICP0Δ144/314. To generate pSVEICP0(144-278), the 0.9-kb EcoRI-HindIII fragment of pSVEICP0(1-278) was cloned into the EcoRI and HindIII sites of pUC19 and designated pUEICP0(1-278). pUEICP0(1-278) was digested with NheI and NcoI, filled with Klenow fragment, and self-ligated to generate pUEICP0(144-278). The 0.4-kb EcoRI-HindIII fragment of pUEICP0(144-278) was cloned into the EcoRI and HindIII sites of pSVSPORT1, resulting in pSVEICP0(144-278).
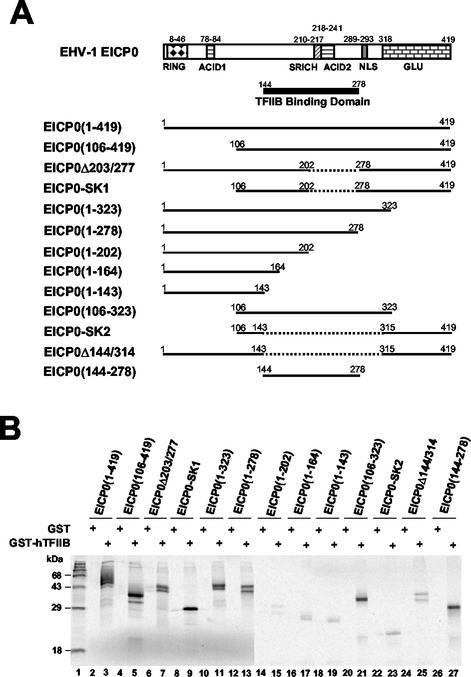
FIG. 7.
GST-pulldown assays with various deletion mutants of the EICP0 protein. (A) Schematic diagram and relative TFIIB-binding activities of the deletion mutants of the EICP0 protein. The top diagram represents the 419-aa EICP0 protein of EHV-1. RING, Ring finger domain; ACID1 and ACID2, acidic regions 1 and 2; SRICH, serine-rich region; GLU, glutamine-rich region. The numbers refer to the number of amino acids from the N terminus of each protein. (B) Equal amounts of each radiolabeled species were incubated with GST (lanes 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, and 26) or GST-TFIIB (lanes 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, and 27) proteins and then precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. The bands were quantitated by PhosphorImager analysis. The numbers on the left represent 14C-methylated protein markers in kilodaltons. +, Present.
(iii) Chloramphenicol acetyltransferase (CAT) reporter and effector plasmids.
To generate pIE(−169/+73)-CAT, the SphI fragment of pIE(−802/+73)-CAT (56) was cloned into the SphI site of pCAT-basic (Promega). To generate pIE(−69/+73)-CAT, the MscI-XbaI fragment of pIE(−802/+73)-CAT was cloned into pCAT-basic. pCAT-basic was previously digested with SalI and treated with Klenow fragment to create a blunt end, and then the DNA was cut a second time with XbaI. Plasmid pIE(−359/+73)-CAT has been described previously (56). Plasmids pgK-CAT, pgK(−153/+14)-CAT, and pgK(−83/+14)-CAT have been described previously (35). Plasmids pSVIE, pTK-CAT, and pIR5-CAT have been described previously (56). Plasmid pEICP22(E)-CAT has been described previously (28).
(iv) Human GST-TFIIB deletion and GST-TBP mutants.
To facilitate the mapping of the domain of TFIIB that mediates its interaction with the EICP0 protein, a panel of GST-TFIIB deletion mutants was constructed (Fig. 8A). Plasmid pGSTKG-TFIIB, which contains the entire TFIIB coding sequence cloned in frame with the GST gene and expresses the fusion protein GST-hIIB(1-316), was described previously (30). To generate plasmid pGST4T1-TFIIB, pGBKTFIIB was digested with SmaI and SalI, and the released TFIIB gene was cloned into the SmaI and SalI sites of the pGEX-4T1 (Promega). Plasmid GST-TFIIBΔ4-37 was generated by a two-step cloning strategy. First, pGSTKG-TFIIB was digested with AccI and SalI, followed by blunt ending the 5′ end with Klenow enzyme and inserting a ClaI linker d(pCCCATCGATGGG) (New England Biolabs) at codon 4 to generate pGSTKG-TFIIBaa4C. Plasmid pGSTKG-TFIIBaa4C was subsequently digested with ClaI and HindIII and ligated with a PCR-amplified segment of TFIIB spanning codons 38 to 311. The forward primer was 5′-CCATCGATGGCTTGGTTGTAGGTGACCGGG-3′ and the reverse primer was 5′-CCCAAGCTTTTATAGCTGTGGTAGTTTGTC-3′. pGST-TFIIBΔ4-66 is a derivative of pGSTKG-TFIIBaa4C. Plasmid pGSTKG-TFIIBaa4C was digested with ClaI and HindIII and ligated with a PCR-amplified segment of TFIIB spanning codons 67 to 311. The forward primer was 5′-CCATCGATGTTGGAGATTCTCAGAATCCTC-3′, and the reverse primer was 5′-CCCAAGCTTTTATAGCTGTGGTAGTTTGTC-3′. Plasmid pGST-TFIIBΔ4-123 is a subclone of pGSTKG-TFIIBaa4C and was generated by inserting a PCR-amplified segment of TFIIB corresponding to codons 124 to 311. The forward primer was 5′-CCATCGATATGGCAGACAGAATCAATCTAC-3′, and the reverse primer was 5′-CCCAAGCTTTTATAGCTGTGGTAGTTTGTC-3′. Plasmid pGST-TFIIBΔ67-123 was derived from pGST4T1-TFIIBaa67N, which was generated by partially digesting pGST4T1-TFIIB with XhoI, blunt ending the 5′ end, and inserting the NcoI linker d(pCCCATGGG) (New England Biolabs) at codon 67. Plasmid pGST4T1-TFIIBaa67N was subsequently digested with NcoI and religated to generate pGST-TFIIBΔ67-123. Plasmid pGST-TFIIBΔ125-174 was derived from pGSTKG-TFIIBaa174N, which was generated by digesting pGSTKG-TFIIB with AvrII, blunt ending the 5′ end, and inserting the NcoI linker d(pCAGCCATGGCTG) (New England Biolabs). Plasmid pGSTKG-hIIBaa174N was subsequently digested with NcoI and religated to generate pGST-TFIIBΔ125-174. Plasmid pGST-TFIIBΔ176-201 was generated by digesting pGSTKG-TFIIB with AvrII and Eco47III, blunt ending the 5′ end with Klenow enzyme, and religating the resulting DNA fragment. Plasmid pGST-TFIIBΔ202-269 was cloned by digesting pGSTKG-hIIB with Eco47III and PpuMI, blunt ending the 5′ end, and religation. Plasmid pGST-TFIIBΔ271-297 was derived from pGSTKG-TFIIBaa270B, which was cloned by digesting pGSTKG-TFIIB with PpuMI, blunt ending the 5′ end, and inserting the BglII linker d(pGGAAGATCTTCC) (New England Biolabs). Plasmid pGSTKG-TFIIBaa270B was subsequently digested with BglII and religated to generate pGST-TFIIBΔ271-297. Plasmid pGST-TFIIBΔ201-316 was cloned by digesting pGSTKG-TFIIB with Eco47III and HindIII, blunt ending the 5′ end, and religating the resulting DNA fragment. Plasmid pGST-TFIIBΔ271-316 was cloned by digesting pGSTKG-TFIIB with PpuMI and HindIII, blunt ending the 5′ end, and religating the resulting DNA fragment. pGST-TFIIB(1-123) was created by digesting pGSTKG-TFIIB with NcoI and HindIII, filling in the 5′ overhang with Klenow enzyme, and self-ligating the DNA fragment. pGST-TFIIB(67-200) was cloned by digesting pGST-TFIIBΔ4-66 with Eco47III and HindIII and self-ligating the resulting Klenow-treated restriction enzyme fragment. pGST-TFIIB(175-316) was generated by cutting pGST-TFIIBΔ4-66 with ClaI and AvrII and self-ligating the resulting Klenow-treated restriction enzyme fragment.
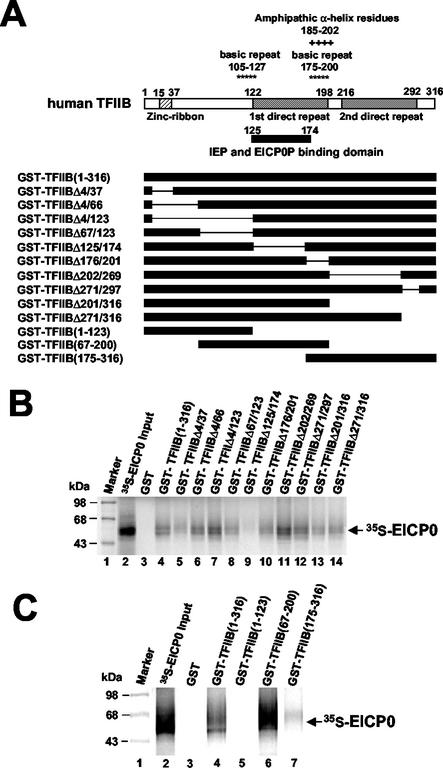
FIG. 8.
The EICP0 protein interacts with human TFIIB. (A) Schematic diagram of the deletion mutants of human TFIIB. The top diagram represents the 316-aa human TFIIB. The numbers refer to the number of amino acids from the N terminus of each protein. (B) Equal amounts of the IVTT 35S-EICP0 protein were incubated with the GST (lane 3) and GST-TFIIB (lanes 4 to 14) proteins and precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. The bands were quantitated by PhosphorImager analysis. (C) Equal amounts of the IVTT 35S-EICP0 protein were incubated with GST (lane 3), GST-TFIIB(1-316) (lane 4), GST-TFIIB(1-123) (lane 5), GST-TFIIB(67-200) (lane 6), or GST-TFIIB(175-316) (lane 7) proteins and then precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. The bands were quantitated by PhosphorImager analysis. The numbers on the left represent 14C-methylated protein markers in kilodaltons.
To generate pGST-hTBP, which expresses GST-human TBP fusion protein, the TBP gene from pQE30-hTBP (gift of Arnold J. Berk, University of California at Los Angeles) was amplified by PCR with the primers hTBP#F1 (5′-CGCGGATCCATGGATCAGAACAACAGCCTC-3′) and hTBP#R1 (5′-CCGGAATTCTTACGTCGTCTTCCTGAATCC-3′), which contained BamHI and EcoRI sites, respectively, and cloned into the BamHI and EcoRI sites of the pGEX-KG plasmid (22).
Purification of the EICP0 protein from inclusion bodies in E. coli.
To generate recombinant EICP0 protein, the pTriEx-1 expression system (Novagen) was used. The EICP0 gene was cloned into the pTriEx-1 vector, resulting in the pTriEICP0(1-419) clone that expresses the EICP0-His protein in BL21DE3(pLysE)pLacI (Novagen) cells. To purify soluble forms of the EICP0 protein, the EICP0 protein inclusion bodies were isolated from the crude cell lysate by centrifugation and washed twice with wash buffer (20 mM Tris-HCl [pH 7.5], 10 mM EDTA, 1% Triton X-100) to remove loosely associated contaminants. The nourishment refolding kit (Novagen) was used to solubilize the recombinant EICP0 protein that accumulates in inclusion bodies in E. coli. The solubilized fraction was dialyzed against a neutral pH buffer (20 mM Tris-HCl [pH 8.5]) containing a reducing agent to encourage correct disulfide bond formation. A second dialysis step removed excess reducing agent. Finally, the soluble EICP0-His protein was purified with a His-Bind kit (Novagen).
Western blot analysis.
Samples were boiled in 2× Laemmli sample buffer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Separated proteins were transferred to nitrocellulose filters (Schleicher & Schuell, Inc.) at 100 V for 1 h. Blots were blocked for 30 min in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% Tween 20) containing 10% nonfat powdered milk and then incubated with an EICP0-specific polyclonal antibody TrpE-ICP0 (2) at a 1:2,000 dilution and/or an IE peptide-specific polyclonal antibody (26) at a 1:2,000 dilution in TBST for 30 min. The blots were washed three times for 10 min each in TBST and incubated with secondary antibody (anti-mouse immunoglobulin G [IgG][Fc]-alkaline phosphatase [AP] conjugate [Promega]) at a dilution of 1:4,000 for an additional 30 min. The membranes were washed in TBST for three 10-min washes, and the proteins were visualized by incubating the membranes in AP buffer (0.1 M Tris-HCl [pH 9.5], 0.1 M NaCl, 5.0 mM MgCl2) containing nitroblue tetrazolium (0.33 mg/ml; Life Technologies) and BCIP (5-bromo-4-chloro-3-indolylphosphate; 0.165 mg/ml; Life Technologies).
DNA transfection and CAT assays.
L-M cells seeded at 3 × 106 cells per tissue culture dish (60 mm) in Eagle minimal essential medium with 5% fetal bovine serum were transfected by the liposome-mediated DNA transfection method at 24 h (28). The reporter plasmid was transfected in the amounts indicated in the figure legends. All effector plasmids were transfected in 0.3-pmol amounts. The total amount of DNA per transfection was adjusted to 8 μg by the addition of pUC19. After a further 5 h, the cells were washed and refed with fresh medium. At 60 h later, total cell extracts were prepared, and CAT activities were assayed as described previously (56).
IVTT.
In vitro transcription/translation (IVTT) reactions were performed by using the TNT-coupled transcription/translation reticulocyte lysate system (Promega) according to the manufacturer's instructions. Briefly, 1.4 μg of plasmids were incubated with 25 μl of rabbit reticulocyte lysate, 1 μl of Sp6 RNA polymerase, and [35S]methionine (40 μCi/ml; specific activity, 1,175 Ci/mmol; New England Nuclear Corp.). Reaction mixtures were incubated for 2 h at 30°C and either stored at −80°C or used immediately for in vitro interaction assays.
GST-pulldown and competition assays.
GST-pulldown assays were performed by using either 20 μl of nuclear extract or 10 μl each of in vitro-transcribed or -translated proteins. To these reactions, 20 μl of glutathione-Sepharose 4B was added, together with either 1 μg of a GST fusion protein or 2 μg of GST. Reaction volumes were adjusted to 500 μl with NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 8.0], 0.5% Nonidet P-40 [Sigma]) and were placed on a wheel at room temperature for 2 h. Beads were washed three times with NETN buffer, resuspended in 20 μl of loading buffer (10% 2-mercaptoethanol, 4% SDS, 20% glycerol, 0.02% [wt/vol] bromophenol blue), and boiled for 4 min. The proteins were separated by SDS-PAGE. Gels were subjected to Western blot analysis or, when radiolabeled proteins were used, the gels were dried, and the bands were quantitated by PhosphorImager analysis (Molecular Dynamics). The competition assays were performed essentially as before except that unlabeled in vitro-synthesized proteins, purified EICP0-His, and GST-IE fusion proteins were employed to compete for the binding of the 35S-labeled EICP0 (35S-EICP0) to TFIIB-agarose conjugate (Santa Cruz Biotechnology).
Laser scanning confocal microscopy.
ETCC cells were seeded on two-chamber glass slides (Nalge Nunc International) and infected with EHV-1 virus (KyA strain) at a multiplicity of infection of 10 PFU/cell. At appropriate times after infection, cells were fixed to the slides in methanol at −20°C for 10 min, rehydrated in phosphate-buffered saline (PBS) for 10 min, blocked with 10% normal goat serum in PBS for 30 min, and reacted first with a 1:100 dilution of the A1.4 monoclonal antibody to the IE protein (5) and then with a 1:200 dilution of a polyclonal antibody to the EICP0 protein (TrpE-ICP0) (2) in PBS with 1% bovine serum albumin (BSA) for 3 h. After a rinse with PBS, the cells were reacted with a TRITC (tetramethyl rhodamine isothiocyanate)-conjugated anti-mouse IgG and a fluorescein isothiocyanate-conjugated anti-rabbit IgG in PBS with 1% BSA for 1 h. Slides were mounted in 10% PBS in mount solution (0.1% p-phenylenediamine and 90% glycerol in PBS). Cells were examined in a laser-scanning confocal microscope system (MRC600; Bio-Rad Laboratories) attached to a Nikon Diaphot microscope (47).
RESULTS
The EHV-1 IE protein antagonizes the trans-activation ability of the EICP0 protein.
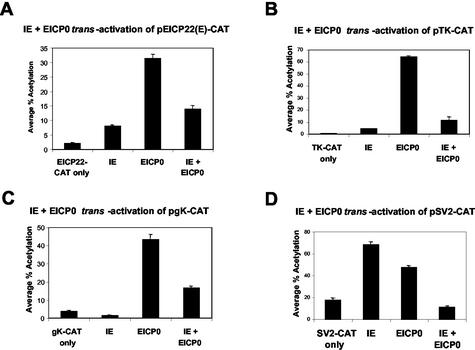
Our previous results suggested that the IE and EICP0 proteins, potent trans-activators of EHV-1 promoters, do not function synergistically but antagonistically (1, 35). To confirm that the IE and EICP0 proteins have an antagonistic relationship, L-M cells were cotransfected with the various EHV-1 and heterologous promoter-CAT reporter plasmids and effector plasmids (pSVIE and pSVICP0K). Both the IE and EICP0 proteins independently trans-activated the EICP22(E) promoter by 4- and 16-fold, respectively (Fig. 1A, bars 2 and 3, respectively). When used together, they did not function synergistically, and the IE protein inhibited the activation of the EICP22 promoter by the EICP0 protein (Fig. 1A, bar 4). This antagonism between the IE and EICP0 proteins was also observed with the early TK (pTK-CAT), true late gK (pgK-CAT) and even the heterologous SV40 promoter-CAT (pSV2-CAT) (Fig. 1B, C, and D, respectively). The amounts of the IE and EICP0 proteins produced in transfected L-M cells were similar, indicating that the reduction in CAT activity was not due to the failure of one or both viral proteins to be synthesized (data not shown). In addition, the antagonism was observed at various ratios of the IE and EICP0 proteins as documented in our previous work (1). These results demonstrated that the EICP0 and IE proteins, the two most powerful EHV-1 trans-activators, do not function synergistically, but antagonistically.
FIG. 1.
The EHV-1 IE and EICP0 proteins function antagonistically. Transient-transfection assays were performed with the various EHV-1 promoter-CAT reporter plasmids, EICP22 (A), thymidine kinase (TK) (B), gK (C), and SV40 (D) promoter-CAT. Transient-transfection assays were performed as described in Materials and Methods. L-M cells were transfected with 0.5 pmol of the reporter plasmids and 0.3 pmol of effector plasmids (pSVIE and pSVICP0K). Each transfection was performed in triplicate. The data are averages and are representative of several independent experiments. Error bars show standard deviations.
Essential domains for the antagonism between the IE and EICP0 proteins.
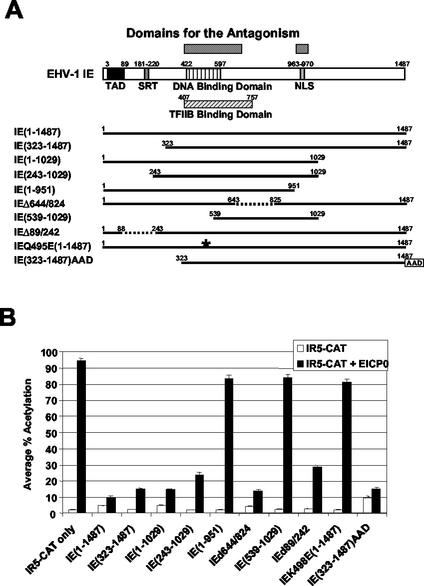
To determine the domains that mediate the antagonism between the IE and EICP0 proteins, transient-transfection assays were performed with a panel of deletion mutants of the IE gene (Fig. 2A) and the EICP0 protein expression vector pSVICP0K. The IE protein trans-activated very weakly the late IR5 promoter (pIR5-CAT) (Fig. 2B, bar 3); however, the EICP0 protein trans-activated this promoter very strongly (Fig. 2B, bar 2). When used together, the IE protein inhibited the ability of the EICP0 protein to trans-activate this late promoter (Fig. 2B, bar 4). IR2 [IE(323-1487)] and IE(243-1029) lack the acidic transcriptional activation domain (TAD). IE(1-951) lacks the nuclear localization signal (NLS), and both IE(539-1029) and IEQ495E(1-1487) lack the DNA-binding activity. These five mutant proteins were not able to trans-activate the IR5 promoter (Fig. 2B, bars 5, 9, 11, 15, and 19, respectively). When these nonfunctional IE protein expression vectors were cotransfected with pSVICP0K, none of the five inhibited the trans-activation function of the EICP0 protein (Fig. 2B, bars 6, 10, 12, 16, and 20, respectively), indicating that the DNA-binding domain, the TFIIB-binding domain, and/or the NLS may be important for the antagonism. Transient-transfection assays were performed with pSVIE(323-1437)AAD in which the TAD of the IE protein was deleted and the acidic activation domain (AAD) of the strong trans-activator HSV-1 VP16 was inserted at the C terminus of the IE(323-1487). The IE(323-1437)AAD fusion protein increased CAT activity by fourfold compared to results obtained with the intact IE protein (compare Fig. 2B, bar 21, with Fig. 2B, bar 3). Interestingly, this protein also inhibited the trans-activation function of the EICP0 protein (Fig. 3B, bar 22).
FIG. 2.
Domains essential for the antagonism between the IE and EICP0 proteins. (A) Schematic diagram of the deletion mutants of the IE protein. The top diagram represents the 1,487-aa IE protein of EHV-1. TAD, acidic transcriptional activation domain; SRT, serine-rich tract. The numbers refer to the number of amino acids from the N terminus of each protein. The asterisk indicates a point mutation in the DNA-binding domain of the IE protein. (B) CAT assays with the IE deletion mutants. L-M cells were transfected with 2.0 pmol of reporter plasmid (pIR5-CAT) and 0.3 pmol of effector plasmids. Each transfection was performed in triplicate. The data are averages and are representative of several independent experiments. Error bars show standard deviations.
FIG. 3.
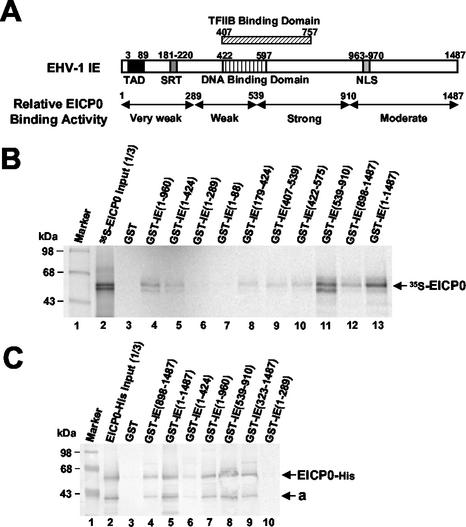
The EICP0 protein interacts with the IE protein. (A) Relative EICP0 protein-binding activity of the IE protein. The top diagram represents the 1,478-aa IE protein. The numbers refer to the number of amino acids from the N terminus of each protein. (B) GST-pulldown assays. Equal amounts of the IVTT 35S-EICP0 protein were incubated with GST (lane 3) and various GST-IE fusion proteins (lanes 4 to 13) and then precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. The bands were quantitated by PhosphorImager analysis (Molecular Dynamics). The numbers on the left represent 14C-methylated protein markers (Pharmacia) in kilodaltons. (C) Equal amounts of purified EICP0-His protein were incubated with GST (lane 3) or various GST-IE fusion proteins (lanes 4 to 10) and then precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. Gels were subjected to Western blot analysis. The “a” on the right indicates an isoform of the EICP0 protein. The numbers on the left represent molecular mass standards in kilodaltons.
The IE protein interacts with the EICP0 protein.
To address how the IE protein antagonizes the trans-activation of EHV-1 promoters by the EICP0 protein, we investigated whether the IE protein interacts with the EICP0 protein. To do this, GST-pulldown assays examined the ability of the various GST-IE fusion proteins to precipitate 35S-EICP0 protein from IVTT reactions. Control experiments showed that each of the mutant forms of GST-IE proteins was produced, as also documented in our previous work (30, 36). As observed during EHV-1 infection (3), the 35S-EICP0 protein was expressed as two bands in the IVTT reactions (Fig. 3B). Both isoforms of the IVTT 35S-EICP0 protein were precipitated with the GST-full-length IE protein (lane 13), but not with GST (lane 3), indicating that the EICP0 protein interacts with the IE protein. GST-IE(539-910) (Fig. 3B, lane 11) precipitated the EICP0 protein with an efficiency that was comparable to that of the GST-full-length IE protein. However, the GST-N-terminal IE mutants, GST-IE(1-88) and GST-IE(1-289), failed to precipitate the EICP0 protein (Fig. 3B, lanes 7 and 6, respectively). The other GST-IE deletion mutants, GST-IE(1-960), GST-IE(1-424), GST-IE(179-424), GST-IE(407-539), GST-IE(422-575), and GST-IE(898-1487) interacted weakly with the EICP0 protein (Fig. 3B, lanes 4, 5, 8, 9, 10, and 12, respectively).
To investigate whether the EICP0 protein directly interacts with the IE protein, GST-pulldown assays were performed with purified EICP0-His (His-tagged EICP0 protein) and GST-IE fusion protein. EICP0-His protein was also expressed as two bands in E. coli (Fig. 3C). In GST-pulldown assays, results similar to these obtained with EICP0 from IVTT reactions were obtained with purified EICP0-His protein (Fig. 3C), demonstrating that the IE protein directly interacted with the EICP0 protein.
The IE and EICP0 proteins colocalize in the nucleus of infected equine cells.
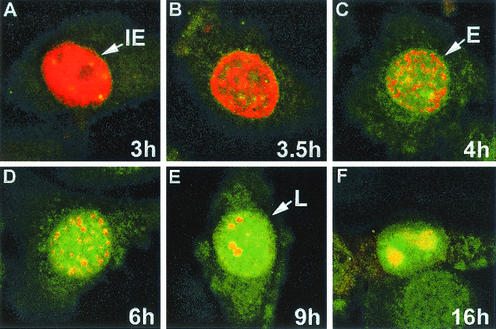
To determine whether the IE and EICP0 proteins colocalize in infected cells, EHV-1-infected equine ETCC cells were analyzed by immunofluorescence (IF) microscopy after labeling with a TrpE-ICP0-specific antibody and IE-specific monoclonal antibody. IE in infection (2 to 3 h postinfection), the IE protein (red) was dispersed throughout the nucleus (Fig. 4A), and thereafter the IE protein began to aggregate in small, dense nuclear structures within the nucleoplasm (Fig. 4B). Early in infection (4 to 6 h postinfection) the IE protein and EICP0 protein (green) partially overlapped (colocalized) in the nuclei of infected cells (Fig. 4C), but at late times (7 to 16 h postinfection) these proteins overlapped extensively in the nucleus of infected cells (Fig. 4D to F). These observations are supportive of the results of the biochemical analyses that indicate that the IE and EICP0 proteins physically interact. It should be noted that the photographs presented are typical results observed in the examination of numerous cells in repeated experiments
FIG. 4.
Photomicrographs of infected equine ETCC cultures. EHV-1 strain KyA-infected ETCC equine cells were fixed at 3 h (A), 3.5 h (B), 4 h (C), 6 h (D), 9 h (E), and 16 h (F) postinfection. The cells were reacted first with a 1:100 dilution of the A1.4 monoclonal antibody to the IE protein and a 1:200 dilution of a polyclonal antibody to the TrpE-EICP0 protein in PBS-1% BSA for 3 h. After a rinsing, the cells were reacted with a TRITC-conjugated anti-mouse IgG and an fluorescein isothiocyanate-conjugated anti-rabbit IgG for 1 h and examined under a confocal microscope. The IE and EICP0 proteins merged at early and late times of infection. IE, immediate-early pattern (2 to 3 h); E, early pattern (4 to 6 h); L, late pattern (7 to 16 h).
The EICP0 protein trans-activates the IE and gK promoters by affecting an area proximal to the transcription initiation site.
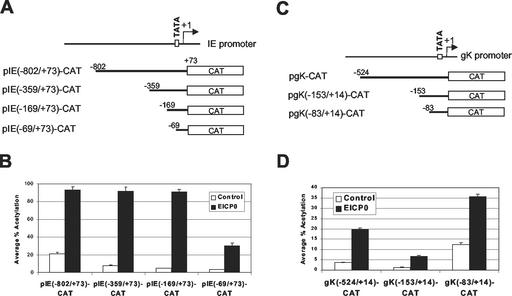
The EICP0 protein is a powerful, promiscuous trans-activator that activates all classes of EHV-1 promoters (1, 2). Our unpublished data (S. K. Kim and D. J. O'Callaghan) showed that the EICP0 protein does not bind to DNA, suggesting that the EICP0 protein trans-activates EHV-1 promoters by protein-protein interactions. To investigate which region of EHV-1 promoters is a target for trans-activation by the EICP0 protein, 5′ truncation constructs of the IE and gK promoters were generated and used in CAT assays (Fig. 5A and B, respectively). As expected, the EICP0 protein trans-activated strongly the native IE and gK promoters (Fig. 5B, bar 2, and D, bar 2, respectively). These results are consistent with our previous results (1, 35). Both the pIE(−69/+79)-CAT and pgK(−83/+14)-CAT reporter plasmids contain a minimal promoter region, but the EICP0 protein trans-activated strongly both of these reporter plasmids (Fig. 5B, bar 8, and D, bar 6, respectively). These results suggest that the EICP0 protein trans-activated IE and gK promoters by affecting an area proximal to the transcription initiation site.
FIG. 5.
The EICP0 protein trans-activates the IE and gK promoters by affecting an area proximal to the transcription initiation site. Upstream deletion constructs of the IE and gK promoters were generated and used for CAT assays. (A) Schematic diagram of upstream deletion constructs of the IE promoter. The top diagram represents the EHV-1 IE promoter. (B) Transient-transfection assays with the pIE-CAT reporter plasmids. L-M cells were transfected with 1.4 pmol of reporter plasmid pIE-CAT and 0.3 pmol of effector plasmid pSVICP0K. Each transfection was performed in triplicate. Data are averages and are representative of several independent experiments. Error bars show standard deviations. (C) Schematic diagram of upstream deletion constructs of the gK promoter. The top diagram represents the EHV-1 gK promoter. (D) Transient-transfection assays with the pgK-CAT reporter plasmids. L-M cells were transfected with 1.4 pmol of reporter plasmid pgK-CAT and 0.3 pmol of effector plasmid pSVICP0K. Each transfection was performed in triplicate. The data are averages and are representative of several independent experiments. Error bars show standard deviations.
The EICP0 protein interacts with human TFIIB.
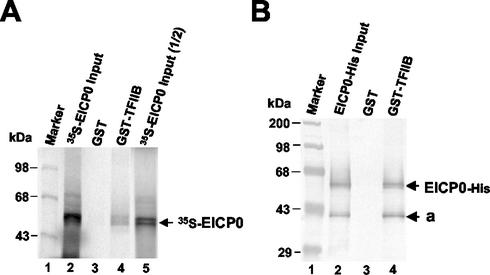
The finding that the EICP0 protein trans-activated EHV-1 promoters harboring only a minimal promoter region (TATA box and cap site) suggested that the EICP0 protein trans-activates EHV-1 promoters by interacting with GTFs. Therefore, possible interactions between the EICP0 protein and GTFs (TFIIB, TFIID, etc.) were tested. In vitro interaction assays were performed in which the basal transcription factor TFIIB synthesized as a GST fusion protein was examined for the ability to precipitate the EICP0 protein from IVTT reactions. Figure 6A, lanes 2 and 5, shows the amount of IVTT 35S-EICP0 protein included in each sample. GST-human TFIIB precipitated both isoforms of the EICP0 protein but not the GST portion, demonstrating that the EICP0 protein specifically interacts with TFIIB.
FIG. 6.
GST-pulldown assays with the IVTT and purified EICP0 proteins. (A) The EICP0 protein was in vitro transcribed-translated and radiolabeled with [35S]methionine as described in Materials and Methods. Equal amounts of each radiolabeled species were incubated with GST (lane 2) or GST-TFIIB (lane 3) proteins and then precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. The bands were quantitated by PhosphorImager analysis. The numbers on the left represent 14C-methylated protein markers in kDa. (B) The purified EICP0 protein was preincubated with the GST (lane 2) or GST-TFIIB (lane 3) proteins and then precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. Gels were subjected to Western blot analysis. The numbers on the left represent molecular mass standards (Gibco-BRL) in kilodaltons.
To investigate whether the EICP0 protein directly interacts with TFIIB, GST-pulldown assays were performed with the purified EICP0-His and GST-TFIIB. Western blot analysis of the GST-TFIIB precipitates revealed that the purified EICP0-His was precipitated with the GST-TFIIB, indicating that the EICP0 protein directly interacts with human TFIIB (Fig. 6B, lane 4).
The TFIIB-binding domain of the EICP0 protein lies within residues 144 to 278.
To map the EICP0 protein sequence(s) required for its interaction with TFIIB, various deletion mutants of the EICP0 protein were generated (Fig. 7A), expressed by IVTT reactions, and used in GST-pulldown assays. Each of the mutant EICP0 proteins was expressed and migrated in gels at the expected size (Fig. 7B). GST-hTFIIB but not GST precipitated the intact EICP0 protein from IVTT reactions (Fig. 7B, lanes 3 and 2, respectively). Any deletion within the EICP0 protein reduced its ability to interact with GST-TFIIB. GST-TFIIB was not able to precipitate N-terminal or C-terminal mutants of the EICP0 protein (Fig. 7B, lanes 14 to 19 and lanes 22 to 25, respectively). Deletion of the EICP0 (aa 203 to 278) resulted in a critical reduction in the interaction with the GST-TFIIB (compare Fig. 7B, lane 13, with lane 15). These results demonstrated that the TFIIB-binding domain of the EICP0 protein lies within the aa 144 to 278.
Residues 125 to 174 of TFIIB mediate its interaction with the EICP0 protein.
Constructs that express mutant forms of GST-hTFIIB were generated, and each was shown to produce the fusion protein of the expected size (data not shown). GST-pulldown assays performed with various hTFIIB deletion mutants (Fig. 8A) revealed that deletion of aa 125 to 174 of TFIIB decreased its interaction with the EICP0 protein (Fig. 8B, lane 9). To confirm that this domain (aa 125 to 174) of TFIIB is required for the interaction with the EICP0 protein, three additional hTFIIB deletion mutants were used in GST-pulldown assays. The results indicated that GST-TFIIB(67-200) strongly interacted with the EICP0 protein as efficiently as did intact GST-TFIIB (Fig. 8C, lane 6). However, the other two mutants [GST-TFIIB(1-123) and GST-TFIIB(175-316)] failed to interact (Fig. 8C, lanes 5 and 7, respectively), confirming that sequences located at aa 125 to 174 of TFIIB are required for its interaction with the EICP0 protein. Our recent results have shown that the IE protein-binding domain of the human TFIIB resides within the region from aa 125 to 174 (Albrecht et al., unpublished). Thus, both the IE and EICP0 regulatory proteins interact with the same domain of TFIIB.
A possible mechanism for the antagonism observed between the IE and EICP0 proteins may involve a physical interaction between the two proteins as shown in Fig. 3. Another possible explanation of this antagonism is that both the IE and EICP0 proteins compete for cellular transcription factors. Our recent data (Albrecht et al., unpublished) and those presented in Fig. 8 indicate that the IE and EICP0 proteins bind to the same domain within TFIIB. To confirm this, competition assays were performed in which TFIIB-agarose was incubated with 35S-EICP0 protein in the presence or absence of increasing amounts of competitors that included purified EHV-1 recombinant proteins or unlabeled EHV-1 proteins generated by IVTT (data not shown). These results demonstrated that the EICP0 protein competed with the IE protein for binding to human TFIIB, suggesting that the IE protein antagonizes the trans-activation ability of the EICP0 protein by competing for TFIIB. In data not shown, Western blot analyses revealed that GST-TFIIB, but not GST alone, precipitated the native EICP0 protein from nuclear extracts derived from EHV-1 KyA-infected L-M cells.
The EICP0 protein interacts with human TBP.
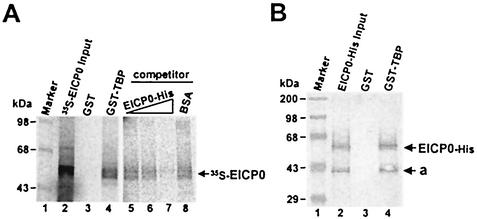
To investigate whether human TBP interacts with the EICP0 protein, GST-pulldown assays were performed. Interestingly, GST-TBP is able to precipitate the IVTT 35S-EICP0 protein (Fig. 9A, lane 4), but the GST portion failed in this regard (Fig. 9A, lane 3). In competition assays, the addition of increasing amounts of purified EICP0 protein gradually decreased the amounts of 35S-EICP0 protein that were precipitated (Fig. 9A, lanes 5 to 7); however, nonspecific competitor BSA did not (Fig. 9A, lane 8). These results indicated that the EICP0 protein specifically interacts with TBP. As expected, the TFIID-agarose conjugate (Santa Cruz Biotech) also precipitated IVTT 35S-EICP0 protein (data not shown). To investigate whether the EICP0 protein directly interacts with TBP, GST-pulldown assays were performed with purified EICP0-His protein and GST-TBP. Western blot analysis revealed that the purified EICP0-His protein also precipitated the GST-TBP, indicating that the EICP0 protein directly interacts with human TBP (Fig. 9B, lane 4).
FIG. 9.
GST-pulldown assays with the IVTT and purified EICP0 proteins. (A) Equal amounts of the IVTT 35S-EICP0 protein were incubated with GST (lane 2) or GST-TBP (lane 3) proteins and then precipitated with glutathione-Sepharose 4B beads. For competition assays, the IVTT 35S-EICP0 protein was incubated with the GST-TBP (lanes 5 to 8) proteins in the presence of specific competitor (purified EICP0-His) or nonspecific competitor (BSA) and then precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. The numbers on the left represent 14C-methylated protein markers in kilodaltons. (B) The purified EICP0-His protein was incubated with GST (lane 3) or GST-TFIIB (lane 4) proteins and precipitated with glutathione-Sepharose 4B beads. The precipitated pellets were electrophoresed through SDS-10% PAGE gels. Gels were subjected to Western blot analysis. The numbers on the left represent molecular mass standards in kilodaltons.
DISCUSSION
The IE and EICP0 proteins are potent trans-activators of EHV-1 promoters. However, they do not function synergistically but antagonistically. In comparison, HSV-1 ICP0 functions synergistically with ICP4 to activate expression of HSV-1 E and L promoters (10, 15). In order to help understand the mechanism(s) by which the IE protein antagonizes the trans-activation ability of the EICP0 protein, we assessed whether these proteins can physically interact and/or if these proteins competitively interact with the GTFs TBP and TFIIB.
Of the many possibilities that may explain the antagonism, a possible mechanism is that the IE and EICP0 proteins physically interact in a fashion that perturbs their regulatory function. Consistent with the results of the laser scanning confocal microscopy studies on EHV-1-infected cells, the in vitro interaction assays indicated that the IE protein interacts with the EICP0 protein. It has been reported that HSV-1 ICP0 interacts directly and specifically with ICP4 and with itself (63). Mullen et al. (46) have also detected an apparently specific intracellular interaction between the ICP4 and ICP0 proteins that leads to colocalization within punctate granules when they are both introduced into the same DNA-transfected Vero cells. HSV-1 ICP4 has an enhancing effect and ICP27 has an inhibitory effect on the nuclear localization of ICP0 (66). The results of our studies suggested that the IE protein may antagonize the trans-activation ability of the EICP0 protein by their interaction.
Another possible explanation of the antagonistic relationship is that the IE and EICP0 proteins compete for binding to cellular transcription factors. GST-pulldown assays indicated that the ICP0 protein directly interacts with TFIIB. The two-hybrid system in Saccharomyces cerevisiae has shown that a physical interaction occurred between the EICP0 protein and TFIIB (data not shown). Our recent studies showed that the IE protein interacts with the domain within TFIIB at aa 125 to 174, as well as with TBP (Albrecht et al., unpublished). Interestingly, the EICP0 protein interacts with the same domain within TFIIB at aa 125 to 174 and is able to compete with the IE protein for interaction with TFIIB in competition assays. The EICP0 protein also specifically and directly interacts with human TBP. Taken together, these results suggest that the IE protein may antagonize the trans-activation ability of the EICP0 protein by competing for binding to these transcription factors. The E2 protein of bovine papillomavirus type 1 binds directly to TFIID and TFIIB and competes with VP16 for factors important for trans-activation in vivo (51).
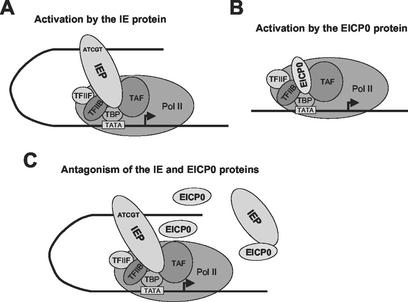
Our previous findings (30, 36) suggested that the IE protein binds to the IE binding consensus 5′-ATCGT-3′ sequence of EHV-1 promoters and then interacts with the basal transcription factors TFIIB and TBP to trans-activate EHV-1 promoters (Fig. 10A). The experiments presented here suggest that the EICP0 protein also interacts with TFIIB and TBP by direct protein-protein interaction to trans-activate EHV-1 promoters (Fig. 10B). We propose a model (Fig. 10C) to explain the mechanism of the antagonism between the IE and EICP0 proteins. Our results suggest two possibilities. First, the antagonism is attributed to interactions between the IE and EICP0 proteins. Second, the antagonistic relationship between the IE and EICP0 proteins is mediated by competition for TFIIB and TBP. Transient-transfection assays with the IE deletion mutants and the EICP0 protein expression vector indicated that the DNA-binding domain (overlaps with TFIIB-binding domain) and the NLS of the IE protein may be important for its antagonism with the EICP0 protein. These results suggested that the IE protein enters the nucleus and binds to the target DNA to antagonize the trans-activation ability of the EICP0 protein. The EICP0 protein does not bind to DNA (Kim and O'Callaghan, unpublished). The herpesvirus C3HC4 polypeptides do not bind to either DNA or RNA (13). Thus, DNA-bound IE protein stably gains access to the assembly of a PIC, which results in squelching of the EICP0 protein and prevents its participation in protein-protein interactions.
FIG. 10.
Models of how the IE and EICP0 proteins may activate transcription. (A) The IE protein binds to the IE protein-binding consensus 5′-ATCGT-3′ sequence of an EHV-1 promoter and then interacts with the basal transcription factors TFIIB and TBP to trans-activate EHV-1 promoters. (B) The EICP0 protein directly interacts with TFIIB and TBP to trans-activate EHV-1 promoters. (C) The mechanism of the antagonism observed between the IE and EICP0 proteins. Two possibilities were proposed according to our data. First, the IE and EICP0 proteins antagonize by an interaction between the two proteins. Second, the IE and EICP0 proteins antagonize by competing for binding to TFIIB and TBP. The arrows indicate the transcription initiation sites.
Transient-transfection assays showed that the EICP0 protein trans-activated EHV-1 promoters harboring only a minimal promoter region (TATA box and cap site), suggesting that the EICP0 protein trans-activates EHV-1 promoters by interacting with GTFs. The TFIIB-binding domain of the EICP0 protein lies within aa 143 to 278 (Fig. 7). Our previous results showed that the trans-activation ability of the EICP0 protein was severely inhibited by the loss of aa 167 to 282, which includes a serine-rich region (aa 210 to 217) and the acidic region 2 (aa 218 to 241) (1). The acidic activation domain of VP16 can directly interact with TFIIB and increase its stable assembly into a PIC (40, 41). Interaction between TFIIB and the acidic activation domain of VP16 is required for transcriptional activation (52). These results implied that the acidic region 2 (aa 218 to 241) of the EICP0 protein may be important for the interaction with TFIIB and for transcriptional activation.
The large T antigen (Tag) of SV40 is also a promiscuous trans-activator that can activate transcription from a variety of different promoters (51). The findings that Tag can interact with numerous transcription factors, any of which may be limiting for a given promoter, is consistent with Tag's promiscuity (31). HSV-1 ICP0, which is a powerful and promiscuous trans-activator, interacts with viral regulatory molecules (ICP4 and ICP27), cellular proteins (EF-1δ, cyclin D3, proteasomes, etc.), and the cellular transcription factor BMAL1 (4, 14, 32-34, 39, 60, 63). HSV-1 ICP0 dynamically interacts with the proteasome and possesses in vitro ubiquitin E3 ligase activity (60), which may target degradation of specific cellular proteins. It is possible that the EICP0 protein also interacts with the proteasome and possesses ubiquitin E3 ligase activity that may target degradation of specific proteins. However, the IE protein is present in significant amounts during all stages of EHV-1 infection, and the degradation of this protein as a result of the activities of the EICP0 protein appears unlikely. The EICP0 protein is a powerful, promiscuous trans-activator that activates all classes of EHV-1 promoters (1, 2). The protein contains a cysteine-rich zinc RING finger near the N terminus that is essential for activation of the E and L promoters (1). Our findings that the EICP0 protein specifically and directly interacts with basal transcription factors TFIIB and TBP, as well as the IE protein also indicates the EICP0 protein's promiscuity of protein interactions.
During a productive lytic infection, the genes of EHV-1 are coordinately expressed and temporally regulated in an IE, E, and L fashion (5, 18, 19). The IE protein turns on the expression of the E genes (28, 44, 48, 56, 65) but by itself is not able to activate the expression of some of the γ1 L genes and γ2 true L genes (28, 56). The EICP0 protein is the only regulatory protein capable of independently activating γ2 L gene expression (1, 2, 35), whereas the sole IE protein acts in concert with the EICP22 and EICP27 proteins to turn on the expression of the γ1 and some γ2 L genes (28, 56, 65). Taken together, these observations provide the basis for our hypothesis that the EICP0 protein is a regulatory factor that contributes to the switch from E to γ1 and γ2 L gene expression. The antagonism between the IE and EICP0 proteins may help govern the temporal regulation of gene expression. The IE protein may prevent the EICP0 protein from trans-activating L genes inappropriately at E times.
The studies presented in here have identified that the EICP0 protein directly interacts with TFIIB, TBP, and the IE protein, suggesting that the IE protein may antagonize the trans-activation ability of the EICP0 protein by their interaction and/or by competition for TFIIB and TBP. There is considerable information but no vital in vivo data regarding the antagonism. Experiments to further investigate the mechanism of the antagonism between the IE and EICP0 proteins are in progress.
Acknowledgments
We thank Arnold J. Berk for providing the plasmid pQE30-hTBP. We thank Suzanne Zavecz for excellent technical assistance.
Support for this investigation was obtained from Public Health Service research grant AI-22001 from the National Institutes of Health.
REFERENCES
- 1.Bowles, D. E., S. K. Kim, and D. J. O'Callaghan. 2000. Characterization of the trans-activation properties of equine herpesvirus 1 EICP0 protein. J. Virol. 74:1200-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowles, D. E., V. R. Holden, Y. Zhao, and D. J. O'Callaghan. 1997. The ICP0 protein of equine herpesvirus 1 is an early protein that independently transactivates expression of all classes of viral promoters. J. Virol. 71:4904-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruatowski, S. 1994. The basics of basal transcription by RNA polymerase II. Cell 77:1-3. [DOI] [PubMed] [Google Scholar]
- 4.Bruni, R., B. Fineschi, O. Ogle, and B. Roizman. 1999. A novel cellular protein, p60, interacting with both herpes simplex virus 1 regulatory proteins ICP22 and ICP0 is modified in a cell-type-specific manner and is recruited to the nucleus after infection. J. Virol. 73:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caughman, G. B., J. Staczek, and D. J. O'Callaghan. 1985. Equine herpesvirus type 1 infected cell polypeptides: evidence for immediate-early/early/late regulation of viral gene expression. Virology 145:49-61. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., Z. Xiuxuan, and S. Silverstein. 1991. Mutational analysis of the sequence encoding ICP0 from herpes simplex virus type 1. Virology 180:207-220. [DOI] [PubMed] [Google Scholar]
- 7.Cheung, A. K. 1991. Cloning of the latency gene and the early protein 0 gene of pseudorabies virus. J. Virol. 65:5260-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi, T., P. Lieberman, K. Ellwood, and M. Carey. 1995. A general mechanism for transcriptional synergy by eukaryotic activators. Nature 377:254-257. [DOI] [PubMed] [Google Scholar]
- 9.Choy, B., and M. R. Green. 1993. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature 366:531-536. [DOI] [PubMed] [Google Scholar]
- 10.Ciufo, D. M., M. A. Mullen, and G. S. Hayward. 1994. Identification of a dimerization domain in the C-terminal segment of the IE110 transactivator protein from herpes simplex virus. J. Virol. 68:3267-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derbigny, W. A., S. K. Kim, G. B. Caughman, and D. J. O'Callaghan. 2000. The EICP22 protein of equine herpesvirus 1 physically interacts with the immediate-early protein and with itself to form dimers and higher-order complexes. J. Virol. 74:1425-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derbigny, W. A., S. K. Kim, H. K. Jang, and D. J. O'Callaghan. 2002. EHV-1 EICP22 protein sequences that mediate its physical interaction with the immediate-early protein are not sufficient to enhance the trans-activation activity of the IE protein. Virus Res. 84:1-15. [DOI] [PubMed] [Google Scholar]
- 13.Everett, R. D., P. N. Barlow, A. Milner, B. Luisi, A. Orr, G. Hope, and D. Lyon. 1993. A novel arrangement of zinc-binding residues and secondary structure in the C3HC4 motif of an alpha herpes protein family. J. Mol. Biol. 234:1038-1047. [DOI] [PubMed] [Google Scholar]
- 14.Everett, R. D., M. R. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., C. M. Preston, and N. D. Stow. 1991. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication, p. 49-76. In E. K. Wagner (ed.), Herpesvirus transcription and its regulation. CRC Press, Inc., Boca Raton, Fla.
- 16.Garko-Buczynski, K. A., R. H. Smith, S. K. Kim, and D. J. O'Callaghan. 1998. Complementation of a replication-defective mutant of equine herpesvirus type 1 by a cell line expressing the immediate-early protein. Virology 248:83-94. [DOI] [PubMed] [Google Scholar]
- 17.Geisberg, J. V., W. S. Lee, A. J. Berk, and R. P. Ricciardi. 1994. The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc. Natl. Acad. Sci. USA 91:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray, W. L., R. P. Baumann, A. T. Robertson, D. J. O'Callaghan, and J. Staczek. 1987. Characterization and mapping of equine herpesvirus type 1 immediate-early, early, and late transcripts. Virus Res. 8:233-244. [DOI] [PubMed] [Google Scholar]
- 19.Gray, W. L., R. P. Baumann, A. T. Robertson, G. B. Caughman, D. J. O'Callaghan, and J. Staczek. 1987. Regulation of equine herpesvirus type 1 gene expression: characterization of immediate early, early, and late transcription. Virology 158:79-87. [DOI] [PubMed] [Google Scholar]
- 20.Grondin, B., and N. A. DeLuca. 2000. Herpes simplex virus type 1 ICP4 promotes transcription preinitiation complex formation by enhancing the binding of TFIID to DNA. J. Virol. 74:11504-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grundy, F. J., R. P. Baumann, and D. J. O'Callaghan. 1989. DNA sequence and comparative analysis of the equine herpesvirus type 1 immediate-early gene. Virology 172:223-236. [DOI] [PubMed] [Google Scholar]
- 22.Guan, K., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 23.Ha, I., S. Roberts, E. Maldonado, X. Sun, L. Kim, M. Green, and D. Reinberg. 1993. Multiple functional domains of human transcription factor IIB: distinct interactions with two general transcription factors and RNA polymerase II. Genes Dev. 7:1021-1032. [DOI] [PubMed] [Google Scholar]
- 24.Hahn, S. 1998. The role of TAFs in RNA polymerase II transcription. Cell 95:579-582. [DOI] [PubMed] [Google Scholar]
- 25.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. 62:456-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harty, R. N., and D. J. O'Callaghan. 1991. An early gene maps within and is 3′ coterminal with the immediate-early gene of equine herpesvirus 1. J. Virol. 65:3829-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holden, V. R., R. R. Yalamanchili, R. N. Harty, and D. J. O'Callaghan. 1992. ICP22 homolog of equine herpesvirus type 1: expression from early and late promoters. J. Virol. 66:664-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holden, V. R., Y. Zhao, Y. Thompson, G. B. Caughman, R. H. Smith, and D. J. O'Callaghan. 1995. Characterization of the regulatory function of the ICP22 protein of equine herpesvirus type 1. Virology 210:273-282. [DOI] [PubMed] [Google Scholar]
- 29.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang, H. K., R. A. Albrecht, K. A. Buczynski, S. K. Kim, W. A. Derbigny, and D. J. O'Callaghan. 2001. Mapping the sequences that mediate interaction of the equine herpesvirus 1 immediate-early protein and human TFIIB. J. Virol. 75:10219-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston, S. D., X.-M. Yu, and J. E. Mertz. 1996. The major transcriptional transactivation domain of simian virus 40 large T antigen associates nonconcurrently with multiple components of the transcriptional preinitiation complex. J. Virol. 70:1191-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawaguchi, Y., C. van Sant, and B. Roizman. 1997. Herpes simplex virus regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1α regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects transitional machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, S. K., D. E. Bowles, and D. J. O'Callaghan. 1999. The γ2 late glycoprotein K promoter of equine herpesvirus 1 is differentially regulated by the IE and EICP0 proteins. Virology 256:173-179. [DOI] [PubMed] [Google Scholar]
- 36.Kim, S. K., R. H. Smith, and D. J. O'Callaghan. 1995. Characterization of DNA binding properties of the immediate-early gene product of equine herpesvirus type 1. Virology 213:46-56. [DOI] [PubMed] [Google Scholar]
- 37.Kim, S. K., V. R. Holden, and D. J. O'Callaghan. 1997. The ICP22 protein of equine herpesvirus 1 cooperates with the IE protein to regulate viral gene expression. J. Virol. 71:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi, N., P. J. Horn, S. M. Sullivan, S. J. Triezenberg, T. G. Boyer, and A. J. Berk. 1998. DA-complex assembly activity required for VP16C transcriptional activation. Mol. Cell. Biol. 18:4023-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, Y. S., and M. R. Green. 1991. Mechanism of action of an acidic transcriptional activator in vitro. Cell 64:971-981. [DOI] [PubMed] [Google Scholar]
- 41.Lin, Y. S., E. Maldonado, D. Reingerg, and M. R. Green. 1991. Binding of general transcription factor TFIIB to an acidic activating region. Nature 353:569-571. [DOI] [PubMed] [Google Scholar]
- 42.Mackem, S., and B. Roizman. 1981. Regulation of herpesvirus macromolecular synthesis: temporal order of transcription of protein synthesis. J. Virol. 40:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manet, E., C. Allera, H. Gruffat, I. Mikaelian, A. Rigolet, and A. Sergeant. 1993. The acidic activation domain of the Epstein-Barr virus transcription factor R interacts in vitro with both TBP and TFIIB and is cell-specifically potentiated by a proline-rich region. Gene Expr. 3:49-59. [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumura, T., R. H. Smith, and D. J. O'Callaghan. 1993. DNA sequence and transcription analyses of the region of the equine herpesvirus type 1 Kentucky A strain genome encoding glycoprotein C. Virology 193:910-923. [DOI] [PubMed] [Google Scholar]
- 45.Moriuchi, H., M. Moriuchi, and J. I. Cohen. 1994. The RING finger domain of the varicella-zoster virus open reading frame 61 protein is required for its transregulatory functions. Virology 205:238-248. [DOI] [PubMed] [Google Scholar]
- 46.Mullen, M.-A., D. M. Ciufo, and G. S. Hayward. 1994. Mapping of intracellular localization domains and evidence for colocalization interactions between the IE110 and IE175 nuclear transactivator proteins of herpes simplex virus. J. Virol. 68:3250-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Callaghan, D. J., C. F. Colle III, C. C. Flowers, R. H. Smith, J. N. Benoit, and C. A. Bigger. 1994. Identification and initial characterization of the IR6 protein of equine herpesvirus 1. J. Virol. 68:5351-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Callaghan, D. J., and N. Osterrieder. 1999. Equine herpesviruses, p. 508-515. In R. G. Webster and A. Granoff (ed.), Encyclopedia of virology, 2nd ed. Academic Press, Inc., San Diego, Calif.
- 49.O'Callaghan, D. J., W. P. Cheevers, G. A. Gentry, C. C. Randall. 1968. Kinetics of cellular and viral DNA synthesis in equine abortion (herpes) virus infection L-M cells. Virology 36:104-114. [DOI] [PubMed] [Google Scholar]
- 50.Perdue, M. L., M. C. Kemp, C. C. Randall, D. J. O'Callaghan. 1974. Studies of the molecular anatomy of the L-M strain of equine herpesvirus type 1: protein of nucleocapsid and intact virion. Virology 59:201-216. [DOI] [PubMed] [Google Scholar]
- 51.Rice, P. W., and C. N. Cole. 1993. Efficient transcriptional activation of many simple modular promoters by simian virus 40 large T antigen. J. Virol. 67:6689-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts. S. G. E., I. Ha, E. Maldonado, D. Reinberg, and M. R. Green. 1993. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature 363:741-744. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Saurin, A. J., K. L. B. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21:208-214. [PubMed] [Google Scholar]
- 55.Smith, C. A., P. Bates, R. Rivera-Gonzalenz, B. Gu, and N. A. DeLuca. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 67:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, R. H., G. B. Caughman, and D. J. O'Callaghan. 1992. Characterization of the regulatory functions of the equine herpesvirus 1 immediate-early gene product. J. Virol. 66:936-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, R. H., V. R. Holden, and D. J. O'Callaghan. 1995. Nuclear localization and transcriptional activation activities of truncated versions of the immediate-early gene product of equine herpesvirus 1. J. Virol. 69:3857-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith, R. H., Y. Zhao, and D. J. O'Callaghan. 1994. The equine herpesvirus type 1 immediate-early gene product contains an acidic transcriptional activation domain. Virology 202:760-770. [DOI] [PubMed] [Google Scholar]
- 59.Stevenson, D., K. L. Colman, and A. J. Davison. 1992. Characterization of the varicella-zoster virus gene 61 protein. J. Gen. Virol. 73:521-530. [DOI] [PubMed] [Google Scholar]
- 60.Van Sant, C., R. Hagglund, P., Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe, S., E. Ono, Y. Shimizu, and H. Kida. 1995. Pseudorabies virus early protein 0 transactivates the viral gene promoters. J. Gen. Virol. 76:2881-2885. [DOI] [PubMed] [Google Scholar]
- 62.Wirth, U. V., C. Fraefel, B. Vogt, C. Vlcek, V. Paces, and M. Schwyer. 1992. Immediate-early RNA 2.9 and early 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 66:2763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao, F., and P. A. Schaffer. 1994. Physical interaction between the herpes simplex virus type 1 immediate early regulatory proteins ICP0 and ICP4. J. Virol. 68:8158-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao, Y., V. R. Holden, R. N. Harty, and D. J. O'Callaghan. 1992. Identification and transcriptional analysis of the UL3 and UL4 genes of equine herpesvirus 1, homolog of the ICP27 and glycoprotein K genes of herpes simplex virus. J. Virol. 66:5363-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao, Y., V. R. Holden, R. H. Smith, and D. J. O'Callaghan. 1995. Regulatory function of the equine herpesvirus 1 ICP27 gene product. J. Virol. 69:2786-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, Z., W. Cai, and P. A. Schaffer. 1994. Cooperativity among herpes simplex virus type 1 immediate-early regulatory proteins ICP4 and ICP27 affect the intracellular localization of ICP0. J. Virol. 68:3027-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]