Abstract
Several HLA alleles are associated with attenuated human immunodeficiency virus disease progression. We explored the relationship between the expression of particular major histocompatibility complex (MHC) class I alleles and viremia in simian immunodeficiency virus SIVmac239-infected macaques. Of the common MHC class I alleles, animals that expressed Mamu-A*01 exhibited the best control of viral replication.
It is becoming increasingly clear that an individual's HLA genes can affect the outcome of human immunodeficiency virus (HIV) infection. Carrington et al. demonstrated that HLA-B*35 and -Cw*04 are present at a high frequency in individuals who progress rapidly to AIDS (5). Conversely, there is a heterozygous advantage in HIV infection (5). Furthermore, two specific alleles, HLA-B*57 and HLA-B*27, are associated with slow progression to disease (10, 12, 15). HLA-B*57-positive individuals develop a broad repertoire of cytotoxic T-lymphocyte (CTL) responses, especially against the Gag protein (15). HLA-B*27 binds an immunodominant epitope, KK10, derived from the Gag protein. In some patients, this response has selected for viral escape variants associated with progression to disease (8, 9).
Unique properties of the simian immunodeficiency virus (SIV)-infected rhesus macaque make it an ideal model for HIV infection (11). Both SIV and HIV-1 are primate lentiviruses, sharing similar genomic organizations, biological properties, and tissue tropisms (11). Both viruses use the CD4 molecule as the primary receptor and chemokine receptors as coreceptors (6). Persistent disease is the result of infection with HIV and SIV. Individuals experience ongoing viral replication, and set point viremia represents the equilibrium between the ability of the virus to replicate and the ability of the host's immune responses to limit replication. Virus replication is held in check for an extended period of time (usually years for humans), until viremia increases followed by the onset of AIDS (21). The concentration of viral RNA (vRNA) at the set point in plasma has been inversely correlated with progression to disease (14). Rhesus macaques infected with SIVmac239 develop symptoms characteristic of human AIDS, including depletion of CD4+ T cells followed by development of opportunistic infections.
We wanted to determine whether a major histocompatibility complex (MHC) effect existed in a cohort of animals experimentally infected with the molecular clone SIVmac239. Previous studies have shown that SIVmac251-infected animals that express the class I Mamu-A*01 molecule (18) control viremia more effectively than Mamu-A*01-negative animals. We therefore wanted to explore the role of Mamu-A*01 in controlling replication of SIVmac239.
To determine the effect of different MHC class I alleles on viral replication, we screened for the expression of the following eight common MHC class I alleles in our cohort of 53 SIVmac239-infected animals by PCR-sequence-specific primers (SSP): Mamu-A*01, Mamu-A*02, Mamu-A*08, Mamu-A*11, Mamu-B*01, Mamu-B*03, Mamu-B*04, and Mamu-B*17 (Table 1). These alleles were chosen because they purportedly bind SIV-derived epitopes (3, 4, 7). The 3′-terminal region of PCR-SSP primers targeted a nucleotide polymorphism unique to these eight Indian rhesus MHC class I alleles. Approximately 75 ng of genomic DNA was isolated from 200 μl of EDTA-anticoagulated whole blood or buffy coat for each sample in a total reaction volume of 25 μl. Final reaction mixtures contained 1× PCR buffer (Invitrogen, Carlsbad, Calif.) composed of 60 mM Tris-HCl (pH 9.5), 2 mM MgCl2, 15 mM ammonium sulfate, 410 μM each deoxynucleoside triphosphate (dNTP; Promega, Madison, Wis.), 0.5 μM each PCR-SSP primer, 0.3 μM each internal control primer, and 0.961 U of Platinum Taq polymerase (Gibco BRL, Life Technologies, Gaithersburg, Md.). The thermal cycling conditions were as follows: an initial 1 min of denaturation at 96°C, followed by 5 cycles of 96°C denaturation for 25 s, 70°C annealing for 50 s, and a 45-s extension at 72°C; and, finally, 4 cycles of 96°C denaturation for 25 s, 55°C annealing for 60 s, and a 120-s extension at 72°C. Subsequently, PCR products were electrophoresed on 2% agarose gels (0.5× Tris-borate-EDTA [TBE]) at a constant voltage and analyzed for the presence of the required internal control product and each of the specific allele amplicons relative to a 100-bp DNA ladder (Gibco BRL).
TABLE 1.
Viruses used, MHC class I alleles, set point viremia, and vaccination information for each SIV-infected animal in our cohorta
| Animal | Virus strain | Allele type
|
No. of set point copies of vRNA/ml of plasma | Survivorship (no. of days postinfection)b | Vaccination statusc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A*01 | A*02 | A*08 | A*11 | B*01 | B*03 | B*04 | B*17 | |||||
| 95061 | SIVmac239/PBL | + | + | − | − | − | − | − | + | 400 | 908* | V |
| 1937 | SIVmac239/CEMx174 | + | − | − | + | − | − | − | + | 747 | 728* | C |
| 95096 | SIVmac239/CEMx174 | + | − | − | + | − | − | − | + | 2,364 | 728* | C |
| 95058 | SIVmac239stop/PBL | + | − | − | − | − | − | − | − | 11,380 | 620 | V |
| 85013 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 62,653 | 407 | C |
| 93062 | SIVmac239/PBL | − | − | − | − | − | − | − | − | 66,264 | 309 | V |
| 2130 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 79,271 | 195* | C |
| 95084 | SIVmac239/CEMx174 | + | + | − | − | − | − | − | − | 81,303 | 460 | C |
| 1975 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 83,522 | 593* | V |
| 2122 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 86,764 | 195* | V |
| 96135 | SIVmac239/PBL | + | − | + | − | − | − | − | − | 88,000 | 295 | V |
| 96118 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 88,464 | 764 | V |
| 2095 | SIVmac239/PBL | + | − | − | − | − | − | − | + | 102,959 | 195* | C |
| 80035 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 131,158 | 311 | V |
| 97085 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 158,859 | 106 | V |
| 96114 | SIVmac239/PBL | + | − | + | − | − | + | − | − | 185,285 | 153 | V |
| 96111 | SIVmac239/PBL | + | + | − | − | + | − | − | − | 193,189 | 398 | C |
| 95086 | SIVmac239/PBL | + | − | + | − | − | − | − | − | 199,904 | 308 | C |
| 94004 | SIVmac239/PBL | + | − | + | − | − | + | − | − | 205,941 | 307 | V |
| 92080 | SIVmac239/PBL | − | − | − | + | − | − | − | + | 222,473 | 427 | C |
| 93057 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 222,473 | 477 | C |
| 2129 | SIVmac239/PBL | + | − | − | + | − | − | − | + | 279,859 | 195* | C |
| 98018 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 281,625 | 195* | C |
| 2127 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 291,335 | 195* | C |
| 2124 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 315,125 | 195* | V |
| 2126 | SIVmac239/PBL | + | + | − | − | − | − | − | − | 328,656 | 195* | V |
| 97073 | SIVmac239/PBL | − | − | + | − | − | − | − | − | 337,034 | 539* | V |
| 96113 | SIVmac239/PBL | − | − | − | − | − | + | − | − | 387,650 | 526 | C |
| 92077 | SIVmac239/CEMx174 | + | − | − | − | − | − | − | − | 410,000 | 219 | C |
| 87081 | SIVmac239stop/PBL | − | − | − | + | − | − | − | − | 411,205 | 296 | V |
| 95045 | SIVmac239stop/PBL | + | − | + | − | − | + | − | − | 519,350 | 378 | V |
| 95114 | SIVmac239stop/PBL | + | − | − | − | + | − | − | − | 698,450 | 424 | C |
| 2065 | SIVmac239/PBL | + | − | + | − | − | + | − | + | 829,674 | 195* | C |
| 96020 | SIVmac239/PBL | − | + | + | − | − | + | − | − | 881,531 | 398 | C |
| 90069 | SIVmac239/PBL | − | − | − | − | − | − | − | − | 957,299 | 539* | C |
| 96031 | SIVmac239stop/PBL | + | + | − | − | − | − | − | − | 1,013,700 | 1,143 | C |
| 95115 | SIVmac239stop/PBL | + | − | + | − | + | + | − | − | 1,142,204 | 633 | C |
| 87082 | SIVmac239/PBL | − | + | + | − | − | − | − | − | 1,178,314 | 227 | V |
| 96104 | SIVmac239/PBL | − | − | − | − | + | − | − | − | 1,367,915 | 420 | C |
| 81035 | SIVmac239/PBL | − | − | − | − | + | − | − | − | 1,928,291 | 214 | V |
| 96123 | SIVmac239/PBL | + | − | + | − | − | + | − | − | 2,022,816 | 316 | V |
| 96093 | SIVmac239/PBL | − | − | − | − | − | − | − | − | 2,573,018 | 448 | C |
| 96072 | SIVmac239/PBL | − | + | − | − | − | − | − | + | 2,669,679 | 467 | C |
| 95003 | SIVmac239stop/PBL | − | − | + | − | + | + | − | − | 2,845,750 | 489 | C |
| 96016 | SIVmac239/PBL | + | − | − | − | − | − | − | − | 3,909,697 | 271 | V |
| 80025 | SIVmac239/CEMx174 | + | − | − | − | + | − | − | − | 4,100,000 | 101 | C |
| 87108 | SIVmac239/CEMx174 | + | − | − | − | − | − | − | − | 4,100,000 | 120 | C |
| 97086 | SIVmac239/PBL | − | + | + | − | − | + | − | − | 6,397,434 | 553* | C |
| 92050 | SIVmac239/PBL | − | + | − | − | − | + | − | − | 6,547,283 | 269 | C |
| 96081 | SIVmac239/PBL | − | − | + | − | − | + | − | − | 63,942,700 | 180 | C |
| 90131 | SIVmac239/PBL | − | − | + | − | + | − | − | − | 177,000,000 | 115 | C |
| 95112 | SIVmac239stop/PBL | − | − | + | − | + | − | − | − | 149,935,800 | 153 | C |
| 83108d | SIVmac239/PBL | − | − | − | − | + | + | − | − | died wk 5 | 24 | V |
Animals were infected with either SIVmac239/nef stop, SIVmac239/nef open, or SIVmac239/nef open expanded on CEMx174 cells. Using PCR-SSP, we screened for the presence of eight different MHC class I alleles. We measured viral loads between 12 and 16 weeks when viral replication stabilized in these animals (set point).
*, animal still alive as of writing.
V, vaccines; C, controls.
Animal 83108 was not included in viral load analysis because no set point viremia was available. The animal died of non-AIDS-related complications 5 weeks postinfection.
All macaques were infected with a molecularly cloned virus, SIVmac239 (Table 1). Three sets of viruses were used: SIVmac239/nef open and SIVmac239/nef stop, which were both expanded on rhesus peripheral blood lymphocytes (PBLs); and SIVmac239/nef open, which was expanded on CEMx174 cells only. SIVmac239/nef stop differs from SIVmac239/nef open by a stop codon present in the nef gene (13, 19). There is selection for full-length Nef protein in vivo, and the Nef open reading frame is restored within a few weeks after infection. The third virus stock was expanded in vitro on CEMx174 cells rather than on rhesus PBLs. Animals were infected intrarectally with SIVmac239/nef open (3,000 50% tissue culture infectious doses [TCID50]) and SIVmac239/nef stop (1,000 TCID50). Animals infected with SIVmac239/nef open expanded on CEMx174 cells only received 20 ng of p27 SIVmac239 intrarectally. A subset of animals was vaccinated according to DNA-modified vaccinia virus ankara (MVA) regimens containing different constructs (Table 1).
We stratified the animals based on their set point viremia. Viral loads were quantitated with the SIV branched DNA assay (Chiron Diagnostics, Emeryville, Calif.) and the Taqman kinetic reverse transcription (RT)-PCR assay. Plasma SIV RNA concentrations were measured every week for the first 4 weeks and then biweekly thereafter. Viral set points were measured between weeks 12 and 16, when plasma vRNA concentrations were constant for at least 2 weeks.
Viral load differences between groups of animals infected with SIVmac239 were tested for statistical significance with t tests after log transforming the data to improve normality and homoscedasticity. (Animal 83108 was not included in the analysis, because this animal was euthanized 5 weeks postinfection due to non-AIDS-related complications. No set point viremia was therefore available for this animal.) In addition, Levene's test for homoscedasticity was conducted, and if a difference was found to be significant, the Welch correction for unequal variances was employed. Finally, to further examine the robustness of the results, a nonparametric test, the Mann-Whitney U test, was performed. The P values for the nonparametric tests were calculated by exact methods. All P values are two tailed.
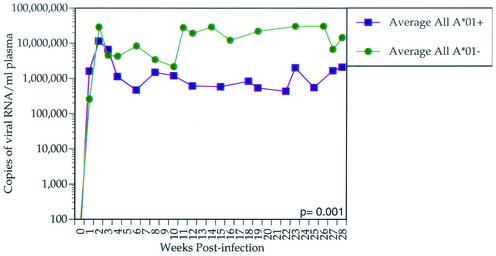
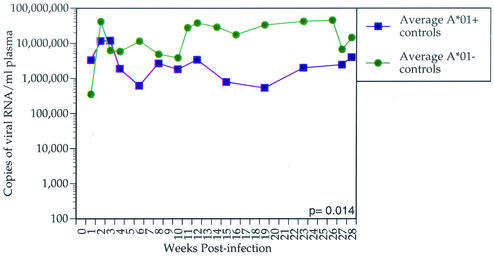
We found that the majority of animals with the lowest set point vRNA levels expressed Mamu-A*01. Between weeks 12 and 28 postinfection, Mamu-A*01-positive animals controlled viral replication more effectively than Mamu-A*01-negative animals (P = 0.001; t test) (Fig. 1). To ensure that the Mamu-A*01 effect is not due to a subset of vaccinated Mamu-A*01-positive animals, we removed these animals from the analysis. Indeed, vaccine-naive Mamu-A*01-positive animals restricted viral replication more efficiently than vaccine-naive Mamu-A*01-negative animals (P = 0.014; t test) (Fig. 2). We compared the virus loads between vaccinated and nonvaccinated Mamu-A*01-positive animals. We did not find a significant difference in set point viremia between these groups by either independent t test (P = 0.58) or Mann-Whitney test (P = 0.43). In addition to viral load analysis, we performed survival analysis by Cox regression (proportional hazards model) with A*01 status as a covariate. The hazard ratio of Mamu-A*01-positive animals to Mamu-A*01-negative animals is 0.76, which is not significant (P = 0.43). Nevertheless, analysis of survivorship as a function of vRNA revealed that survivorship is significantly inversely related to set point viremia (P < 0.001; Cox proportional hazards regression).
FIG. 1.
Average plasma SIVmac239 vRNA concentration for all Mamu-A*01-positive animals versus Mamu-A*01-negative animals. The average plasma vRNA concentration for each of the two groups of animals is shown for the first 28 weeks postinfection. Mamu-A*01-positive animals significantly control viral replication more effectively than Mamu-A*01-negative animals (P = 0.001).
FIG. 2.
Average plasma SIVmac239 vRNA concentration for control Mamu-A*01-positive animals versus Mamu-A*01-negative animals. The average plasma vRNA concentration for each of the two groups of animals is shown for the first 28 weeks postinfection. Control Mamu-A*01-positive animals significantly control viral replication more effectively than Mamu-A*01-negative animals (P = 0.014).
We have previously shown that Mamu-A*01-restricted epitopes dominate the anti-SIV CTL responses in Mamu-A*01-positive animals early in infection (16). Additionally, we have identified 14 different Mamu-A*01-restricted CTL epitopes, indicating that this MHC class I molecule engenders a broad repertoire of CTL responses (1). This may partially explain why Mamu-A*01-positive animals control viral replication more successfully than Mamu-A*01-negative animals. Furthermore, one of these responses, directed against the Tat28-35SL8 epitope, exerts selective pressure on the virus, as evidenced by viral escape from this response early in infection (2). This epitope has also been shown to be recognized by CTL with high functional avidity, requiring a low peptide concentration to trigger a CTL response (17).
The association of Mamu-A*01 with enhanced control of viral replication has been shown for infection with the heterogeneous biological isolate SIVmac251 (18), but not for infection with SHIV 89.6p. Our studies extend those findings to SIVmac239 replication. SHIV 89.6 is a chimeric virus created by using Env, as well as Tat, Vpu, and Rev from the HIV 89.6 isolate with the backbone of the molecular clone SIVmac239 (20). Mamu-A*01-restricted responses directed against SIV Env, Tat, Vpr, and Rev may therefore be partially responsible for the Mamu-A*01 effect in both SIVmac239 infection, shown here, and SIVmac251 infection (18). Tat28-35SL8 and other Mamu-A*01-restricted responses (1) may contribute to the effect of diminishing viral replication in Mamu-A*01-positive animals. Understanding how particular MHC genes affect viremia may provide insight into the correlates of protection from lentiviral infection.
Acknowledgments
We thank Millie Schultz, Elizabeth Meek, and Christopher Fischer for technical help, in addition to Kevin Kunstman and Steve Wolinsky for plasma vRNA concentration analysis. We also thank Jody Helgeland for assistance with blood processing, Jacque Mitchen for coordinating all animal procedures, and Carol L. Emerson for performing all animal procedures.
This work was supported by NIH grants RR1537, AI46366, AI45461, and RR00167 (awarded to David I. Watkins). David I. Watkins is a recipient of an Elizabeth Glaser Scientist Award.
REFERENCES
- 1.Allen, T. M., B. R. Mothe, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8+ lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule Mamu-A*01: implications for vaccine design and testing. J. Virol. 75:738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 3.Allen, T. M., J. Sidney, M. F. del Guercio, R. L. Glickman, G. L. Lensmeyer, D. A. Wiebe, R. DeMars, C. D. Pauza, R. P. Johnson, A. Sette, and D. I. Watkins. 1998. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 160:6062-6071. [PubMed] [Google Scholar]
- 4.Allen, T. M., and D. I. Watkins. 1999. SIV and SHIV CTL epitopes identified in macaques, p. IV-45-IV-54. In B. T. Korber, C. Brander, B. F. Haynes, R. A. Koup, C. L. Kuiken, J. P. Moore, B. D. Walker, and D. I. Watkins (ed.), HIV Molecular Immunology Database. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 5.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Z., A. Gettie, D. D. Ho, and P. A. Marx. 1998. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in west Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology 246:113-124. [DOI] [PubMed] [Google Scholar]
- 7.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 8.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, M. M. Addo, S. He, J. S. Mukherjee, M. N. Phillips, M. Bunce, S. A. Kalams, R. P. Sekaly, B. D. Walker, and C. Brander. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 10.Hill, A. V. 1998. The immunogenetics of human infectious diseases. Annu. Rev. Immunol. 16:593-617. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, R. P. 1996. Macaque models for AIDS vaccine development. Curr. Opin. Immunol. 8:554-560. [DOI] [PubMed] [Google Scholar]
- 12.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, M. G., S. Bellah, K. McKinnon, J. Yalley-Ogunro, P. M. Zack, W. R. Elkins, R. C. Desrosiers, and G. A. Eddy. 1994. Titration and characterization of two rhesus-derived SIVmac challenge stocks. AIDS Res. Hum. Retrovir. 10:213-220. [DOI] [PubMed] [Google Scholar]
- 14.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 15.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mothé, B. R., H. Horton, D. K. Carter, T. M. Allen, M. E. Liebl, P. Skinner, T. U. Vogel, S. Fuenger, K. Vielhuber, W. Rehrauer, N. Wilson, G. Franchini, J. D. Altman, A. Haase, L. J. Picker, D. B. Allison, and D. I. Watkins. 2002. Dominance of CD8 responses specific for epitopes bound by a single major histocompatibility complex class I molecule during the acute phase of viral infection. J. Virol. 76:875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 18.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 20.Reimann, K. A., J. T. Li, R. Veazey, M. Halloran, I.-W. Park, G. B. Karlsson, J. Sodroski, and N. L. Letvin. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sewell, A. K., D. A. Price, A. Oxenius, A. D. Kelleher, and R. E. Phillips. 2000. Cytotoxic T lymphocyte responses to human immunodeficiency virus: control and escape. Stem Cells 18:230-244. [DOI] [PubMed] [Google Scholar]