Abstract
Thogoto virus (THOV) is a tick-transmitted orthomyxovirus with a genome of six negative-stranded RNA segments. The sixth segment encodes two different transcripts: a spliced transcript that is translated into the matrix protein (M) and an unspliced transcript. Here, we report that the unspliced transcript encodes an elongated form of M named ML. A THOV isolate deficient in ML expression was an efficient interferon inducer, whereas ML-expressing wild-type strains were poor interferon inducers. These results were confirmed with recombinant THOVs rescued from cDNAs. Expression of ML efficiently suppressed activation of the beta interferon promoter by double-stranded RNA. These results indicate that ML is an accessory protein that functions as a potent interferon antagonist by blocking transcriptional activation of alpha/beta interferons.
Mammalian cells react to virus infections with the production of alpha/beta interferons (IFN-α/β) (16, 22) that are most commonly induced by double-stranded RNA (dsRNA), a by-product of viral replication (17). IFN-α/β stimulates the expression of antiviral factors. In particular, the products of the Mx genes have been shown to be active against orthomyxoviruses (13, 30). Therefore, the orthomyxoviruses, like most other viruses, have evolved strategies with which to counteract the IFN system (9, 11). The influenza A virus (FLUAV) NS1 protein was the first example of an orthomyxovirus gene product with IFN-antagonistic activity. A recombinant FLUAV that lacks the NS1 protein, FLUAV(delNS1), induces a much stronger IFN response in infected cells than the wild-type virus (10). The functional basis for this effect is most probably the ability of NS1 to bind dsRNA and to prevent the activation of the IFN-β promoter, PKR, and 2′,5′-oligoadenylate synthetase (20, 31, 35).
Thogoto virus (THOV) is the prototype of tick-transmitted orthomyxoviruses (28). It was independently isolated from ticks that were collected in Africa, Asia, and southern Europe (25). The genome of THOV consists of six single-stranded RNA segments of negative polarity that are encapsidated by the viral nucleoprotein (NP) and associate with the viral RNA polymerase complex to form ribonucleoprotein complexes (6, 14). Each segment codes for a single structural protein: the three subunits of the viral RNA polymerase complex (PB2, PB1, and PA), the viral surface glycoprotein, NP, and the matrix protein (M) (14). THOV is structurally and genetically similar to influenza viruses but is different in its ability to infect ticks, as well as mammals (6). This extended host range requires specific adaptations to allow the virus to replicate in both cell types.
The genome of THOV encodes the basic set of orthomyxovirus proteins but no additional proteins like the NS2/NEP of FLUAV, an M2 ion channel, or an NS1-like protein. We have previously shown that the sixth segment of THOV encodes two transcripts: a spliced mRNA that codes for the M protein and an unspliced mRNA with no assigned gene product (19). Here, we compared the nucleotide sequences of different wild-type isolates of THOV with our laboratory strain and found that wild-type strains lack one nucleotide within the intron region of segment 6 compared to the sequence of the laboratory strain. Therefore, the unspliced transcripts of the wild-type strains contain a reading frame that encodes a C-terminally extended M protein. We named the new gene product ML (for matrix protein long). Our data demonstrate that the newly identified ML gene product of THOV is a potent inhibitor of the IFN-α/β system.
(This research was conducted by K. Hagmaier and S. Jennings in partial fulfillment of the requirements for a Ph.D. degree from the Faculty of Biology of the University of Freiburg, Freiburg, Germany.)
Induction of IFN-α/β by THOV.
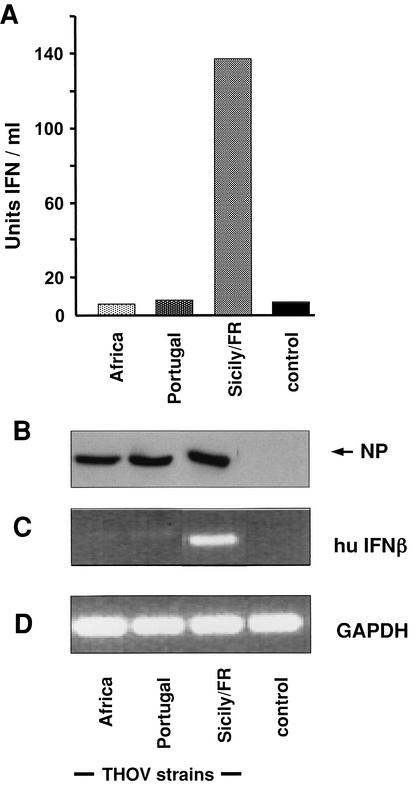
We tested the induction of IFN-α/β by different strains of THOV, namely, the original THOV isolate from Africa, IIA (12), an isolate from Portugal, PoTi503 (7), and a derivative of the Sicilian isolate SiAr126 (1), designated Sicily/FR, that has been used in our laboratory since 1993 (13). 293 cells were infected with 3 PFU of THOV per cell for 24 h. Supernatants of the infected cells were harvested, and infectious virus was inactivated by acidification. The amount of acid-stable IFN in the cell supernatants was then determined by using a reporter plasmid coding for firefly luciferase (FF-Luc) under the control of an IFN-stimulated response element promoter as described (18, 36). Reporter gene expression (FF-Luc) directly correlated with the amount of IFN-α/β present in the supernatants of the virus-infected cells as determined by an IFN-α standard (data not shown). Infection with the THOV isolates from Africa and Portugal did not induce measurable levels of IFN (Fig. 1A). Surprisingly, however, infection of the cells with strain Sicily/FR induced large amounts of IFN. Western blot analysis of the lysates of virus-infected cells with an antibody directed against the viral NP (Fig. 1B) confirmed that the cells were infected to similar levels.
FIG. 1.
IFN-α/β induction in THOV-infected cells. 293 cells were infected with 3 PFU of THOV strain IIA (Africa), PoTi503 (Portugal), or Sicily/FR per cell or left untreated. At 24 h postinfection, supernatants of the cells were harvested and RNA was isolated from the cells. (A) Accumulation of IFN-α/β in virus-depleted supernatants was determined. (B) Replication of the different THOV strains was monitored by detection of NP accumulation by Western blot analysis of the cell lysates. The RNA was analyzed by RT-PCR using primer pairs specific for IFN-β (C) or GAPDH (D). The PCR products were analyzed by electrophoresis on an ethidium bromide-stained agarose gel. hu, human.
To investigate whether the difference in IFN production was on the transcriptional level, we determined the accumulation of IFN-β transcripts. Total RNA was isolated from 293 cells infected with the different THOV strains 24 h postinfection, and IFN-β transcripts, as well as transcripts of human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were amplified by reverse transcription (RT)-PCR with primers described previously (21, 31). Figure 1C shows that cells infected with strain Sicily/FR contained IFN-β transcripts, whereas cells infected with the THOV strains from Portugal and Africa had no detectable IFN-β mRNA. GAPDH mRNA could be amplified in all RT-PCRs, indicating that all of the preparations contained similar amounts of RNA (Fig. 1D). Thus, IFN-β secretion in response to THOV Sicily/FR infection correlated with the presence of IFN-β transcripts in infected cells, suggesting that the transcriptional activation of the gene for IFN-β is affected.
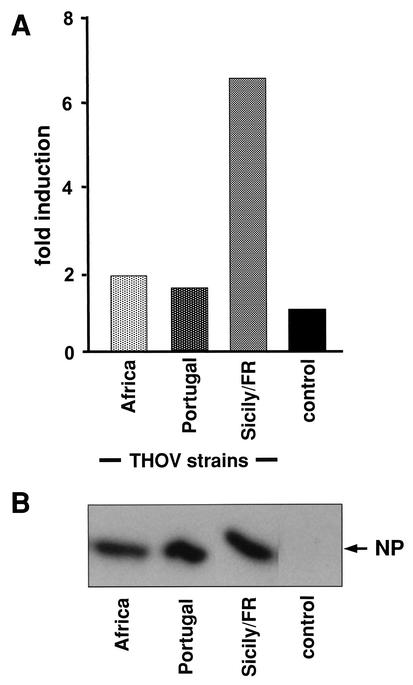
We directly determined activation of the IFN-β promoter by using a reporter construct coding for FF-Luc under the control of the IFN-β promoter (38). 293 cells were transfected with FF-Luc reporter plasmid and a control plasmid, pRL-SV40, coding for Renilla luciferase (REN-Luc) under the control of the constitutive simian virus 40 promoter. After 6 h, the cells were infected with 3 PFU of THOV per cell for 20 h and lysed and aliquots of the cell extracts were used to measure luciferase activities. Infection with strain Sicily/FR resulted in high FF-Luc activity, whereas the African and Portuguese strains showed much lower reporter activation (Fig. 2A). Western blot analysis confirmed equal infection of the cells by the three virus strains (Fig. 2B). These results demonstrate that the various THOV strains differ drastically in the ability to induce IFN-α/β gene expression in infected cells.
FIG. 2.
Induction of the IFN-β promoter in THOV-infected cells. 293 cells were transfected with an IFN-β promoter-FF-Luc plasmid together with a control plasmid encoding REN-Luc under the control of a constitutive promoter. At 6 h posttransfection, the cells were infected with 3 PFU of THOV strain IIA (Africa), PoTi503 (Portugal), or Sicily/FR per cell or left untreated. At 20 h postinfection, cells were lysed and analyzed for reporter gene expression (A) and accumulation of the viral NP (B) by Western blot analysis. FF-Luc activity was normalized to REN-Luc activity and is indicated as fold induction compared to that of the uninfected control.
THOV strain Sicily/FR is a mutant virus.
The sixth segment of THOV codes for two transcripts: a full-length mRNA and a spliced mRNA (19). The spliced transcript encodes the viral M protein, but the coding capacity of the unspliced transcript was unclear because its open reading frame was not terminated by a stop codon and no corresponding gene product could be detected in virus-infected cells. A recently discovered mechanism of mRNA surveillance called “nonstop mRNA decay“ (8, 32) may explain why there was no translation product of the unspliced mRNA.
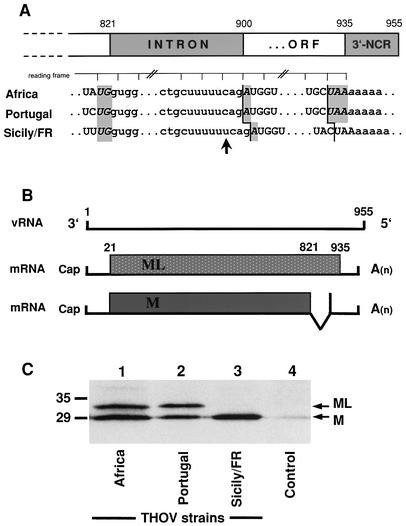
In order to analyze the unspliced transcript of the other THOV strains, we determined the sequences of the sixth segment of the three viruses by RT-PCR as described previously (33). Sequence analysis showed a high degree of similarity among the three viruses. The sequence of Sicily/FR differed from that of the Portuguese and African viruses by only 6.5 and 10%, respectively (data not shown). The predicted amino acid sequence of the M protein that is encoded by the spliced mRNA of the Sicily/FR strain showed even greater similarity, with 100 and 97.4% identity to the Portuguese and African strains, respectively. The intron region of segment 6, however, displayed an unexpected difference: while strain Sicily/FR contained a stretch of six U nucleotides within the intron region at positions 893 to 898, as expected (19), this region contained only five U nucleotides in the Portuguese and African strains (Fig. 3A). This lack of one U results in a frame shift that creates a terminated reading frame on the unspliced transcript. It potentially codes for a protein with a predicted molecular mass of 32 kDa that corresponds to an M protein with a C-terminal extension of 38 amino acids (Fig. 3B).
FIG. 3.
Wild-type THOV segment 6 encodes two proteins. (A) Nucleotide sequence alignment of regions of segment 6 from THOV strains IIA (Africa), PoTi503 (Portugal), and Sicily/FR. The intron sequence is indicated by lowercase letters. The stop codons UGA and UAA of the spliced and unspliced mRNAs, respectively, are indicated by gray boxes. The ruler at the top indicates the reading frame of the unspliced transcript. The arrow marks the additional uridine residue in the Sicilian strain sequence. (B) Coding strategy of segment 6. The genomic viral RNA is transcribed into a full-length transcript and a spliced transcript. The spliced mRNA codes for M, and the unspliced mRNA codes for an M protein with a C-terminal extension of 38 amino acids called ML. (C) Western blot analysis of the M proteins of the different THOV strains. Lysates of virus-infected cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blot analysis with M-specific antiserum. Positions of molecular mass markers (sizes are in kilodaltons) are indicated on the left, and the positions of the viral M and ML proteins are indicated on the right. ORF, open reading frame; 3′-NCR, 3′ noncoding region.
We tested the abilities of the African and Portuguese isolates to express this protein by Western blot analysis of infected cells using M-specific antibodies. Indeed, we detected an additional 32-kDa protein in cells infected with the African or Portuguese strain but not in cells infected with strain Sicily/FR (Fig. 3C). Since this protein is a C-terminally elongated version of M, we decided to call it ML (for matrix protein long). A database search showed that the unique region of ML does not contain any known functional motifs such as transmembrane domains or dsRNA-binding motifs.
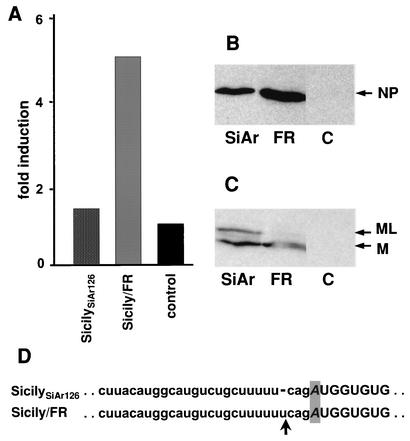
To examine the origin of the sequence variation in the sixth segment, we compared strain Sicily/FR to the original Sicilian THOV isolate, SiAr126, which was kindly provided by Robert E. Shope, Reference Center for Arboviruses, University of Texas Medical Branch at Galveston. By Western blot analysis using the M-specific antibody, we detected only one gene product in strain Sicily/FR-infected cells, as expected (19), whereas strain SiAr126 showed a second protein band corresponding to ML (Fig. 4C). The nucleotide sequence of the corresponding segments was determined. No other difference was found between the sixth segments of the two Sicilian strains, except for the additional U in the intron region of the Sicily/FR virus (Fig. 4D) that destroys the reading frame of the ML transcript. In a test for activation of the IFN-β promoter, strain SiAr126 did not activate IFN-β promoter activity, similar to the Portuguese and African isolates, whereas strain Sicily/FR was a good IFN inducer, as expected (Fig. 4A). Western blot analysis of NP production showed slightly higher expression of the viral NP in Sicily/FR-infected cells (Fig. 4B), although the cells were infected with the same amount of virus.
FIG. 4.
THOV Sicily/FR is a laboratory-adapted mutant of SiAr126. 293 cells were transfected with an IFN-β promoter-FF-Luc reporter plasmid together with a REN-Luc expression plasmid under the control of a constitutive promoter. At 6 h posttransfection, cells were infected with 3 PFU of THOV SiAr126 or Sicily/FR per cell or left untreated. At 20 h postinfection, cells were lysed and analyzed for reporter gene expression. (A) FF-Luc activity was normalized to REN-Luc activity and is indicated as fold induction compared to that of the uninfected control. Western blot analysis of the cell lysates was performed to detect accumulation of the viral NP (B) and the viral matrix proteins (M and ML) (C). (D) Nucleotide sequences of part of the sixth segment of the two Sicilian variants as determined after RT-PCR amplification from total RNA isolated from infected-cell lysates. The intron sequence is indicated by lowercase letters. The adenine nucleotide representing the third nucleotide of the M stop codon at the splice acceptor site is indicated by a gray box, and the arrow marks the additional uridine residue in the Sicily/FR sequence.
A sequence comparison of the two Sicilian variants indicates that the Sicily/FR strain has acquired the additional U nucleotide during repeated cell culture passage in our laboratory. The stretch of five U nucleotides within the intron region of segment 6 resembles the polyadenylation signal at the 5′ end of the THOV genomic segments (37). We speculate that the additional U may have been introduced by stuttering of the viral RNA polymerase during replication, as previously described for the polyadenylation reaction of orthomyxoviruses (29, 39).
ML is an IFN antagonist.
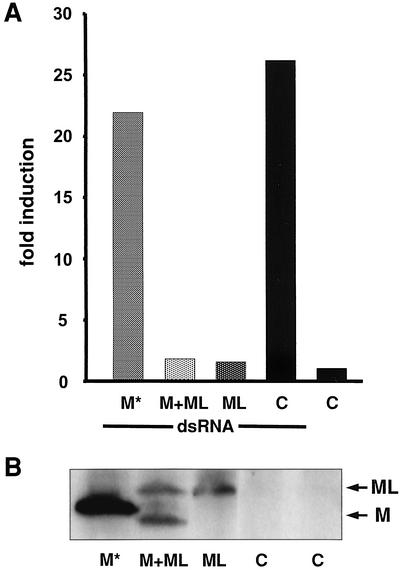
To investigate whether the ML protein is responsible for the observed differences in IFN production in THOV-infected cells, we tested the effect of recombinant ML on IFN-β promoter activation in a virus-free system. cDNAs encoding either M or ML were cloned into eukaryotic expression vector pCAGGS.MCS under the control of the chicken β-actin promoter (24). For the expression of M, we constructed pCAGGS-M-HA, containing a cDNA sequence that corresponds to the spliced transcript of Sicily/FR segment 6. For the expression of ML, we constructed a full-length cDNA of Sicily/FR segment 6 in which one of the six T nucleotides in the intron was deleted by in vitro mutagenesis in order to restore the open reading frame of ML. The resulting construct, pCAGGS-ML, encodes both M and ML. For expression of ML only, we destroyed the splice acceptor site around position 900 in the ML open reading frame, resulting in plasmid pCAGGS-MLΔSA. Efficient expression of the recombinant proteins in transfected 293 cells was confirmed by Western blot analysis with an M-specific antibody. As expected, viral proteins M and ML were both expressed from pCAGGS-ML (Fig. 5B, M+ML) while only the ML protein was detected in cell lysates after transfection of pCAGGS-MLΔSA (Fig. 5B, ML). pCAGGS-M-HA encoded a recombinant M protein with a slightly higher molecular weight because it contained a C-terminal hemagglutinin tag of nine amino acids (Fig. 5B, M). To analyze the IFN-antagonistic activity of the recombinant proteins, cells were transfected with the different M and ML expression plasmids and treated with poly(I · C) for 24 h. Figure 5A shows specific activation of the IFN-β promoter by poly(I · C) in mock-transfected cells. The induction of the IFN-β promoter was not influenced by expression of M, but the expression of ML drastically reduced promoter activation, independently of whether ML was expressed alone or together with M. These data suggest that ML is an IFN antagonist and that it is the expression of ML by the African and Portuguese isolates of THOV that is responsible for the low induction of IFN.
FIG. 5.
The ML protein of THOV suppresses the dsRNA-mediated induction of the IFN-β promoter. 293 cells were transfected with an IFN-β promoter reporter plasmid together with a REN-Luc expression plasmid under the control of a constitutive promoter. Expression plasmids pCAGGS-M-HA, -M5xT, and -M5xTΔSA, encoding hemagglutinin-tagged M (M*), M and ML, or ML only, were cotransfected. As a control, the empty plasmid, C, was transfected. At 24 h posttransfection, the cells were either treated with poly(I · C) or left untreated for another 24 h. Cell lysates were then prepared and analyzed for reporter gene expression by luciferase activity assays (A) and for accumulation of the viral matrix proteins (M and ML) by Western blot analysis (B). FF-Luc activity was normalized to REN-Luc activity and is indicated as fold induction compared to that of the untreated control.
ML is sufficient to suppress THOV-induced IFN production.
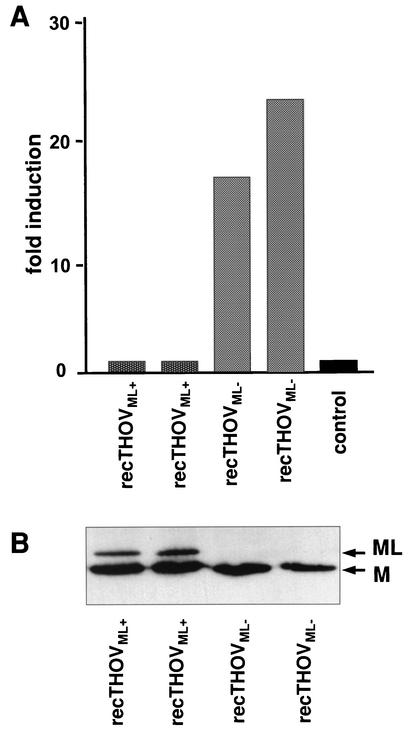
In order to exclude the influence of other, unknown genetic differences between THOV strains on the observed effects, we generated recombinant THOVs (recTHOV) that differ only in a single U nucleotide in the intron region of segment 6 (33). The absence or presence of the additional U determines whether the virus is able to express ML (recTHOVML+) or not (recTHOVML−). Western blot analysis of two independently generated recombinant virus clones of each genotype confirmed that recTHOVML+, but not recTHOVML−, has the ability to produce ML (Fig. 6B). The ability of the two viruses to activate the IFN-β promoter was determined in reporter assays. recTHOVML− strongly activated the IFN-β promoter, just like the Sicily/FR strain, whereas infection with recTHOVML+ did not lead to promoter activation (Fig. 6A). This result confirms that ML is a potent IFN antagonist that enables the virus to replicate without inducing IFN-α/β gene expression.
FIG. 6.
A recombinant THOV with a defective gene for ML activates IFN expression. 293 cells were transfected with a reporter plasmid encoding FF-Luc under the control of the IFN-β promoter together with the REN-Luc expression plasmid. At 6 h posttransfection, the cells were infected with 3 PFU of two independent clones of each recombinant THOV expressing ML (recTHOVML+) or not expressing ML (recTHOVML−) or left untreated. At 20 h postinfection, cells were lysed and analyzed for reporter gene expression. (A) FF-Luc activity normalized to REN-Luc activity and indicated as fold induction compared to that of the uninfected control. (B) Western blot analysis of virus particles isolated from cell supernatants performed to detect accumulation of viral matrix proteins M and ML.
In summary, we have identified a new Thogoto viral protein, ML, that is able to suppress the induction of IFN-α/β in order to evade the innate immune response. Thus, THOV ML can be added to the growing list of viral proteins that have been characterized as suppressors of IFN-α/β induction (9, 11), like the E3L protein of vaccinia virus (4), the NS1 proteins of influenza viruses (10, 31), VP35 of Ebola virus (2), or the NSs proteins of Bunyaviruses (3, 36).
The genetic organization of THOV is very similar to that of influenza viruses, except that the coding capacity is limited to six RNA segments. Previous studies identified six structural proteins to be sufficient for THOV replication (33, 34). In contrast to FLUAV, THOV lacks the coding capacity for additional proteins like the NS1 IFN antagonist, the NS2/NEP that acts as a nuclear export factor (23, 26), or an M2-like ion channel. Instead, THOV has been found to express one additional gene product by extension of the viral M protein. The recent identification of as yet unknown viral gene products in the genomes of orthomyxoviruses, like the CM2 protein of influenza C virus (15, 27), the PB1-F2 protein of FLUAV (5), or the ML protein of THOV (this study), has revealed an unexpectedly high coding capacity of the orthomyxovirus genome. The two recombinant THOVs that differ only in the capacity to express the ML gene product will be the basis for future studies to elucidate the role of this protein in virus multiplication and pathogenesis.
Nucleotide sequence accession numbers.
The nucleotide sequences of segment 6 of the different THOV strains reported in this paper have been submitted to the GenBank database and assigned accession numbers AF236794 for Sicily/FR, AF527529 for the African isolate (IIA), AF527530 for the Portuguese isolate (PoTi503), and AF527531 for the Sicilian isolate (SiAr126).
Acknowledgments
K.H. and S.J. contributed equally to this work.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Ko 1579/3-5) to G.K.
We thank Otto Haller for constant support and suggestions and Michael Frese, Othmar Engelhardt, and Peter Staeheli for discussions and critical comments on the manuscript. We are grateful to Patricia A. Nuttall, Armindo R. Filipe, and Robert E. Shope for providing isolates of THOV and to Takashi Fujita and Stephen Goodbourn for reporter plasmids.
REFERENCES
- 1.Albanese, M., C. Bruno-Smiraglia, G. Di Cuonzo, A. Lavagnino, and S. Srihongse. 1972. Isolation of Thogoto virus from Rhipicephalus bursa ticks in western Sicily. Acta Virol. 16:267.. [PubMed] [Google Scholar]
- 2.Basler, C. F., X. Wang, E. Mühlberger, V. Volchkov, J. Paragas, H.-D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouloy, M., C. Janzen, P. Vialat, H. Khun, J. Pavlovic, M. Huerre, and O. Haller. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, H.-W., L. H. Uribe, and B. L. Jacobs. 1995. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J. Virol. 69:6605-6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 6.Clerx, J. P. M., F. Fuller, and D. H. Bishop. 1983. Tick-borne viruses structurally similar to orthomyxoviruses. Virology 127:205-219. [DOI] [PubMed] [Google Scholar]
- 7.Filipe, A. R., and C. H. Calisher. 1984. Isolation of Thogoto virus from ticks in Portugal. Acta Virol. 28:152-155. [PubMed] [Google Scholar]
- 8.Frischmeyer, P. A., A. van Hoof, K. O'Donnell, A. L. Guerrerio, R. Parker, and H. C. Dietz. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295:2258-2261. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 11.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 12.Haig, D. A., J. Woodall, and D. Danskin. 1965. Thogoto virus: a hitherto undescribed agent isolated from ticks in Kenya. J. Gen. Microbiol. 38:389-394. [DOI] [PubMed] [Google Scholar]
- 13.Haller, O., M. Frese, D. Rost, P. A. Nuttall, and G. Kochs. 1995. Tick-borne Thogoto virus infection in mice is inhibited by the orthomyxovirus resistance gene product Mx1. J. Virol. 69:2596-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haller, O., and G. Kochs. 2002. Thogotovirus, p. 615-619. In C. A. Tidona and G. Darai (ed.), The Springer index of viruses. Springer Verlag GmbH, Berlin, Germany.
- 15.Hongo, S., K. Sugawara, Y. Muraki, Y. Matsuzaki, E. Takashita, F. Kitame, and K. Nakamura. 1999. Influenza C virus CM2 protein is produced from a 374-amino-acid protein (P42) by signal peptidase cleavage. J. Virol. 73:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isaacs, A., and J. Lindenmann. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. 147:258-267. [PubMed] [Google Scholar]
- 17.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 18.King, P., and S. Goodbourn. 1998. STAT1 is inactivated by a caspase. J. Biol. Chem. 273:8699-8704. [DOI] [PubMed] [Google Scholar]
- 19.Kochs, G., F. Weber, S. Gruber, A. Delvendahl, C. Leitz, and O. Haller. 2000. Thogoto virus matrix protein is encoded by a spliced mRNA. J. Virol. 74:10785-10789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 21.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct IFN-alpha genes by positive feedback through IFN regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller, U., U. Steinhoff, L. F. L. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 23.Neumann, G., M. T. Hughes, and Y. Kawaoka. 2000. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 19:6751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niwa, H., K. Yamamura, and J. Miyazali. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 25.Nuttall, P. A., M. A. Morse, L. D. Jones, and A. Portela. 1995. Adaptation of members of the Orthomyxoviridae family to transmission by ticks, p. 416-425. In A. J. Gibbs, C. H. Calisher, and F. Garcia-Arenal (ed.), Molecular basis of virus evolution. Cambridge University Press, Cambridge, England.
- 26.O'Neill, R., J. Talon, and P. Palese. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17:288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pekosz, A., and R. A. Lamb. 1998. Influenza C virus CM2 integral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence. Proc. Natl. Acad. Sci. USA 95:13233-13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pringle, C. R. 1996. Virus taxonomy 1996—a bulletin from the Xth International Congress of Virology in Jerusalem. Arch. Virol. 141:2251-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson, J. S., M. Schubert, and R. M. Lazzarini. 1981. Polyadenylation sites for influenza virus mRNA. J. Virol. 38:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staeheli, P., O. Haller, W. Boll, J. Lindenmann, and C. Weissmann. 1986. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44:147-158. [DOI] [PubMed] [Google Scholar]
- 31.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Hoof, A., P. A. Frischmeyer, H. C. Dietz, and R. Parker. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295:2262-2264. [DOI] [PubMed] [Google Scholar]
- 33.Wagner, E., O. G. Engelhardt, S. Gruber, O. Haller, and G. Kochs. 2001. Rescue of recombinant Thogoto virus from cloned cDNA. J. Virol. 75:9282-9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner, E., O. G. Engelhardt, F. Weber, O. Haller, and G. Kochs. 2000. Formation of virus-like particles from cloned cDNA of Thogoto virus. J. Gen. Virol. 81:2849-2853. [DOI] [PubMed] [Google Scholar]
- 35.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents the activation of NFκB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber, F., O. Haller, and G. Kochs. 1996. Nucleoprotein viral RNA and mRNA of Thogoto virus: a novel “cap-stealing” mechanism in tick-borne orthomyxoviruses? J. Virol. 70:8361-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the IFN-alpha/beta system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng, H., H. A. Lee, P. Palese, and A. García-Sastre. 1999. Influenza A virus RNA polymerase has the ability to stutter at the polyadenylation site of a viral RNA template during RNA replication. J. Virol. 73:5240-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]