Abstract
Earlier studies have shown that translation elongation factor 1δ (EF-1δ) is hyperphosphorylated in various mammalian cells infected with representative alpha-, beta-, and gammaherpesviruses and that the modification is mediated by conserved viral protein kinases encoded by herpesviruses, including UL13 of herpes simplex virus type 1 (HSV-1), UL97 of human cytomegalovirus, and BGLF4 of Epstein-Barr virus (EBV). In the present study, we attempted to identify the site in EF-1δ associated with the hyperphosphorylation by the herpesvirus protein kinases. Our results are as follows: (i) not only in infected cells but also in uninfected cells, replacement of the serine residue at position 133 (Ser-133) of EF-1δ by alanine precluded the posttranslational processing of EF-1δ, which corresponds to the hyperphosphorylation. (ii) A purified chimeric protein consisting of maltose binding protein (MBP) fused to a domain of EF-1δ containing Ser-133 (MBP-EFWt) is specifically phosphorylated in in vitro kinase assays by purified recombinant UL13 fused to glutathione S-transferase (GST) expressed in the baculovirus system. In contrast, the level of phosphorylation by the recombinant UL13 of MBP-EFWt carrying an alanine replacement of Ser-133 (MBP-EFS133A) was greatly impaired. (iii) MBP-EFWt is also specifically phosphorylated in vitro by purified recombinant BGLF4 fused to GST expressed in the baculovirus system, and the level of phosphorylation of MBP-EFS133A by the recombinant BGLF4 was greatly reduced. (iv) The sequence flanking Ser-133 of EF-1δ completely matches the consensus phosphorylation site for a cellular protein kinase, cdc2, and in vitro kinase assays revealed that purified cdc2 phosphorylates Ser-133 of EF-1δ. (v) As observed with EF-1δ, the casein kinase II β subunit (CKIIβ) was specifically phosphorylated by UL13 in vitro, while the level of phosphorylation of CKIIβ by UL13 was greatly diminished when a serine residue at position 209, which has been reported to be phosphorylated by cdc2, was replaced with alanine. These results indicate that the conserved protein kinases encoded by herpesviruses and a cellular protein kinase, cdc2, have the ability to target the same amino acid residues for phosphorylation. Our results raise the possibility that the viral protein kinases mimic cdc2 in infected cells.
The family Herpesviridae can be divided into three subfamilies (the Alphaherpesvirinae, the Betaherpesvirinae, and the Gammaherpesvirinae), and to date, approximately 130 herpesviruses have been identified from various animal species (41). Despite the wide range of biological properties and pathogenic manifestations of the herpesviruses, their genomes contain a significant number of conserved genes (41). This conservation suggests that the products of these genes play essential roles in the life cycle of herpesviruses. Herpesviruses contain viral genes that are predicted to encode protein kinases (4, 44). Among them, a subset exemplified by UL13 of herpes simplex virus type 1 (HSV-1) is conserved in all of the Herpesviridae (4, 44). Conceivably herpesviruses universally utilize their products both to regulate their own replicative processes and to modify cellular machinery through the phosphorylation of target viral and cellular proteins.
HSV-1 UL13, a subject of this study, is a serine/threonine protein kinase that is packaged into the virion (6, 34). Studies with UL13 mutants showed that the viral protein kinase affects the accumulation of an α protein, ICP0, and a subset of γ2 proteins, including UL26, UL26.5, UL38, UL41, and Us11 (37), suggesting that UL13 plays a role in viral gene expression in infected cells. Several lines of evidence listed below indicate that the function of UL13 is closely linked to that of the other viral regulatory proteins ICP22 and Us1.5, both of which are encoded by the α22 gene. First, ICP22 and Us1.5 are hypophosphorylated in cells infected with UL13 mutant viruses, suggesting that the UL13 protein kinase phosphorylates these viral regulatory proteins (38). Second, the phenotype of UL13 deletion mutants cannot be differentiated from that of ICP22 and Us1.5 deletion mutant viruses with respect to the accumulation of ICP0 and the subset of γ2 proteins (37). Third, both UL13 and ICP22 and Us1.5 are involved in the HSV-1-induced activation and modification of cellular enzymes, including the protein kinase cdc2 (1) and the large subunit of RNA polymerase II (22). Although this series of observations suggests that UL13 expresses its regulatory function by phosphorylating ICP22 and Us1.5, the direct linkage between phosphorylation of ICP22 and Us1.5 by UL13 and the proposed regulatory functions of these proteins described above remains to be determined.
Other than ICP22 and Us1.5, the phosphorylation and posttranslational processing of several viral proteins, including VP22, VP13 and -14, ICP0, and gE-gI complex, are affected in cells infected with UL13 mutant viruses (5, 6, 30, 33, 40), suggesting that these viral proteins are substrates of UL13 protein kinase. The phosphorylation of tegument proteins such as VP22 and VP13 and -14 by UL13 is implied to promote tegument disassembly in the early events of HSV-1 infection, which occur between the viral penetration of cells and the onset of viral protein synthesis (5, 28). Taken together, UL13 seems to play an important role in various stages of the HSV-1 life cycle by phosphorylating target proteins. In contrast to the wealth of knowledge regarding viral targets of UL13, little is known about cellular targets of UL13. The only cellular target of UL13 identified to date is cellular elongation factor 1δ (EF-1δ) (16), another subject of this study.
The relevant background regarding interaction between EF-1δ and UL13 includes the following: (i) EF-1δ is a subunit of EF-1, a complex of proteins that mediate the elongation of polypeptide chains during translation of mRNA (25, 27, 39, 45). EF-1α transports aminoacyl-tRNA for binding to ribosomes concurrent with hydrolysis of GTP, whereas EF-1δ is a component of the EF-1βγδ complex responsible for GDP-GTP exchange on EF-1α (25, 27, 39, 45). EF-1δ, therefore, plays a key role in protein synthesis by regulating the activity of EF-1α. EF-1δ is phosphorylated by several cellular protein kinases, including cdc2, casein kinase II (CKII), protein kinase C, and multipotential S6 kinase (3, 29, 35, 46). Studies on the phosphorylation of EF-1δ suggest that these modifications of the protein alter translational efficiency (3, 26, 46, 47).
(ii) The roots of this investigation rest on the observation that ICP0, a promiscuous transactivator, interacts with EF-1δ and that the carboxyl-terminal domain of ICP0 that binds to EF-1δ affects translational efficiency in vitro (12). In the course of studying the interaction between ICP0 and EF-1δ, we also found that HSV-1 infection induces extensive hyperphosphorylation of EF-1δ and that the viral protein kinase UL13 mediates the modification of EF-1δ (16). Since the amino acid sequence of UL13 protein kinase is conserved in all members of the Herpesviridae subfamilies (4, 44), it was predicted that the EF-1δ modification in infected cells is a conserved function that is expressed by all subfamilies of herpesviruses and that the conserved viral protein kinases universally mediate modification. This hypothesis is supported by the two lines of evidence listed below. First, representative alpha-, beta-, and gammaherpesviruses commonly induce EF-1δ modification in cells from various mammalian species (13). Second, human cytomegalovirus UL97 and Epstein-Barr virus (EBV) BGLF4, beta- and gammaherpesvirus homologues of UL13, mediate the hyperphosphorylation of EF-1δ (11, 13).
Based on the universality described above, it is conceivable that EF-1δ plays important and conserved roles in herpesvirus infection. However, the physiological function of the hyperphosphorylation of EF-1δ by the conserved herpesvirus protein kinases remains to be elucidated. As a first step toward clarifying this issue, we attempted to identify the site on EF-1δ responsible for the hyperphosphorylation of the protein mediated by HSV-1 UL13. In the present study, we report that (i) a serine residue at position 133 of EF-1δ (Ser-133) is responsible for posttranslational processing of the protein both in mock-infected cells and in cells infected with HSV-1, (ii) HSV-1 UL13, EBV BGLF4, and the cellular protein kinase cdc2 phosphorylate Ser-133 in vitro, and (iii) UL13 phosphorylates in vitro a serine residue at position 209 of the CKII β subunit (CKIIβ), which has been reported to be targeted by cdc2 for phosphorylation (21). These results indicated that the conserved protein kinases of herpesviruses and cdc2 have the ability to phosphorylate the same amino acid residues of target proteins.
MATERIALS AND METHODS
Cells and viruses.
The monkey kidney epithelial cell line COS-7 was maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum. Spodoptera frugiperda Sf9 cells were maintained in Sf-900 II (Invitrogen) supplemented with 10% fetal calf serum. A wild-type strain, HSV-1(F), and a UL13-negative recombinant virus, R7356, were kindly provided by B. Roizman. The properties of these viruses were described previously (9, 37).
Plasmids.
To construct pGEM-EF-1δ, the entire coding sequence of EF-1δ was amplified by PCR from pBH1003 (12) and inserted into pGEM3Z (Promega). To generate pGEM-EF-1δ(F), the oligonucleotide 5′-GCCCCGGGGACTACAAGGACGACGATGACAAGCCCGGGGC-3′, annealed with its complement, was digested with AvaI and inserted into the AvaI site of pGEM-EF-1δ. Then the EcoRI-XbaI fragment of pGEM-EF-1δ(F) was inserted into EcoRI and XbaI sites of pME18S to yield pME-EF-1δ(F). pME18S was kindly provided by K. Maruyama. Deletion mutants of pME-EF-1δ(F) were constructed by cloning into pME18S the DNA fragments amplified by PCR with appropriate primer pairs. pGEM-EF-1δS133A(F) or pGEM-EF-1δS133E(F) in which Ser-133 of EF-1δ was replaced with an alanine or glutamic acid codon, respectively, was generated using a QuikChange Site-Directed Mutagenesis kit with complementary oligonucleotides containing the specific nucleotide substitution according to the manufacturer's instructions (Stratagene). The EcoRI-XbaI fragment of pGEM-EF-1δS133A(F) or pGEM-EF-1δS133E(F) was inserted into the EcoRI and XbaI sites of pME18S to yield pME-EF-1δS133A(F) or pME-EF-1δS133E(F), respectively. pME-EF-1δS118/119A(F) in which serine codons at positions 118 and 119 of EF-1δ were replaced with alanine codons was constructed in the same way as pME-EF-1δS133A(F) or pME-EF-1δS133E(F). pBS-UL13(F) was constructed by cloning the entire coding sequence of HSV-1 UL13 amplified by PCR into pBS-Flag-Stop (14) in frame with the Flag epitope. The EcoRI-NotI fragment of pBS-UL13(F) was inserted into the EcoRI and NotI sites of pAcGHLT-A (Pharmingen) in frame with glutathione S-transferase (GST) to yield pAcGHLT-UL13. pAcGHLT-UL13K176 M in which the lysine codon at position 176 (Lys-176) was replaced with a methionine codon was generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene) as described above. To construct pMAL-EF-1δ or pMAL-EF-1δS133A, DNA fragments encoding EF-1δ codons 107 to 146 amplified by PCR from pGEM-EF-1δ(F) or pGEM-EF-1δS133A(F), respectively, was cloned into pMAL-c (New England BioLabs) in frame with maltose binding protein (MBP). pMAL-CKIIβ was constructed by cloning DNA fragments encoding the entire coding sequence of CKIIβ amplified from an EBV-transformed human peripheral blood lymphocyte cDNA library (Clontech) by PCR into pMAL-c in frame with MBP. pMAL-CKIIβS209A, in which the serine codon at position 209 (Ser-209) of CKIIβ was replaced with the alanine codon, was constructed using the QuikChange Site-Directed Mutagenesis kit (Stratagene) as described above.
Generation of recombinant baculoviruses.
Recombinant baculoviruses Bac-GST, Bac-GST-BGLF4, and Bac-GST-BGLF4K102I were described elsewhere (11; K. Kato et al., submitted for publication). To generate Bac-GST-UL13 or Bac-GST-UL13K176M, pAcGHL-UL13 or pAcGHL-UL13K176M was cotransfected with linearized baculovirus DNA BaculoGold (Pharmingen) into Sf9 cells using Lipofectin (Invitrogen) as described previously (17). The recombinant baculoviruses were propagated in Sf9 cells.
Purification of GST fusion protein from baculovirus-infected cells.
Sf9 cells (107) infected with each baculovirus (either Bac-GST-UL13, Bac-GST-UL13K176 M, Bac-GST-BGLF4, or Bac-GST-BGLF4K102I) in 2 ml of ice-cold buffer C (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P-40, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride) were lysed by sonication. After the insoluble materials were removed by centrifugation, the supernatants were reacted with 120 μl of a 50% slurry of glutathione-Sepharose beads (Amersham Pharmacia) for 2 h to overnight. The beads were extensively washed with buffer C, washed once with kinase buffer for UL13 (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 15 mM MgCl2, 0.1% Nonidet P-40, and 1 mM dithiothreitol) or for BGLF4 (50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 50 mM MgCl2, 0.1% Nonidet P-40, and 1 mM dithiothreitol), and were eluted in each kinase buffer containing 10 mM glutathione. The eluted proteins (GST-UL13, GST-UL13K176M, GST-BGLF4, and GST-BGLF4K102I) were used for further experiments.
Production and purification of MBP fusion proteins expressed in Escherichia coli.
MBP fusion proteins were expressed in E. coli XL-1 blue transformed with either pMAL-EF-1δ, pMAL-EF-1δS133A, pMAL-CKIIβ, or pMAL-CKIIβS209A and were purified on amylose resin (New England BioLabs) according to the same procedure used for the purification of GST fusion proteins expressed in E. coli (12), except that phosphate-buffered saline containing 1% Tween 20 was used instead of phosphate-buffered saline containing 1% Triton X-100.
In vitro kinase assays.
Purified MBP fusion proteins captured on amylose beads were rinsed twice with washing buffer (50 mM Tris-HCl [pH 8.0] and 1 mM dithiothreitol) and were subjected to in vitro kinase assays. The assays were performed to determine whether certain MBP fusion proteins could serve as substrates for GST-UL13, GST-BGLF4, or cdc2. cdc2 was purchased from New England BioLabs. Kinase buffer for UL13, BGLF4, or cdc2 (50 μl) containing 10 μM ATP, 10 μCi of [γ-32P]ATP, and purified GST fusion protein or cdc2 was added to the beads (15 μl) that had captured MBP fusion proteins, and samples were reacted for 30 min at 30°C. After incubation, the samples were extensively washed with TNE buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, and 1 mM EDTA) and subjected to electrophoresis on denaturing gels or phosphatase treatment. The gels were then stained with Coomassie brilliant blue (CBB) and exposed to X-ray film.
Roscovitine (Calbiochem) was used in certain in vitro kinase assays as follows. Kinase buffer for UL13 or cdc2 (50 μl) containing 10 μM ATP, 10 μCi of [γ-32P]ATP, purified GST fusion protein or cdc2, and 0, 6.5, or 26 μM roscovitine was added to the amylose beads (15 μl) that had captured MBP fusion proteins. Then the samples were processed as described above.
Phosphatase treatment.
After the in vitro kinase assays, the MBP fusion proteins captured on the beads were washed three times with TNE buffer and twice with buffer 2 (New England BioLabs). Then buffer 2 (50 μl) containing 50 U of alkaline phosphatase (New England BioLabs) was added to the beads, and the samples were incubated for 30 min at 37°C, after which they were processed as described above.
Transfection, infection, and immunoblotting.
COS-7 cells were transfected with appropriate expression plasmids according to the DEAE-dextran method (15). At 24 h posttransfection, transfected COS-7 cells were mock infected or infected with 10 PFU of HSV-1(F) or R7356 per cell. At 24 h after infection, COS-7 cells were harvested and subjected to immunoblotting with a rabbit polyclonal antibody to EF-1δ (12) or a mouse monoclonal antibody to Flag epitope (M2; Sigma) as described previously (17). The rabbit polyclonal antibody to UL13 used to detect GST-UL13 or GST-UL13K176 M was described elsewhere (7).
RESULTS
Mapping of the site of EF-1δ associated with the posttranslational modification of the protein in mock-infected or infected cells.
The objectives of the series of experiments in this section were to map the site on EF-1δ involved in the hyperphosphorylation of the protein mediated by HSV-1 UL13 in infected cells. As reported before, EF-1δ consists of two predominant forms, a hypophosphorylated form and a hyperphosphorylated form (12, 16, 29, 42). In denaturing gels, EF-1δ is detected as a fast-migrating form (apparent Mr of 38,000) and a slowly migrating form (apparent Mr of 40,000), both of which can be radiolabeled with 32Pi (16). The pattern of bands of EF-1δ radiolabeled by 32Pi in mock-infected or infected cells is exactly the same as that of EF-1δ detected by immunoblotting (16), indicating that the phosphorylation status of the protein can be monitored by immunoblotting.
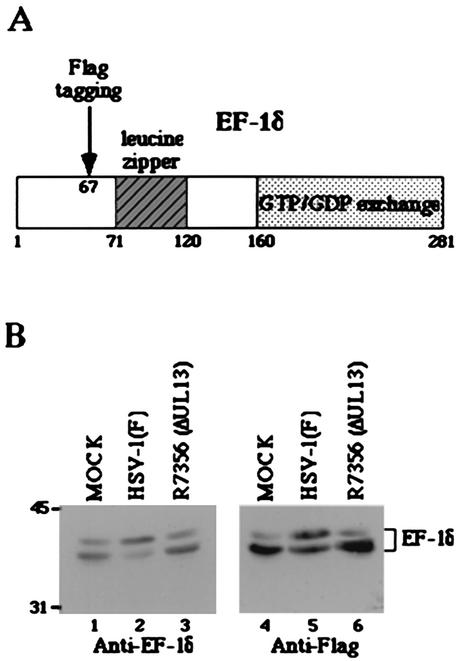
In the first series of experiments, we constructed an expression plasmid, pME-EF-1δ(F), in which EF-1δ is tagged with Flag epitope (Fig. 1A) and tested whether the transiently expressed recombinant EF-1δ (EF-1δ[F]) behaves like endogenous EF-1δ in COS-7 cells mock infected or infected with HSV-1(F). EF-1δ was tagged with Flag epitope to differentiate the mutants of EF-1δ(F) used in the mapping procedures from endogenous EF-1δ. COS-7 cells transfected with pME-EF-1δ(F) were mock infected or infected with 10 PFU of wild-type HSV-1(F) or UL13 deletion virus R7356 per cell, harvested at 24 h after infection, solubilized, electrophoretically separated on denaturing gels, and transferred to nitrocellulose sheets and reacted with the rabbit polyclonal antibody to EF-1δ or the monoclonal antibody to Flag epitope (M2). As shown in Fig. 1B (right panel), transiently expressed EF-1δ(F) harvested from COS-7 cells forms two predominant bands in the denaturing gel. In mock-infected cells, the fast-migrating band that corresponds to the hypophosphorylated form of EF-1δ was dominant (lane 4), whereas the relative amounts of protein in the slowly migrating band that corresponds to the hyperphosphorylated form of EF-1δ significantly increased after infection with wild-type HSV-1(F) (lane 5). Since UL13 mediates the hyperphosphorylation of EF-1δ, the electrophoretic pattern of EF-1δ in mock-infected cells cannot be differentiated from that in cells infected with UL13 deletion mutant virus R7356 (lane 6). These results were almost identical to what has been observed with endogenous EF-1δ (left panel), indicating that the behavior of the two isoforms of the recombinant EF-1δ(F) in mock-infected or infected cells was similar to that of endogenous EF-1δ and that EF-1δ (F) can be useful for the mapping procedures described below.
FIG. 1.
(A) Schematic diagram of the sequence of EF-1δ. The domain containing the leucine zipper motif or responsible for GTP/GDP exchange is shaded. The position for insertion of the Flag epitope is also shown. (B) Immunoblot of electrophoretically separated cell lysates from COS-7 cells mock infected or infected with 10 PFU of the indicated virus per cell. (Left panel) The infected COS-7 cells were harvested at 24 h after infection and subjected to immunoblotting with a rabbit antiserum raised against GST-EF-1δ. (Right panel) COS-7 cells were transfected with pME-EF-1δ(F). At 24 h after transfection, COS-7 cells were mock infected or infected with the virus indicated, incubated for an additional 24 h, and then harvested and subjected to immunoblotting with the mouse monoclonal antibody to the Flag epitope. Molecular weights (in thousands) are shown on the left.
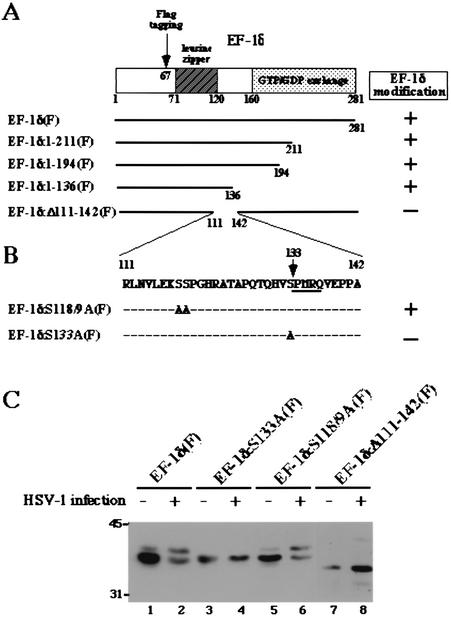
In the second series of experiments, we constructed a series of 3′ sequential deletion mutants and an internal deletion mutant of EF-1δ(F) (Fig. 2A) and tested whether the relative amount of the hyperphosphorylated form of these mutants increased after HSV-1 infection. The results were that the relative amount of hyperphosphorylated form of all 3′ deletion mutants of EF-1δ(F) increased after HSV-1 infection (Fig. 2A, data not shown). In contrast, an internal deletion mutant of EF-1δ, EF-1δ:Δ111-142(F), seemed not to be processed posttranslationally in mock-infected or infected cells, forming only a single band in the denaturing gel (Fig. 2C, lanes 7 and 8). These results suggest that the site of EF-1δ associated with hyperphosphorylation in both mock-infected and infected cells was located between codons 111 and 142 of the protein.
FIG. 2.
Mapping of the site of EF-1δ associated with posttranslational processing. (A) Schematic diagram of the sequence of EF-1δ. Deletion mutants of EF-1δ(F) are shown below. The levels of posttranslational processing induced by HSV-1 infection are also indicated. (B) Amino acid sequence of EF-1δ between codons 111 and 142. Alanine substitution mutants of EF-1δ(F) are shown below. The levels of posttranslational processing induced by HSV-1 infection are also indicated. (C) Immunoblot of electrophoretically separated cell lysates from COS-7 cells reacted with the monoclonal antibody to the Flag epitope. COS-7 cells were transfected with either pME-EF-1δ(F), pME-EF-1δS133A(F), pME-EF-1δS118/119A(F), or pME-EF-1δΔ111-142(F). At 24 h after transfection, COS-7 cells were mock infected or infected with wild-type HSV-1(F), incubated for an additional 24 h, and then harvested and subjected to immunoblotting. Molecular weights (in thousands) are shown on the left.
In the third series of experiments, we mutagenized pME-EF-1δ(F) by replacing with alanines the serine codons at positions 118 and 119 (Ser-118/119) or the single serine codon at position 133 (Ser-133) (Fig. 2B) and tested whether the mutants carrying amino acid substitutions are posttranslationally processed in mock-infected or infected cells. As shown in Fig. 2C, the expression profile of the two isoforms of mutant EF-1δ:S118/119A(F) cannot be differentiated from that of wild-type EF-1δ(F) in either mock-infected or infected cells (lanes 1, 2, 5, and 6), whereas another mutant, EF-1δ:S133A(F), was not posttranslationally processed in mock-infected or infected cells (lanes 3 and 4). These results indicated that Ser-133 is the site involved in the hyperphosphorylation of EF-1δ by the cellular protein kinase(s) in uninfected cells and by the viral protein kinase UL13 in infected cells. Interestingly the sequence flanking the identified site (SPMR) completely matches the consensus phosphorylation site for the cellular protein kinase cdc2 (SPX[R/K/H]) (23), and furthermore, it has been reported that the hyperphosphorylation of EF-1δ is mediated by cdc2 (29). These features led us to hypothesize that cdc2 and UL13 phosphorylate the same site, Ser-133, in EF-1δ.
Generation and purification of recombinant GST-UL13 and its kinase-negative mutant GST-UL13K176 M.
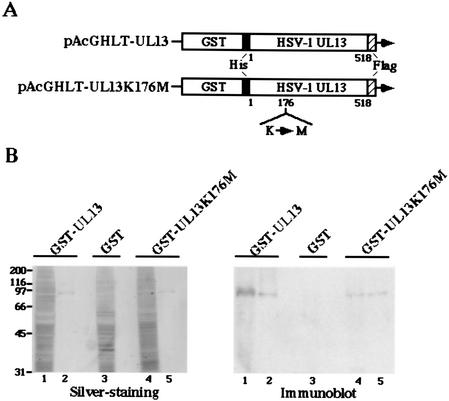
To test the hypothesis described above, a system to demonstrate the specific protein kinase activity of UL13 in vitro had to be developed. Because attempts to express the UL13 protein in a variety of prokaryotic cells or in insect cells were unsuccessful (31), all in vitro data concerning UL13 to date have been obtained with purified UL13 from infected cell lysate by immunoprecipitations using rabbit polyclonal antibody to UL13 (16, 30, 33). However, under these conditions, the possibility existed that the protein kinase activity detected in such experiments is responsible for contaminating the kinase(s), which could be physically associated with UL13 or fortuitously pulled down by the antibody. To overcome these problems, we constructed a recombinant baculovirus (Bac-GST-UL13) expressing UL13 fused to GST and purified GST-UL13 from Sf9 cells infected with Bac-GST-UL13 by using glutathione-Sepharose beads as described in Materials and Methods. To eliminate the possibility that the protein kinase activity detected in experiments using GST-UL13 is responsible for contaminating the kinase(s) during the purification procedures, we attempted to generate a mutant that has no intrinsic protein kinase activity but has probably retained its overall structure. To this end, we constructed a recombinant baculovirus (Bac-UL13K176M) expressing a mutant UL13 fused to GST (GST-UL13K176M) in which the Lys-176 of UL13 was replaced with methionine by site-directed mutagenesis. We chose Lys-176 for mutagenesis because it corresponds to an invariant lysine in known protein kinases (10) and because mutations of the corresponding lysine in UL13 homologues of other herpesviruses were shown to result in a loss or reduction of kinase activity (8, 18). The purified GST-UL13 and GST-UL13K176M were then electrophoretically separated in a denaturing gel and either silver stained or immunoblotted with rabbit antiserum against UL13. As shown in Fig. 3B, the purified GST-UL13 or GST-UL13K176M contained one major purified band with an Mr of approximately 90,000 as detected by silver staining (Fig. 3B, left panel) and these proteins were reacted with antiserum against UL13 (Fig. 3B, right panel). The purified GST-UL13 and GST-UL13K176M were also subjected to in vitro kinase assays to examine their enzymatic activity, and it was shown that the wild-type GST-UL13 was labeled with [γ-32P]ATP by autophosphorylation, while the mutant was not (data not shown). These results indicated that the enzymatically active GST-UL13 and its kinase-negative mutant GST-UL13K176M with a single amino acid substitution were successfully purified and that the Lys-176 in UL13 is required for kinase activity.
FIG. 3.
(A) Schematic diagram of the transfer plasmids pAcGHLT-UL13 and pAcGHLT-UL13K176M, used for construction of the recombinant baculoviruses Bac-GST-UL13 and Bac-GST-UL13K176M, respectively. (B) A silver-stained gel (left panel) and an immunoblot (right panel) of purified GST-UL13 (lane 2) or GST-UL13K176M (lane 5) from Sf9 cells infected with the recombinant virus Bac-GST-UL13 or Bac-GST-UL13K176M. Total cell extracts (lane 1 or 4) were subjected to affinity chromatography on glutathione-Sepharose and eluted (lane 2 or 5) as described in Materials and Methods. The proteins and total cell lysate from Sf9 cells infected with Bac-GST (lane 3) were separated on denaturing gels and subjected to silver staining (left panel) or transferred onto a nitrocellulose sheet and reacted with rabbit antiserum raised against UL13 (right panel). Molecular weights (in thousands) are shown on the left.
UL13 and cdc2 phosphorylate Ser-133 of EF-1δ in vitro.
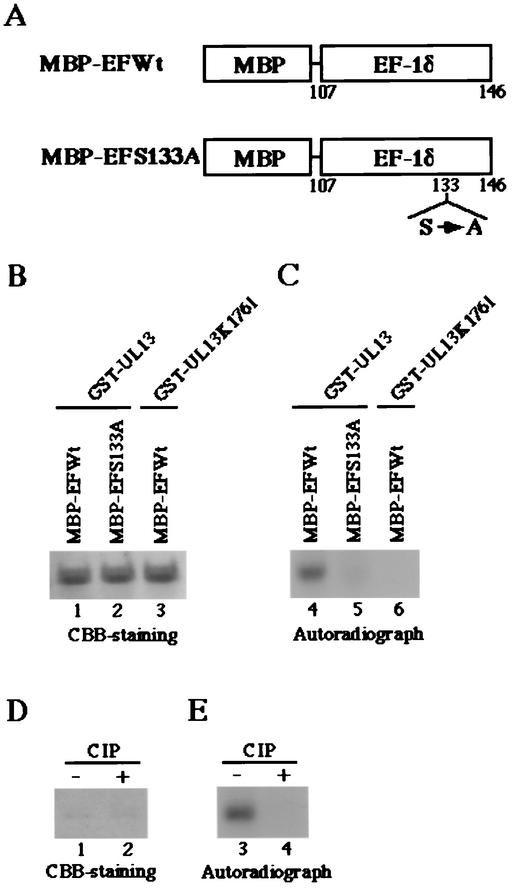
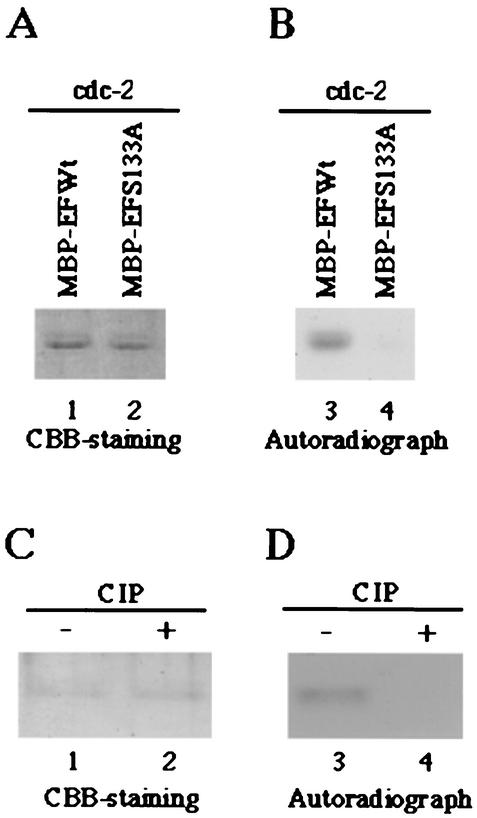
To test whether cdc2 and UL13 target Ser-133 in EF-1δ for phosphorylation, we generated and purified chimeric proteins consisting of MBP fused to an EF-1δ domain of amino acids 107 to 146 (MBP-EFWt) or to a mutated domain of EF-1δ in which Ser-133 was replaced with alanine (MBP-EFS133A) as described in Materials and Methods (Fig. 4A). These MBP fusion proteins captured on amylose resin served as substrates in in vitro kinase assays in the presence of the purified wild-type GST-UL13, the kinase-negative mutant GST-UL13K176 M, or cdc2.
FIG. 4.
Photographic and autoradiographic images of purified MBP fusion proteins subjected to in vitro kinase assay with the purified GST-UL13 or GST-UL13K176M. (A) Schematic representation of MBP fusion protein containing a domain of wild-type EF-1δ between codons 107 and 146 (MBP-EFWt) or a mutant of the domain with a replacement of Ser-133 by alanine (MBP-EFS133A). (B) The purified MBP-EFWt (lanes 1 and 3) or MBP-EFS133A (lane 2) was incubated in kinase buffer containing [γ-32P]ATP and the purified GST-UL13 (lanes 1 and 2) or GST-UL13K176M (lane 3), separated on a denaturing gel, and stained with CBB. (C) Autoradiograph of the gel described for panel B. (D) The purified MBP-EFWt incubated in kinase buffer containing [γ-32P]ATP and the purified GST-UL13, was mock treated (lane 1) or treated with alkaline phosphatase (CIP) (lane 2), separated on a denaturing gel, and stained with CBB. (E) Autoradiograph of the gel described for panel D.
The results were as follows: (i) In the autoradiographic image of the wild-type MBP-EFWt protein, which was incubated in kinase buffer containing GST-UL13 and [γ-32P]ATP, a protein band with an apparent Mr of 47,000 was clearly labeled (Fig. 4C, lane 4). By contrast, the level of labeling was greatly impaired when the mutant MBP-EFS133A protein was reacted with GST-UL13 (Fig. 4C, lane 5) and the MBP-EFWt protein was not labeled in the presence of the kinase-negative mutant GST-UL13K176 M (Fig. 4C, lane 6). The expression of each MBP fusion protein and the identity of the radiolabeled band of MBP-EFWt were verified by CBB staining as shown in Fig. 4B.
(ii) To examine whether the labeling of the MBP-EFWt protein in the presence of GST-UL13 was due to phosphorylation, the labeled MBP-EFWt was washed with TNE buffer to eliminate GST-UL13 and was then incubated with alkaline phosphatase. As shown in Fig. 4E, the labeling of MBP-EFWt by the reaction with UL13 was eliminated by phosphatase treatment, indicating that MBP-EFWt was labeled by phosphorylation. The expression of each MBP fusion protein and the identity of the radiolabeled band of MBP-EFWt were verified by CBB staining as shown in Fig. 4D.
(iii) The MBP-EFWt protein was labeled in the presence of purified cdc2 (Fig. 5B, lane 3), while the level of labeling was greatly reduced when MBP-EFS133A was reacted with the purified cdc2 (Fig. 5B, lane 4). The labeling of MBP-EFWt by the reaction with cdc2 was due to phosphorylation, based on the observation that the labeling was eliminated by phosphatase treatment (Fig. 5C and D).
FIG. 5.
Photographic and autoradiographic images of purified MBP fusion proteins subjected to in vitro kinase assay with the purified cdc2. (A) The purified MBP-EFWt (lane 1) or MBP-EFS133A (lane 2) was incubated in kinase buffer containing [γ-32P]ATP and the purified cdc2, separated on a denaturing gel, and stained with CBB. (B) Autoradiograph of the gel described for panel A. (C) The purified MBP-EFWt incubated in kinase buffer containing [γ-32P]ATP and the purified cdc2 was mock treated (lane 1) or treated with alkaline phosphatase (CIP) (lane 2), separated on a denaturing gel, and stained with CBB. (D) Autoradiograph of the gel described for panel C.
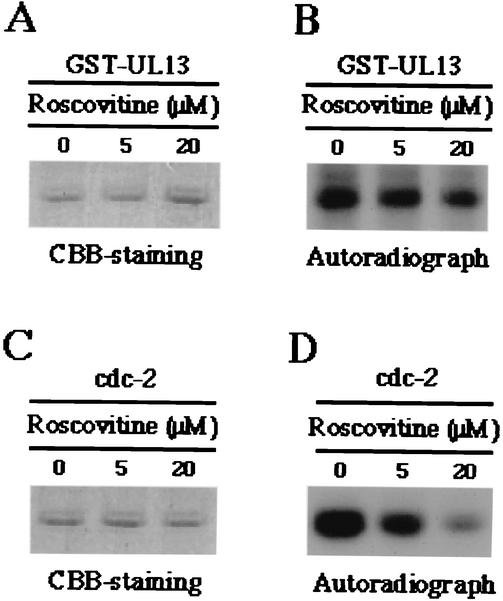
(iv) To further eliminate the possibility that the protein kinase activity detected using purified GST-UL13 is responsible for contaminating insect cdc2 during the purification procedure of GST-UL13, purified GST-UL13 or cdc2 was treated with roscovitine, a specific inhibitor of cdk2, cdc2, and cdk5 (24), followed by addition of the substrate (MBP-EFWt). The purified GST-UL13 or cdc2 was exposed to final concentration of roscovitine of 0, 5, or 20 μM (Fig. 6). The results shown in Fig. 6 indicated that the addition of roscovitine to the in vitro kinase reactions resulted in a dramatic decrease in activity of cdc2 in a dose-dependent manner (Fig. 6C and D) as reported previously (2, 24), while it had little effect on the activity of GST-UL13 (Fig. 6A and B).
FIG. 6.
Photographic and autographic images of the purified MBP fusion protein (MBP-EFWt) subjected to in vitro kinase assay with the purified GST-UL13 or cdc2 in the presence of various concentrations of roscovitine. (A) The purified MBP-EFWt was incubated in kinase buffer containing [γ-32P]ATP, the purified GST-UL13, and indicated concentration of roscovitine; separated on a denaturing gel; and stained with CBB. (B) Autoradiograph of the gel described for panel A. (C and D) Experiments were done exactly as described for panels A and B, respectively, except that the purified cdc2 was used instead of the purified GST-UL13.
These series of results indicated that UL13 and cdc2 specifically phosphorylate the same site, Ser-133, of EF-1δ in vitro.
EBV BGLF4 phosphorylates Ser-133 of EF-1δ in vitro.
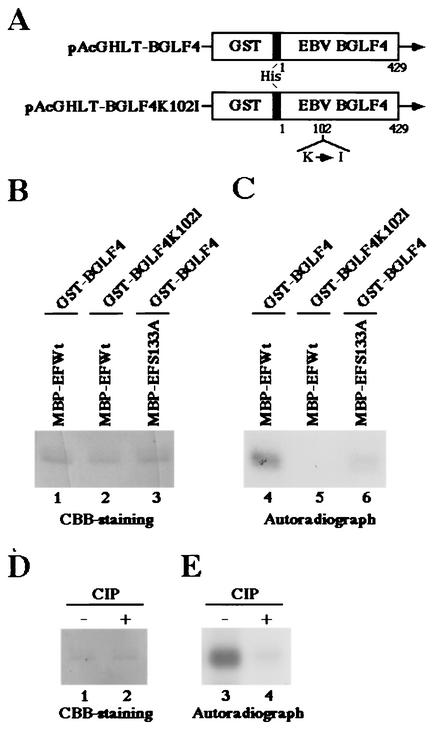
Previously we demonstrated that UL13 homologues of beta- and gammaherpesviruses also mediate the hyperphosphorylation of EF-1δ (11, 13). Taken together with the observations described above, it was hypothesized that the conserved viral protein kinases universally target Ser-133 of EF-1δ for phosphorylation like HSV-1 UL13. To test this hypothesis, we performed in vitro kinase assays using purified recombinant BGLF4, which is an EBV counterpart of UL13. The recombinant BGLF4, GST-BGLF4, and its kinase-negative mutant, GST-BGLF4K102I, were expressed by a baculovirus system and purified from infected Sf9 cells as described elsewhere (11; K. Kato et al., submitted). As shown in Fig. 7C, MBP-EFWt was labeled by the reaction with the purified GST-BGLF4 (lane 4) but not by the kinase-negative mutant GST-BGLF4K102I (lane 5). The level of labeling was greatly reduced when MBP-EFS133A protein was reacted with GST-BGLF4 (Fig. 7C, lane 6). Furthermore, the labeling of MBP-EFWt by the reaction with GST-BGLF4 was eliminated by phosphatase treatment (Fig. 7E). The expression of each MBP protein and the identity of the radiolabeled band were verified by CBB staining as shown in Fig. 7B and D. These results indicated that EBV BGLF4 specifically phosphorylates Ser-133 of EF-1δ in vitro.
FIG. 7.
Photographic and autographic images of purified MBP fusion proteins subjected to in vitro kinase assay with the purified GST-BGLF4 or GST-BGLF4K102I. (A) Schematic diagram of the transfer plasmids pAcGHLT-BGLF4 and pAcGHLT-BGLF4K102I, used for construction of the recombinant baculoviruses Bac-GST-BGLF4 and Bac-GST-BGLF4K102I, respectively. (B) The purified MBP-EFWt (lanes 1 and 2) or MBP-EFS133A (lane 3) was incubated in kinase buffer containing [γ-32P]ATP and the purified GST-BGLF4 (lanes 1 and 3) or GST-BGLF4K102I (lane 2), separated on a denaturing gel, and stained with CBB. (C) Autoradiograph of the gel described for panel B. (D) The purified MBP-EFWt incubated in kinase buffer containing [γ-32P]ATP and the purified GST-BGLF4 was mock treated (lane 1) or treated with alkaline phosphatase (CIP) (lane 2), separated on a denaturing gel, and stained with CBB. (E) Autoradiograph of the gel described for panel D.
UL13 phosphorylates Ser-209 of CKIIβ, which is targeted by cdc2 for phosphorylation.
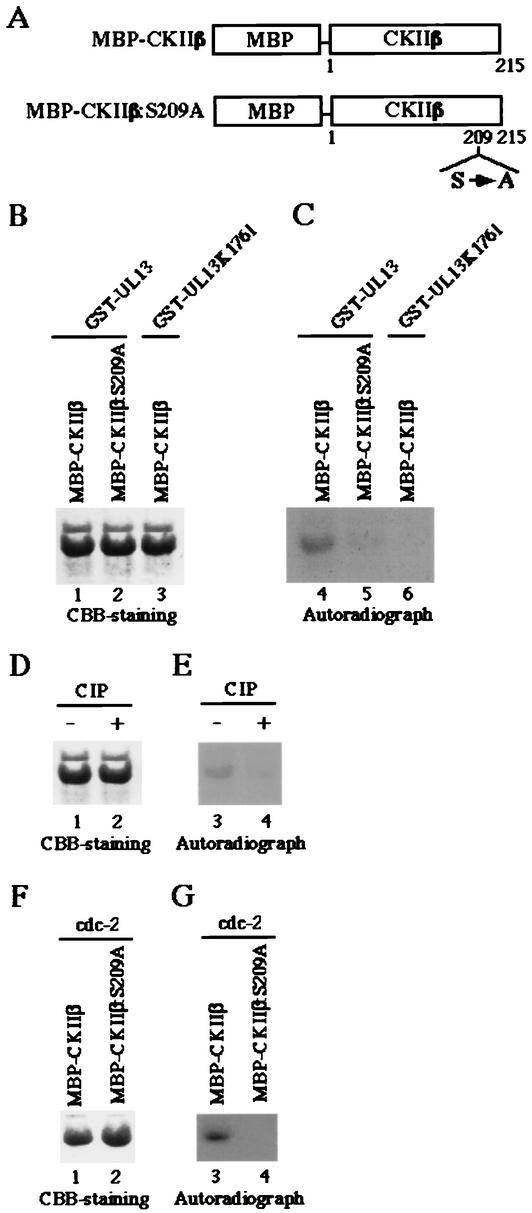
The observation that UL13 homologues and cdc2 target the same phosphorylation site of EF-1δ raised the possibility that UL13 homologues phosphorylate the same sites on other proteins targeted by cdc2. To test this hypothesis, we generated and purified chimeric protein consisting of MBP fused to the entire coding sequence of CKIIβ, which has been reported to be phosphorylated by cdc2 on a single serine residue at position 209 (Ser-209), or to a mutant of CKIIβ in which Ser-209 was replaced by alanine (Fig. 8A). These MBP fusion proteins (MBP-CKIIβ and MBP-CKIIβ:S209A) captured on amylose resin served as substrates in in vitro kinase assays using the purified GST-UL13, GST-UL13K176M, or cdc2.
FIG. 8.
(A) Schematic representation of MBP fusion protein containing a full-length wild-type CKIIβ (MBP-CKIIβ) or a mutant with a replacement of Ser-209 by alanine (MBP-CKIIβ:S209A). (B) The purified MBP-CKIIβ (lanes 1 and 3) or MBP-CKIIβ:S209A (lane 2) was incubated in kinase buffer containing [γ-32P]ATP and the purified GST-UL13 (lanes 1 and 2) or GST-UL13K176M (lane 3), separated on a denaturing gel, and stained with CBB. (C) Autoradiograph of the gel described for panel B. (D) The purified MBP-CKIIβ incubated in kinase buffer containing [γ-32P]ATP and the purified GST-UL13 was mock treated (lane 1) or treated with alkaline phosphatase (CIP) (lane 2), separated on a denaturing gel, and stained with CBB. (E) Autoradiograph of the gel described for panel D. (F) The purified MBP-CKIIβ (lane 1) or MBP-CKIIβ:S209A (lane 2) was incubated in kinase buffer containing [γ-32P]ATP and the purified cdc2, separated on a denaturing gel, and stained with CBB. (G) Autoradiograph of the gel described for panel F.
The results are as follows: (i) Consistent with previous reports (21), MBP-CKIIβ was phosphorylated by cdc2 (Fig. 8G, lane 3), while MBP-CKIIβ:S209A was not (Fig. 8G, lane 4), indicating that cdc2 specifically phosphorylates Ser-209 of CKIIβ. The expression of each MBP protein and the identity of the radiolabeled band were verified by CBB staining as shown in Fig. 8F.
(ii) In the reaction with purified GST-UL13, MBP-CKIIβ was labeled (Fig. 8C, lane 4), whereas it was not labeled by the kinase-negative mutant GST-UL13L176M (Fig. 8C, lane 6). The level of labeling was greatly reduced when MBP-CKIIβ:S209A was reacted with GST-UL13 (Fig. 8C, lane 5). Furthermore, the labeling of MBP-EFWt via the reaction with GST-BGLF4 was eliminated by phosphatase treatment (Fig. 8E). The expression of each MBP protein and the identity of the radiolabeled band were verified by CBB staining as shown in Fig. 8B and D.
Taken together, these results indicated that UL13 and cdc2 target the same serine residue of CKIIβ for phosphorylation as observed with EF-1δ.
DISCUSSION
Analyses of the function of HSV-1 UL13 have been hampered by difficulty in demonstrating specific protein kinase activity directly in vitro. The expression and purification of the enzymatically active recombinant UL13 using a variety of prokaryotic cells or in insect cells, which would greatly improve the in vitro assay of the activity of UL13, have been unsuccessful (31). Here we succeeded in developing a system to express large amounts of recombinant UL13 (GST-UL13) in insect cells using a recombinant baculovirus and to obtain highly purified UL13 with enzymatic activity. We also generated a kinase-negative mutant of UL13 (GST-UL13K176M) as a control to eliminate the possibility that the kinase activity detected using the purified recombinant UL13 is responsible for contaminating the kinase(s) during the purification. The use of purified recombinant UL13 and its mutant in in vitro kinase assays enabled us to examine the specific activity of UL13 and led to the key finding that the conserved protein kinases exemplified by HSV-1 UL13 and the cellular protein kinase cdc2 phosphorylate the same serine residues of target cellular proteins, including EF-1δ and CKIIβ.
The salient features of our results can be summarized as follows: (i) HSV-1 UL13 and cdc2 target the same serine residue (Ser-133) of EF-1δ for phosphorylation. In vitro kinase assays using the purified UL13 and cdc2 revealed that both protein kinases have the ability to phosphorylate Ser-133 of EF-1δ in vitro. Based on the following observations, it seems highly likely that Ser-133 of EF-1δ is in fact phosphorylated by UL13 and cdc2 in vivo. First, the protein kinase responsible for the hyperphosphorylation of EF-1δ in uninfected and infected cells has been reported to be cdc2 and UL13, respectively (16, 29). Here we observed that an amino acid substitution of Ser-133 of EF-1δ with alanine abolished the hyperphosphorylation of EF-1δ not only in cells infected with HSV-1 but also in uninfected cells. Second, a mutant of EF-1δ carrying a glutamic acid substitution for Ser-133, which is known to mimic constitutive phosphorylation (20, 49), appeared to form a conformation similar to the hyperphosphorylated form of EF-1δ, based on the observation that the electrophoretic mobility of the mutant is similar to that of the hyperphosphorylated form of EF-1δ in a denaturing gel (data not shown). Third, the flanking sequence of Ser-133 completely matches the consensus phosphorylation site for cdc2 (23) and it has been reported that cdc2 mediated the hyperphosphorylation of EF-1δ on a serine residue (29). This series of observations supports our hypothesis that HSV-1 UL13 and cdc2 phosphorylate the same serine residue (Ser-133) of EF-1δ not only in vitro but also in vivo.
(ii) One would argue that the defect of phosphorylation of MBP-EFS133A by UL13 and cdc2 protein kinases results from steric hindrance of EF-1δ caused by the replacement of Ser-133 by alanine. Here we also demonstrated that UL13 and cdc2 phosphorylate the cdc2 target site (Ser-209) of CKIIβ by the same mutational analyses as for EF-1δ. These results would eliminate the slim possibility that the replacement of Ser-133 by alanine by chance causes conformational change of EF-1δ and that the lack of phosphorylation of the mutated substrate by UL13 and cdc2 was due to steric hindrance of the target protein caused by the mutation.
(iii) Earlier studies have shown that representative UL13 homologues other than HSV-1 UL13 mediate the hyperphosphorylation of EF-1δ (11, 13). In this report, we showed that BGLF4, an EBV UL13 homologue, has the ability to phosphorylate Ser-133 of EF-1δ like HSV-1 UL13. These observations, by extension, suggest that the conserved herpesvirus protein kinases (UL13 homologues) and cdc2 universally target the same serine residue of EF-1δ.
(iv) The observation that UL13 homologues and cdc2 phosphorylate the same serine residue of EF-1δ raised the interesting possibility that the UL13 homologues mimic cdc2 in infected cells. If this is the case, UL13 homologues would have the ability to phosphorylate the same amino acid residues of other proteins targeted by cdc2. In the present study, we obtained evidence that this is in fact the case. In one instance, we specifically related the phosphorylation of CKIIβ to HSV-1 UL13. Thus, in vitro kinase assays revealed that HSV-1 UL13 phosphorylates Ser-209 of CKIIβ, reported to be the cdc2 phosphorylation site. Furthermore, our preliminary experiments demonstrated that EBV BGLF4 and cdc2 phosphorylate the same serine residue of an EBV regulatory protein, EBNA-LP, in vitro (K. Kato and Y. Kawaguchi, unpublished observation). These results eliminate the slim possibility that the conserved herpesvirus protein kinases by chance retained the motifs necessary to bind and phosphorylate the cdc2 target motif of EF-1δ.
Taken together, these results support our conclusion that the conserved protein kinases encoded by herpesviruses have the ability to phosphorylate the cdc2 recognition site of target proteins. Consistently, the potential viral substrates of HSV-1 UL13 reported to date, including ICP0, ICP22, UL47, gI, and gE, possess the consensus phosphorylation site of cdc2 (2).
The relevant issues can be summarized as follows: (i) cdc2 has been reported to be activated by HSV-1 infection and both UL13 and ICP22 expression is required for the effect (1). Advani et al. demonstrated that immunoprecipitates obtained with a mouse monoclonal antibody to cdc2 from cells infected with HSV-1 at 12 h postinfection possess much stronger protein kinase activity to phosphorylate histon H1 in vitro than do those from mock-infected cells. These results suggested that cdc2 was activated in cells infected with HSV-1. Although these studies were not able to eliminate completely the possibility that the kinase activity detected in these experiments is responsible for contaminating the kinase(s) that could be associated with cdc2 or fortuitously pulled down by the monoclonal antibody, their conclusion was supported by other observations that HSV-1 induces modification of cdc2 regulators, including wee-1 and cdc-25C, which is predicted to activate cdc2 (1). Consistent with this, Schang et al. reported that specific inhibitors known to block the activity of cdk2 and cdc2 reduced viral transcription and viral replication (43). Furthermore, studies with a dominant-negative mutant of cdc2 revealed that the activity of cdc2 regulates the expression of the γ2 protein Us11 (2). This series of observations indicates that cdc2 plays an important role in the life cycle of HSV-1. Here we reported a series of findings that raised the possibility that conserved herpesvirus protein kinases exemplified by UL13 mimic cdc2 in infected cells. One could argue that this is not necessary in HSV-1-infected cells because the infection activates cdc2. One explanation would be that UL13 makes up for the deficit of cdc2. cdc2 is known to target a variety of cellular proteins (32), and in addition, more than one-third of HSV-1 proteins possess a consensus target site of cdc2 (2). Since, in productively infected cells, large numbers of viral proteins are synthesized, it is conceivable that the activity of cdc2 is not strong enough even though it is stimulated by HSV-1 infection and though UL13 may cover the shortage. Alternatively, it is beneficial for the virus to mimic cdc2, independent of the condition of target host cells, because HSV-1 can infect resting cells and differentiated neural cells in vivo where the expression of cdc2 could be limited.
(ii) Other than HSV-1, VZV gI, EBV EBNA-LP, and HHV8 K-bZIP proteins are phosphorylated by cdc2 (19, 36, 48). Based on the observations made in this study, it is possible that these viral proteins are also phosphorylated by conserved protein kinases encoded by the respective herpesviruses.
In conclusion, we have provided the evidence that the conserved protein kinases encoded by herpesviruses and a cellular protein kinase, cdc2, have the ability to phosphorylate the same amino acid residues on target proteins. We should note, however, that it remains to be elucidated whether these observations in vitro in fact are valid in vivo. Further study to resolve this issue is of importance and presently under way in these laboratories.
Acknowledgments
We thank B. Roizman for the recombinant virus R7356 and HSV-1(F) and K. Maruyama for pME18S.
This study was supported in part by Grants for Scientific Research (Y.K. and Y.Y.) and Grants for Scientific Research in Priority Areas (Y.K. and Y.Y.) from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Advani, S. J., R. Brandimarti, R. R. Weichselbaum, and B. Roizman. 2000. The disappearance of cyclins A and B and the increase in activity of the G2/M-phase cellular kinase cdc2 in herpes simplex virus 1-infected cells require expression of the α22/US1.5 and UL13 viral genes. J. Virol. 74:8-15. [PMC free article] [PubMed] [Google Scholar]
- 2.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2000. The role of cdc2 in the expression of herpes simplex virus genes. Proc. Natl. Acad. Sci. USA 97:10996-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, Y. W., and J. A. Traugh. 1997. Phosphorylation of elongation factor 1 and ribosomal protein S6 by multipotential S6 kinase and insulin stimulation of translational elongation. J. Biol. Chem. 272:28252-28257. [DOI] [PubMed] [Google Scholar]
- 4.Chee, M. S., G. L. Lawrence, and B. G. Barrell. 1989. Alpha-, beta- and gammaherpesviruses encode a putative phosphotransferase. J. Gen. Virol. 70:1151-1160. [DOI] [PubMed] [Google Scholar]
- 5.Coulter, L. J., H. W. Moss, J. Lang, and D. J. McGeoch. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 74:387-395. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham, C., A. J. Davison, A. Dolan, M. C. Frame, D. J. McGeoch, D. M. Meredith, H. W. Moss, and A. C. Orr. 1992. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J. Gen. Virol. 73:303-311. [DOI] [PubMed] [Google Scholar]
- 7.Daikoku, T., S. Shibata, F. Goshima, S. Oshima, T. Tsurumi, H. Yamada, Y. Yamashita, and Y. Nishiyama. 1997. Purification and characterization of the protein kinase encoded by the UL13 gene of herpes simplex virus type 2. Virology 235:82-93. [DOI] [PubMed] [Google Scholar]
- 8.de Wind, N., J. Domen, and A. Berns. 1992. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J. Virol. 66:5200-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 10.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 11.Kato, K., Y. Kawaguchi, M. Tanaka, M. Igarashi, A. Yokoyama, G. Matsuda, M. Kanamori, K. Nakajima, Y. Nishimura, M. Shimojima, H. T. Phung, E. Takahashi, and K. Hirai. 2001. Epstein-Barr virus-encoded protein kinase BGLF4 mediates hyperphosphorylation of cellular elongation factor 1delta (EF-1delta): EF-1delta is universally modified by conserved protein kinases of herpesviruses in mammalian cells. J. Gen. Virol. 82:1457-1463. [DOI] [PubMed] [Google Scholar]
- 12.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi, Y., T. Matsumura, B. Roizman, and K. Hirai. 1999. Cellular elongation factor 1δ is modified in cells infected with representative alpha-, beta-, or gammaherpesviruses. J. Virol. 73:4456-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi, Y., K. Nakajima, M. Igarashi, T. Morita, M. Tanaka, M. Suzuki, A. Yokoyama, G. Matsuda, K. Kato, M. Kanamori, and K. Hirai. 2000. Interaction of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP) with HS1-associated protein X-1: implication of cytoplasmic function of EBNA-LP. J. Virol. 74:10104-10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1998. Eukaryotic elongation factor 1δ is hyperphosphorylated by the protein kinase encoded by the UL13 gene of herpes simplex virus 1. J. Virol. 72:1731-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenyon, T. K., J. Lynch, J. Hay, W. Ruyechan, and C. Grose. 2001. Varicella-zoster virus ORF47 protein serine kinase: characterization of a cloned, biologically active phosphotransferase and two viral substrates, ORF62 and ORF63. J. Virol. 75:8854-8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitay, M. K., and D. T. Rowe. 1996. Cell cycle stage-specific phosphorylation of the Epstein-Barr virus immortalization protein EBNA-LP. J. Virol. 70:7885-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J. S., K. M. Collins, A. L. Brown, C. H. Lee, and J. H. Chung. 2000. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404:201-204. [DOI] [PubMed] [Google Scholar]
- 21.Litchfield, D. W., F. J. Lozeman, M. F. Cicirelli, M. Harrylock, L. H. Ericsson, C. J. Piening, and E. G. Krebs. 1991. Phosphorylation of the beta subunit of casein kinase II in human A431 cells. Identification of the autophosphorylation site and a site phosphorylated by p34cdc2. J. Biol. Chem. 266:20380-20389. [PubMed] [Google Scholar]
- 22.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marin, O., F. Meggio, G. Draetta, and L. A. Pinna. 1992. The consensus sequences for cdc2 kinase and for casein kinase-2 are mutually incompatible. A study with peptides derived from the beta-subunit of casein kinase-2. FEBS Lett. 301:111-114. [DOI] [PubMed] [Google Scholar]
- 24.Meijer, L., A. Borgne, O. Mulner, J. P. Chong, J. J. Blow, N. Inagaki, M. Inagaki, J. G. Delcros, and J. P. Moulinoux. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243:527-536. [DOI] [PubMed] [Google Scholar]
- 25.Merrick, W. C. 1992. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 56:291-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minella, O., P. Cormier, J. Morales, R. Poulhe, R. Belle, and O. Mulner-Lorillon. 1994. cdc2 kinase sets a memory phosphorylation signal on elongation factor EF-1 delta during meiotic cell division, which perdures in early development. Cell. Mol. Biol. (Noisy-Le-Grand) 40:521-525. [PubMed] [Google Scholar]
- 27.Morales, J., P. Cormier, O. Mulner-Lorillon, R. Poulhe, and R. Belle. 1992. Molecular cloning of a new guanine nucleotide-exchange protein, EF1 delta. Nucleic Acids Res. 20:4091.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison, E. E., Y. F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulner-Lorillon, O., O. Minella, P. Cormier, J. P. Capony, J. C. Cavadore, J. Morales, R. Poulhe, and R. Belle. 1994. Elongation factor EF-1 delta, a new target for maturation-promoting factor in Xenopus oocytes. J. Biol. Chem. 269:20201-20207. [PubMed] [Google Scholar]
- 30.Ng, T. I., W. O. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37-48. [DOI] [PubMed] [Google Scholar]
- 31.Ng, T. I., C. Talarico, T. C. Burnette, K. Biron, and B. Roizman. 1996. Partial substitution of the functions of the herpes simplex virus 1 U(L)13 gene by the human cytomegalovirus U(L)97 gene. Virology 225:347-358. [DOI] [PubMed] [Google Scholar]
- 32.Nigg, E. A. 1993. Targets of cyclin-dependent protein kinases. Curr. Opin. Cell Biol. 5:187-193. [DOI] [PubMed] [Google Scholar]
- 33.Ogle, W. O., T. I. Ng, K. L. Carter, and B. Roizman. 1997. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology 235:406-413. [DOI] [PubMed] [Google Scholar]
- 34.Overton, H. A., D. J. McMillan, L. S. Klavinskis, L. Hope, A. J. Ritchie, and P. Wong-kai-in. 1992. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology 190:184-192. [DOI] [PubMed] [Google Scholar]
- 35.Palen, E., R. C. Venema, Y. W. Chang, and J. A. Traugh. 1994. GDP as a regulator of phosphorylation of elongation factor 1 by casein kinase II. Biochemistry 33:8515-8520. [DOI] [PubMed] [Google Scholar]
- 36.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc. Natl. Acad. Sci. USA 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riis, B., S. I. Rattan, B. F. Clark, and W. C. Merrick. 1990. Eukaryotic protein elongation factors. Trends Biochem. Sci. 15:420-424. [DOI] [PubMed] [Google Scholar]
- 40.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 41.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 42.Sanders, J., M. Brandsma, G. M. Janssen, J. Dijk, and W. Moller. 1996. Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1 beta gamma delta in the endoplasmic reticulum. J. Cell Sci. 109:1113-1117. [DOI] [PubMed] [Google Scholar]
- 43.Schang, L. M., A. Rosenberg, and P. A. Schaffer. 1999. Transcription of herpes simplex virus immediate-early and early genes is inhibited by roscovitine, an inhibitor specific for cellular cyclin-dependent kinases. J. Virol. 73:2161-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, R. F., and T. F. Smith. 1989. Identification of new protein kinase-related genes in three herpesviruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J. Virol. 63:450-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Damme, H. T., R. Amons, R. Karssies, C. J. Timmers, G. M. Janssen, and W. Moller. 1990. Elongation factor 1 beta of artemia: localization of functional sites and homology to elongation factor 1 delta. Biochim. Biophys. Acta 1050:241-247. [DOI] [PubMed] [Google Scholar]
- 46.Venema, R. C., H. I. Peters, and J. A. Traugh. 1991. Phosphorylation of elongation factor 1 (EF-1) and valyl-tRNA synthetase by protein kinase C and stimulation of EF-1 activity. J. Biol. Chem. 266:12574-12580. [PubMed] [Google Scholar]
- 47.Venema, R. C., H. I. Peters, and J. A. Traugh. 1991. Phosphorylation of valyl-tRNA synthetase and elongation factor 1 in response to phorbol esters is associated with stimulation of both activities. J. Biol. Chem. 266:11993-11998. [PubMed] [Google Scholar]
- 48.Ye, M., K. M. Duus, J. Peng, D. H. Price, and C. Grose. 1999. Varicella-zoster virus Fc receptor component gI is phosphorylated on its endodomain by a cyclin-dependent kinase. J. Virol. 73:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yokoyama, A., M. Tanaka, G. Matsuda, K. Kato, M. Kanamori, H. Kawasaki, H. Hirano, I. Kitabayashi, M. Ohki, K. Hirai, and Y. Kawaguchi. 2001. Identification of major phosphorylation sites of Epstein-Barr virus nuclear antigen leader protein (EBNA-LP): ability of EBNA-LP to induce latent membrane protein 1 cooperatively with EBNA-2 is regulated by phosphorylation. J. Virol. 75:5119-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]