Abstract
Bovine enteric caliciviruses (BoCVs) have been classified in the Norovirus (Norwalk-like virus) genus of the Caliciviridae, raising questions about zoonotic transmission and an animal reservoir for the human Norwalk-like viruses (NLVs), an important cause of nonbacterial gastroenteritis in humans. We examined the genetic relationship of human NLVs to BoCVs that were identified by using reverse transcription-PCR with primer pairs originally designed to detect human NLVs. Polymerase, capsid, and open reading frame 3 (ORF3) gene sequence analyses of BoCVs that were identified from 1976 to 2000 from throughout the United Kingdom showed that BoCVs formed a distinct third genogroup of closely related viruses distinct from the human genogroup I and II NLVs. Evidence was not obtained to support the concept that BoCVs are circulating in humans and pose a threat to human health.
Viruses classified in the family Caliciviridae have been identified in a number of animal species in association with a range of diseases (13, 32). In animals, caliciviruses are a well-established cause of respiratory, vesicular, and hemorrhagic diseases. In humans, caliciviruses have been associated only with enteric disease and are accepted worldwide as the most common cause of nonbacterial gastroenteritis in human adults (9, 12, 31). Virus particles resembling caliciviruses were identified in the enteric tract of cattle (16, 48), but little progress had been made with their characterization until recently (6, 7, 28, 45). Phylogenetic analyses of a small number of bovine enteric caliciviruses (BoCVs), all but one with limited nucleotide sequence, led Dastjerdi et al. (6), Liu et al. (28), and van der Poel et al. (45) to conclude that the BoCVs were distinct from the animal caliciviruses associated with respiratory, vesicular, and hemorrhagic diseases but genomically close to genogroup I of the Norwalk-like human enteric caliciviruses (NLVs). These observations raised questions of zoonotic transmission of BoCVs and an animal reservoir for human NLVs. Further phylogenetic analyses by Ando et al. (1) by using the partial genome sequence of Bo/Newbury2/76/UK (6) and the complete genome sequence of Bo/Jena/80/DE (28) suggested that the BoCVs belonged to a third genogroup in the Norovirus (Norwalk-like virus) genus. Transmission of animal caliciviruses between animal species is reported for the vesiviruses of swine (vesicular exanthema of swine virus) and sea lions (San Miguel sea lion virus) (2, 40, 41). These two viruses are closely related genomically (33, 34, 46). Clearly, the genomic relationship between BoCVs and the human NLVs needs urgent clarification with a larger number of BoCVs to determine if the BoCVs are a distinct genomic group and whether BoCVs are circulating in humans and are likely to pose a threat to human health. In the present study, we determined and analyzed the genomic sequence of BoCVs obtained from diverse geographical locations throughout the United Kingdom between 1976 and 2000 and examined their relationships to each other, to previously published BoCVs, and to the human NLVs.
The United Kingdom reference BoCV, Newbury agent 2 (Bo/Newbury2/76/UK), was identified in southern England, near Newbury, in 1976 (48), and Dumfries virus (Bo/Dumfries/94/UK) was identified near Dumfries, Scotland, in 1994. Both were available as diarrheic fecal samples from experimentally infected calves (6; D. R. Snodgrass, personal communication) and contained calicivirus-like particles as shown by electron microscopy. Between 1998 and 2000, 476 bovine diarrheic fecal samples from dairy and beef farms were supplied from the Veterinary Laboratory Agency laboratories at Aberystwyth and Carmarthen (western Wales), Shrewsbury (central England), Penrith, Preston, and Thirsk (northern England), Starcross (southwestern England), and Winchester (southern England). Samples were stored at +4°C until tested and were then stored at −70°C. RNA was extracted from fecal samples by using 1,1,2-trichloro-trifluoroethane (Sigma-Aldrich Company Ltd.) and by a method modified from that of Boom et al., with guanidine isothiocyanate and silica (3). To identify positive samples, extracted RNA was reverse transcribed at 37°C for 60 min by using 200 IU of Moloney murine leukemia virus reverse transcriptase (Promega) and 25 pmol of random hexamer primers (Promega). The resulting cDNA was amplified by PCR by using 2 IU of HotStar Taq (Qiagen, Ltd.) in a 50-μl reaction volume containing 70 mM deoxynucleoside trisphosphates and 3 mM MgCl2 with 20 pmol of one of three primer pairs designed to detect human NLVs, namely, NI/E3 (10), GLPSG1/YGDD1, or GLPSG2/YGDD1 (15). A DNA Thermal Cycler (Perkin-Elmer) was programmed for 95°C for 15 min and 40 cycles of 94°C for 15 s, 40°C for 45 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. PCR amplicons were resolved on 2.5% agarose gels. A positive signal for primer pair NI/E3 was a 114-bp amplicon, and positive signals for primer pairs GLPSG1/YGDD1 and GLPSG2/YGDD1 were 155-bp amplicons. Nuclease-free water was used as a negative control for every five fecal samples in both the reverse transcription (RT) and PCR steps and was handled simultaneously. Bo/Dumfries/94/UK was used as a positive virus control for the RT step. cDNA from Bo/Dumfries/94/UK was used as a positive control for the PCR. Positive samples were selected for long RT-PCR to produce amplicons of approximately 2.8 kbp from the 3′ poly-A tail. Positive samples were selected by geographical location, by the primer pair used for the initial amplification from the fecal samples, or by the relationship in a heteroduplex mobility assay (30; paper in preparation) to Bo/Newbury2/76/UK and two human NLVs currently circulating in the United Kingdom, Hu/Grimsby/95/UK and Hu/RHB/93/UK. RT was conducted at 42°C with the primer NA-2 poly-T EcoRI (GCGCTTAAGT(15)NMCAATGC) (LifeTechnologies Ltd.) by using 200 IU of Moloney murine leukemia virus (H-) reverse transcriptase (Promega). PCR was conducted by using the high-fidelity Z-Taq polymerase (BioWhittaker) and primer pairs NI/NA-2 poly-T EcoRI or GLPSG1/NA-2 poly-T EcoRI. Thirty-five cycles of 98°C for 5 s, 50°C for 10 s, and 72°C for 1 min were performed, with final extension at 72°C for 5 min, and PCR amplicons resolved on 1% agarose gels. The ∼2.8-kbp amplicons were gel purified and cloned by using a TOPO-TA XL-PCR cloning kit (Invitrogen Ltd.). Cycle sequencing was performed by using a dye terminator cycle sequencing kit and a CEQ 2000 XL DNA analysis system (Beckman Coulter). Nucleotide sequences for the polymerase, capsid, and open reading frame 3 (ORF3) genes were derived from cloned amplicons. Consensus nucleotide sequences were prepared by using the STADEN sequencing package (43). Multiple alignments of nucleotide and translated amino acid sequences were prepared by using Clustal X version 1.8 (44), and identities were calculated by using GeneDoc (www.psc.edu/biomed/genedoc). Open reading frames (ORFs) were predicted by using the NCBI ORF finder. Phylogenetic analyses were performed with nucleotide and amino acid sequence alignments by parsimony (DNApars and Protpars), Fitch-Margoliash and bootstrap analyses were performed by using PHYLIP (J. Felsenstein, Department of Genetics, University of Washington, Seattle; Phylogeny Inference Package, version 3.5c), and maximum likelihood with quartet puzzling was determined by using TreePuzzle 5.0 (38). Phylogenetic trees were prepared by using TreeView (35). Nucleotide and amino acid sequences of the polymerase, capsid, and ORF3 genes of the UK BoCVs were subjected to a GenBank BLAST search. An amino acid identity plot for the capsid proteins was generated by using SimPlot (29) with a window of 80 amino acids at 20-amino-acid intervals.
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers referred to in the text are human genogroup I NLVs: Hu/NLV/Norwalk/68/US, M87661; Hu/NLV/Chiba/87/JP, AB042808; Hu/NLV/Musgrove/89/UK,AJ277614; Hu/NLV/Seto124/89/JP, AB031013; Hu/NLV/Desert Shield395/90/SA, U04469; Hu/NLV/Thistlehall/90/UK, AJ277621; Hu/NLV/Southampton/91/UK, L07418; Hu/NLV/Birmingham/93/UK, AJ277612; Hu/NLV/Winchester/94/UK, AJ277609; Hu/NLV/Sindlesham/95/UK, AJ277615; Hu/NLV/Valetta/95/Malta, AJ277616; Hu/NLV/Whiterose/96/UK, AJ277610; and Hu/NLV/Hesse3/97/DE, AF093797. The following nucleotide sequence accession numbers are human genogroup II NLVs: Hu/NLV/Hawaii/71/US, U07611; Hu/NLV/Snow Mountain/76/US, U75682; Hu/NLV/Melksham/89/UK, X81879; Hu/NLV/Mexico/89/MX, U22498; Hu/NLV/Hillingdon/90/UK, AJ277607; Hu/NLV/Leeds/90/UK AJ277608; Hu/NLV/Seacroft/90/UK, AJ277620; Hu/NLV/Wortley/90/UK, AJ277618 (given as genogroup I in the GenBank database but genogroup II by J. Green et al. (11); Hu/NLV/Toronto24/91/CAN, U02030; Hu/NLV/Girlington/93/UK, AJ277606; Hu/NLV/Lordsdale/93/UK, X86557; Hu/NLV/Rbh/93/UK, AJ277617 (given as genogroup I in the GenBank database but genogroup II by J. Green et al. (11); Hu/NLV/Camberwell/94/AUS, U46500; Hu/NLV/Bham132/95/UK, AJ277611; Hu/NLV/Grimsby/95/UK, AJ004864; Hu/NLV/Mendoza320/95/ARG, AF190817; Hu/NLV/Parkroyal/95/UK, AJ277613; Hu/NLV/Symgreen/95/UK, AJ277619; Hu/NLV/Alphatron/98-2/1998/NET, AF195847; and Hu/NLV/Amsterdam/98-18/1998/NET, AF195848. The accession number for sapovirus Hu/SLV/Manchester/93/UK is X86560. Accession numbers for BoCVs reported previously are as follows: Bo/Newbury2/76/UK, AF097917 and AF097918; Bo/Jena/80/DE, AJ011099; Bo/NLV/176/98/NET, AF194183; Bo/NLV/CH126/98/NET, AF320625; and Bo/NLV/CH131/98/NET, AF320113. Accession numbers for the BoCVs reported in the present paper are as follows: Bo/Aberystwyth24/00/UK, AY126475; Bo/Aberystwyth58/00/UK, AY126462; Bo/Aberystwyth65/00/UK, AY126463; Bo/Carmarthen42/98/UK, AY126459;Bo/Carmarthen10/99/UK, AY126460; Bo/Carmarthen28/99/UK, AY126461; Bo/Dumfries/94/UK, AY126474; Bo/Newbury2/76/UK, AF097917; Bo/Penrith9/00/UK, AY126469; Bo/Penrith24/00/UK, AY126470; Bo/Penrith33/00/UK, AY126464; Bo/Penrith41/00/UK, AY126471; Bo/Penrith55/00/UK, AY126476; Bo/Shrewsbury24/00/UK, AY126472; Bo/Shrewsbury39/00/UK, AY126465; Bo/Shrewsbury46/00/UK, AY126466; Bo/Shrewsbury54/00/UK, AY126473; Bo/StarCross37/00/UK, AY126467; and Bo/Thirsk10/00/UK, AY126468.
Analysis of the polymerase, capsid and ORF3 gene sequences of the UK reference strain, Bo/Newbury2/76/UK.
A 581-nucleotide fragment of the Bo/Newbury2/76/UK polymerase gene (from nucleotides 4482 to 5064 of the previously described Bo/Jena/80/DE sequence [28]) had 89% amino acid (77% nucleotide) identity to Bo/Jena/80/DE isolated in Germany in 1980. The complete Bo/Newbury2/76/UK capsid gene (nucleotides 5051 to 6610 of the Bo/Jena/80/DE sequence) (Fig. 1) had 68% amino acid (67% nucleotide) identity to Bo/Jena/80/DE, which had two deletions of 3 and 6 nucleotides at positions 871 to 873 (amino acid 291) and 1270 to 1275 (amino acids 424 and 425) compared to Bo/Newbury2/76/UK. The complete Bo/Newbury2/76/UK ORF3 gene (excluding the extended amino terminus [see below]) had 63% amino acid (59% nucleotide) identity to Bo/Jena/80/DE and two deletions of 18 and 3 nucleotides at positions 355 to 372 (amino acids 119 to 124) and 562 to 564 (amino acid 188) compared to the Bo/Jena/80/DE ORF3 gene. The overlap of the Bo/Newbury2/76/UK ORF1 and ORF2 genes was identical in size to that of Bo/Jena/80/DE (nucleotide positions 5051 to 5064) (Fig. 2; Table 1), but the overlap of the Bo/Newbury2/76/UK ORF2 and ORF3 genes was considerably longer (nucleotide positions 6392 to 6610). However, visual inspection showed the presence of a second AUG codon in the Bo/Newbury2/76/UK sequence downstream of the predicted ORF3 initiation codon and in an analogous location to the initiation codon found in Bo/Jena/80/DE. The Bo/Newbury2/76/UK capsid and ORF3 genes were longer than that of Bo/Jena/80/DE, but the 3′ nontranslated region was shorter.
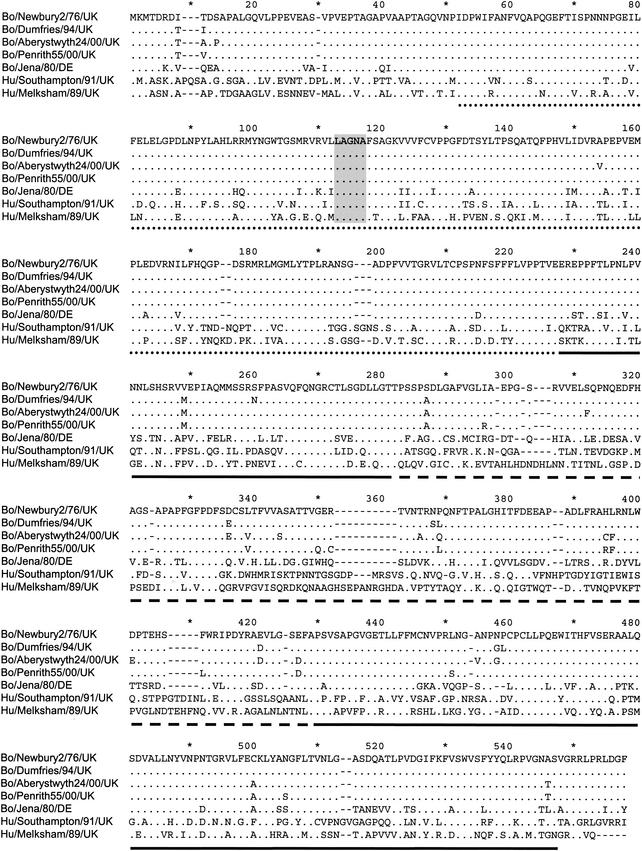
FIG. 1.
Multiple amino acid sequence alignment of the four UK BoCV capsid proteins, the German Bo/Jena/80/DE, and a UK representative of human NLV genogroups I (Hu/NLV/Southampton/91/UK) and II (Hu/NLV/Melksham/89/UK). The dotted line beneath the sequences represents amino acids 50 to 225 of the S domain, the solid lines represent the P1 domain, and the dashed line represents the P2 domain of the Hu/Norwalk/68/US (36). The conserved 5-amino-acid LAGNA motifs located at the 3- and 5-fold axes of symmetry of Hu/Norwalk/68/US are highlighted in gray.
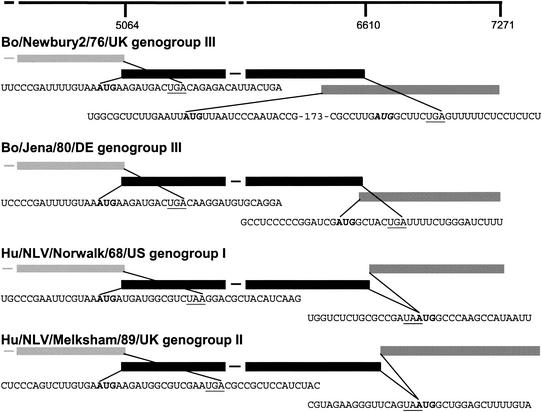
FIG. 2.
Schematic representation and nucleotide sequence of the reference UK BoCV (Bo/Newbury2/76/UK), the German BoCV (Bo/Jena/80/DE), the human NLV genogroup I Hu/NLV/Norwalk/68/US, and the human NLV genogroup II Hu/NLV/Melksham/89/UK viruses in the region of the ORF1-2 and ORF2-3 overlaps, showing start (in bold) and stop (underlined) codons as predicted by the NCBI ORF finder. The boxes on the left depict the 3′ ends of the ORF1s, the pairs of black boxes in the center depicts the 5′ and 3′ ends of the ORF2s, and the boxes on the right depict the complete ORF3s. The second initiation codon of the Bo/Newbury2/76/UK sequence is shown (bold italicized). The 173 bases not shown in the Bo/Newbury2/76/UK ORF2-3 overlap are labeled 173. The scale indicates nucleotide positions in relation to Bo/Jena/80/DE.
TABLE 1.
Comparison of the genomic organizations of the UK BoCVs sequenced in the present study, the German Bo/Jena/80/DE, and human genogroup I and II NLVs, as predicted by the NCBI ORF finderk
| Virus | ORF1-2 overlap (nt) | ORF2 capsid length (aa) | Capsid MW (kDa) | ORF2-3 overlap (nt) | ORF3 length (aa) | 3′ NTR length (nt) |
|---|---|---|---|---|---|---|
| BoCVs | ||||||
| Newbury2/76/UK | 14 | 522 | 56.9 | 209 | 282 | 43 |
| Dumfries/94/UK | 14 | 522 | 56.7 | 209 | 282 | 43 |
| Aberystwyth24/00/UK | 14 | 522 | 57.0 | 209 | 282 | 43 |
| Penrith55/00/UK | 14 | 522 | 57.0 | 209 | 282 | 43 |
| Jena/80/DE | 14a | 519a | 56.6a | 11a | 223a | 70a |
| Human NLVs | ||||||
| Genogroup I | 17b | 530-547c | 56.6-58.8d | 1b | 210-223e | 66-91e |
| Genogroup II | 20-35f | 535-550g | 58.7-60.0h | 1f | 252-268i | 45-85j |
From reference 28.
Based on data for Hu/NLV/Norwalk/68/US (22), Hu/NLV/Chiba/87/JP (42), Hu/NLV/Seto/124/89/JP (23), Hu/NLV/Desert Shield/90/SA (25), and Hu/NLV/Southampton/91/UK (24).
Based on data for Hu/NLV/Norwalk/68/US (22), Hu/NLV/Chiba/87/JP (42), Hu/NLV/Desert Shield/90/SA (25), Hu/NLV/Hesse3/97/DE (39), Hu/NLV/Southampton/91/UK (24), Hu/NLV/Musgrove/89/UK, Hu/NLV/Thistlehall/90/UK, Hu/NLV/Birmingham/93/UK, Hu/NLV/Winchester/94/UK, Hu/NLV/Valetta/95/Malta, Hu/NLV/Sindlesham/95/UK, and Hu/NLV/Whiterose/96/UK (11).
Based on data for Hu/NLV/Norwalk/68/US (22), Hu/NLV/Chiba/87/JP (42), Hu/NLV/Desert Shield/90/SA (25), and Hu/NLV/Southampton/91/UK (24).
Based on data for Hu/NLV/Norwalk/68/US (22), Hu/NLV/Chiba/87/JP (42), and Hu/NLV/Southampton/91/UK (24).
Based on data for Hu/NLV/Hawaii/71/US (26), Hu/NLV/Melksham/89/UK (14), Hu/NLV/Mexico/89/MX (21), Hu/NLV/Toronto/24/91/CAN (27), Hu/NLV/Lordsdale/93/UK (8), Hu/NLV/Camberwell/94/AUS (5), and Hu/NLV/Mendoza/320/95/ARG (20).
Based on data for Hu/NLV/Hawaii/71/US (26), Hu/NLV/Snow Mountain/76/US (19), Hu/NLV/Melksham/89/UK (14), Hu/NLV/Mexico/89/MX (21), viruses Hu/NLV/Hillingdon/90/UK, HU/NLV/Leeds/90/UK, Hu/NLV/Seacroft/90/UK, Hu/NLV/Girlington/93/UK, Hu/NLV/Bham132/95/UK, Hu/NLV/Parkroyal/95/UK, Hu/NLV/Symgreen/95/UK, Hu/NLV/Rbh/93/UK, and Hu/NLV/Wortley/90/UK (11), Hu/NLV/Toronto 24/91/CAN (27), Hu/NLV/Lordsdale/93/UK (8), Hu/NLV/Camberwell/94/AUS (5), Hu/NLV/Grimsby/95/UK (17), Hu/NLV/Mendoza320/95/ARG (20), and viruses Hu/NLV/Alphatron/98-2/98/NET and Hu/NLV/Amsterdam/98-18/98/NET (47).
Based on data for Hu/NLV/Hawaii/71/US (26), Hu/NLV/Snow Mountain/76/US (19), Hu/NLV/Melksham/89/UK (14), Hu/NLV/Mexico/89/MX (21), Hu/NLV/Toronto24/91/CAN (27), Hu/NLV/Lordsdale/93/UK (8), Hu/NLV/Camberwell/94/AUS (5), Hu/NLV/Grimsby/95/UK (17), and Hu/NLV/Mendoza320/95/ARG (20).
Based on data for Hu/NLV/Melksham/89/UK (14), Hu/NLV/Lordsdale/93/UK (8), Hu/NLV/Camberwell/94/AUS (5), and Hu/NLV/Mendoza320/95/ARG (20).
nt, nucleotide; aa, amino acid; NTR, nontranslated region.
Analysis of BoCVs identified from throughout the United Kingdom.
Amplicons of the expected sizes (114 and 155 bp) were obtained by RT-PCR by using primers pairs NI/E3 and/or GLPSG1/YGDD1 (originally designed to detect human genogroup I and II human NLVs) with 38 of the 476 fecal samples collected between 1998 and 2000 from the eight Veterinary Laboratories Agency laboratories and with fecal samples from the experimental calf inoculated with the sample from Dumfries, Scotland, 1994. Additional positive samples were not identified from a subset of 114 samples that were randomly selected from the different geographical locations and tested with primer pair GLPSG2/YGDD1 (originally designed to detect human genogroup II NLVs). Positive samples were obtained from calves aged 6 weeks and under, from one 4-month-old calf (Thirsk10/00/UK), and from two cows (Shrewsbury54/00/UK and StarCross37/00/UK).
Partial polymerase sequences of 76 nucleotides from 18 samples from geographically diverse locations (Aberystwyth24/00/UK, Aberystwyth58/00/UK, Aberystwyth65/00/UK, Carmarthen42/98/UK, Carmarthen10/99/UK, Carmarthen28/99/UK, Dumfries/94/UK, Penrith9/00/UK, Penrith24/00/UK, Penrith33/00/UK, Penrith41/00/UK, Penrith55/00/UK, Shrewsbury24/00/UK, Shrewsbury39/00/UK, Shrewsbury46/00/UK, Shrewsbury54/00/UK, Starcross37/00/UK, and Thirsk10/00/UK), including the samples from the 4-month-old calf and the two cows, showed a close relationship to each other, to Bo/Newbury2/76/UK, and to Bo/Jena/80/DE virus (28) (92 to 100% amino acid and 68 to 100% nucleotide identity). Nucleotide diversity showed no correlation with year of isolation, age, or geographical location within the United Kingdom. Longer partial polymerase sequences of 416 nucleotides of 7 samples, namely, Aberystwyth24/00/UK, Dumfries/94/UK, Penrith9/00/UK, Penrith24/00/UK, Penrith55/00/UK, Shrewsbury24/00/UK, Shrewsbury54/00/UK (identified using the NI/E3 primer pair), and Penrith41/00/UK (identified with the GLPSGI/YGDDI primer pair), also showed a close relationship to each other, to Bo/Newbury2/76/UK, and to Bo/Jena/80/DE (88 to 100% amino acid and 74 to 99% nucleotide identity). In addition, the 61 nucleotides (20 translated amino acids) available from the polymerase gene of the Dutch BoCV, Bo/NLV/176/98/NET (45), which overlapped with the sequences obtained in the present study, had 90 to 100% amino acid (70 to 100% nucleotide) identity to the UK BoCVs.
The complete capsid genes of Newbury2/76/UK, Dumfries/94/UK, Aberystwyth24/00/UK, and Penrith55/00/UK had 95 to 97% amino acid (87 to 89% nucleotide) identity to each other and to two unpublished BoCV sequences from The Netherlands (Bo/NLV/CH126/98/NET and Bo/NLV/CH131/98/NET) but were less closely related to the German Bo/Jena/80/DE virus (68% amino acid identity and 66 to 67% nucleotide identity). Alignment of translated amino acid sequences for the conserved shell (S) domain (36) showed total conservation between the UK BoCVs, apart from one amino acid substitution in the Aberystwyth24/00/UK sequence (Fig. 1), but showed lower identity to the German Bo/Jena/80/DE virus (Fig. 3). In the protruding (P) domain (36), single amino acid substitutions and pairs of substitutions were found between the UK BoCVs which had 93 to 96% amino acid (85 to 88% nucleotide) identities in this region, but substantial variation occurred between the UK BoCVs and Bo/Jena/80/DE (Fig. 3). The complete ORF3 genes (starting from the Bo/Jena/80/DE ORF3 initiation codon) of the four UK BoCVs, Newbury2/76/UK, Dumfries/94/UK, Aberystwyth24/00/UK, and Penrith55/00/UK, had 92 to 96% amino acid (87 to 90% nucleotide) identity to each other but had 61 to 63% amino acid (58 to 59% nucleotide) identity to Bo/Jena/80/DE.
FIG. 3.
Amino acid identity plot generated by using SimPlot (29) with a window of 80 amino acids at 20-amino-acid intervals to compare the complete capsid protein of Bo/Newbury2/76/UK with the capsid protein of Bo/Jena/80/DE and the genogroup I human NLV Hu/Southampton/91/UK and the genogroup II NLV Hu/Melksham/89/UK. The dotted line beneath the plot represents the S domain, the solid line represents the P1 domain, and the dashed line represents the P2 domain of the Hu/NLV/Norwalk/68/US, as determined by X-ray crystallography (36).
The genome organization of Dumfries/94/UK, Aberystwyth24/00/UK, and Penrith55/00/UK was remarkably similar to that of the UK reference Bo/Newbury2/76/UK, showing the same differences and similarities to Bo/Jena/80/DE as Bo/Newbury2/76/UK as well as the additional initiation codon in the ORF2-3 region identified in Bo/Newbury2/76/UK (Table 1). Comparable lengths of sequence were not available from the Dutch BoCVs to establish whether BoCV genome organization falls into two types, Newbury2-like or Jena-like.
Comparison of BoCVs and the human NLVs.
Analysis of the polymerase (76 and 416 nucleotides), complete capsid, and ORF3 gene sequences of the UK BoCVs, plus the German Bo/Jena/80/DE virus, showed that the BoCVs were distinct from representative human genogroup I and II NLVs and the NLVs currently associated with gastroenteritis in the UK human population from 1989 to 2000 (11; personal observations). The highest identities between the BoCVs and the human NLVs were observed with the polymerase gene sequences, but the identities seen between the BoCVs and genogroup I and II NLVs (Table 2) were comparable to those seen between genogroup I and II NLV, which had 62 to 68% amino acid identity in the 416-nucleotide region examined in the present study. The amino acid identities of the complete BoCV capsid proteins to representatives of the seven genetic clusters of human NLV genogroups I and II (41 to 48%) were similar to the identities between the two human NLV genogroups (41 to 46%) (Table 3). The greatest divergence between the BoCVs and the human NLVs was seen with the BoCV ORF3 genes (Table 2). A BLAST search with UK BoCV sequences from all three genes failed to identify human NLVs with identities greater than those between the UK BoCVs and the representative human genogroup I and II NLVs described above. Phylogenetic analyses of the nucleotide and translated amino acid sequences of all three genes by the independent methods of parsimony, distance, and maximum likelihood each showed that the UK BoCVs, the German Bo/Jena/80/DE, and two unpublished capsid sequences from bovine viruses identified in The Netherlands (Bo/NLV/CH126/98/NET and Bo/NLV/CH131/98/NET) formed a distinct genogroup with quartet puzzling support values of 100% for the polymerase and capsid sequences and 76% for the ORF3 sequences (Fig. 4 and 5). Heterogeneity between the BoCVs and the human NLVs was distributed throughout the capsid protein but was concentrated in the P2 domain (Fig. 1 and 3). A 5-amino-acid motif, LAGNA, was conserved in the S domain of the UK BoCVs, the German Bo/Jena/80/DE, 2 Dutch BoCVs (Bo/CH126/1998/NL and Bo/CH131/1998/NL), and 21 genogroup I and 49 genogroup II human NLVs but was absent in the caliciviruses classified in the Vesivirus, Lagovirus and Sapporo-like genera. By using the molecular imaging program RASMOL, the LAGNA motif was located at the 3- and 5-fold axes of symmetry of Hu/Norwalk/68/US, suggesting structural importance (not shown). The UK BoCVs had an overall genomic organization similar to that of the human NLVs but with some consistent differences (Table 1; Fig. 2). The BoCV-predicted ORF1-2 overlaps were at least 3 nucleotides shorter, but the ORF2-3 overlaps were considerably longer than those of the human NLVs because of the additional initiation codon (see above). The termination codon for the UK and German BoCV ORF2 genes was UGA and not the UAA termination codon which has been commonly observed with human NLVs. The UK BoCVs capsid proteins were at least 8 amino acids shorter, their ORF3 genes were longer, and their 3′ untranslated regions were shorter than those of any known human NLV.
TABLE 2.
Percent amino acid (nucleotide) identities of BoCV partial polymerase and ORF3 genes to representative human genogroup I and II NLVs
| Genogroup | Polymerase
|
ORF3 113 aa (339 nt)c | |
|---|---|---|---|
| 35 aa (76 nt)a | 138 aa (416 nt)b | ||
| Id | 72-80 | 77-81 | 38-45 |
| (57-75) | (64-71) | (45-49) | |
| IIe | 64-84 | 62-68 | 31-41 |
| (50-78) | (57-63) | (40-48) | |
Based on nucleotides 4482 to 4556 (numbered according to Bo/Jena/80/DE) of 19 UK BoCVs (Newbury2/76/UK, Aberystwyth24/00/UK, Aberystwyth58/00/UK, Aberystwyth65/00/UK, Carmarthen42/98/UK, Carmarthen10/99/UK, Carmarthen28/99/UK, Dumfries/94/UK, Penrith9/00/UK, Penrith24/00/UK, Penrith33/00/UK, Penrith41/00/UK, Penrith55/00/UK, Shrewsbury24/00/UK, Shrewsbury39/00/UK, Shrewsbury46/00/UK, Shrewsbury54/00/UK, Starcross37/00/UK and Thirsk10/00/UK), and the Bo/Jena/80/DE virus.
Based on nucleotides 4482 to 4896 (numbered according to Bo/Jena/80/DE) of 9 UK BoCVs (Newbury2/76/UK, Aberystwyth24/00/UK, Dumfries/94/UK, Penrith9/00/UK, Penrith24/00/UK, Penrith41/00/UK, Penrith55/00/UK, Shrewsbury24/00/UK, Shrewsbury54/00/UK), and the Bo/Jena/80/DE virus.
Based on 339 nucleotides at the 5′ end of ORF3 (starting from the Bo/Jena/80/DE initiation codon) of 4 UK BoCVs (Newbury2/76/UK, Dumfries/94/UK, Aberystwyth24/00/UK and Penrith55/00/UK) and the Bo/Jena/80/DE virus.
Represented by Hu/Norwalk/68/US (22), Hu/Southampton/91/UK (24), and Hu/DesertShield395/90/SA (25) viruses.
Represented by Hu/NLV/Melksham/89/UK (14), Hu/NLV/Lordsdale/93/UK (8), Hu/NLV/Mexico/89/MX (21), Hu/NLV/Toronto/91/CAN (27), and Hu/NLV/Hawaii/71/US (26) viruses.
aa, amino acid; nt, nucleotide.
TABLE 3.
Percent amino acid identities between the complete capsid proteins of BoCVs and the seven genetic clusters of human genogroup I and II NLVs
| Genetic clustera | NLVs
|
BoCVs
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genogroup I
|
Genogroup II
|
Genogroup III
|
||||||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | ||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||||||
| NLVs | ||||||||||||||||||||
| Genogroup I | ||||||||||||||||||||
| 1. Hu/Norwalk/68/US | 1 | — | 69 | 66 | 72 | 69 | 70 | 64 | 46 | 46 | 45 | 42 | 45 | 41 | 43 | 48 | 47 | 47 | 47 | 47 |
| 2. Hu/Southampton/91/UK | 2 | — | 67 | 74 | 73 | 70 | 63 | 44 | 42 | 41 | 42 | 44 | 41 | 41 | 48 | 48 | 48 | 48 | 46 | |
| 3. Hu/DesertShield395/90/SA | 3 | — | 66 | 66 | 68 | 71 | 44 | 45 | 43 | 43 | 45 | 43 | 44 | 47 | 46 | 46 | 46 | 46 | ||
| 4. Hu/Chiba407/87/JP | 4 | — | 79 | 71 | 64 | 44 | 43 | 41 | 42 | 44 | 41 | 41 | 48 | 48 | 48 | 47 | 47 | |||
| 5. Hu/Musgrove/89/UK | 5 | — | 70 | 67 | 45 | 45 | 43 | 45 | 45 | 42 | 43 | 48 | 47 | 47 | 47 | 46 | ||||
| 6. Hu/Hesse3/97/DE | 6 | — | 64 | 46 | 44 | 43 | 43 | 44 | 42 | 43 | 48 | 47 | 46 | 47 | 46 | |||||
| 7. Hu/Winchester/94/UK | 7 | — | 44 | 43 | 43 | 43 | 44 | 42 | 42 | 46 | 46 | 45 | 45 | 46 | ||||||
| Genogroup II | ||||||||||||||||||||
| 8. Hu/Hawaii/71/US | 1 | — | 75 | 71 | 64 | 75 | 68 | 68 | 45 | 45 | 45 | 45 | 44 | |||||||
| 9. Hu/Melksham/89/UK | 2 | — | 67 | 62 | 78 | 68 | 67 | 44 | 44 | 44 | 44 | 42 | ||||||||
| 10. Hu/Toronto24/91/CAN | 3 | — | 66 | 67 | 71 | 68 | 41 | 42 | 42 | 42 | 42 | |||||||||
| 11. Hu/Lordsdale/93/UK | 4 | — | 62 | 61 | 63 | 44 | 44 | 43 | 44 | 43 | ||||||||||
| 12. Hu/Hillingdon/90/UK | 5 | — | 67 | 67 | 42 | 42 | 42 | 42 | 42 | |||||||||||
| 13. Hu/Seacroft/90/UK | 6 | — | 67 | 41 | 41 | 41 | 41 | 41 | ||||||||||||
| 14. Hu/Leeds/90/UK | 7 | — | 42 | 42 | 42 | 42 | 41 | |||||||||||||
| BoCVs | ||||||||||||||||||||
| Genogroup III | ||||||||||||||||||||
| 15. Bo/Newbury2/76/UK | — | 97 | 95 | 96 | 68 | |||||||||||||||
| 16. Bo/Dumfries/94/UK | — | 96 | 96 | 68 | ||||||||||||||||
| 17. Bo/Aberystwyth24/00/UK | — | 96 | 68 | |||||||||||||||||
| 18. Bo/Penrith55/00/UK | — | 68 | ||||||||||||||||||
| 19. Bo/Jena/80/DE | — | |||||||||||||||||||
From reference 1. From reference 8 for Hu/NLV/Lordsdale/93/UK; 11 for Hu/NLV/Musgrove/89/UK, Hu/NLV/Leeds/90/UK, Hu/NLV/Hillingdon/90/UK, Hu/NLV/Seacroft/90/UK, Hu/NLV/Winchester/94/UK; 14 for Hu/NLV/Melksham/89/UK; 22 for Hu/NLV/Norwalk/68/US; 24 for Hu/NLV/Southampton/91/UK; 25 for Hu/NLV/DesertShield/90/SA; 26 for Hu/NLV/Hawaii/71/US; 27 for Hu/NLV/Toronto24/1991/CAN; 39 for Hu/Hesse3/97/DE; 42 for Hu/NLV/Chiba/87/JP.
FIG. 4.
Phylogenetic analysis generated by using the TreePuzzle maximum likelihood method for the translated amino acid sequences of (A) the long polymerase sequence (138 amino acids) and (B) 113 amino acids from the 5′ end of the ORF3 gene (starting from the Bo/Jena/80/DE ORF3 initiation codon) of the UK BoCVs, the German Bo/NLV/Jena/80/DE, and representatives of the human NLV genogroups I and II, with the Sapporo-like virus Hu/Manchester/93/UK used as the outgroup. The numbers at the nodes (quartet puzzling values) indicate the frequencies of occurrence for 1,000 replicate trees. The asterisk indicates the sample identified with the GLPSG1/YGDD1 primer pair.
FIG. 5.
Phylogenetic analysis generated by using the TreePuzzle maximum likelihood method for the translated amino acid sequences of the complete capsid protein gene, the UK BoCVs, the German Bo/Jena/80/DE, the two unpublished Dutch BoCVs Bo/CH126/98/NET and Bo/CH131/98/NET, and representatives of the seven clusters in the human NLV genogroups I and II, with the Sapporo-like virus Hu/Manchester/93/UK used as the outgroup. The numbers at the nodes (quartet puzzling values) indicate the frequencies of occurrence for 1,000 replicate trees. The superscripts represent the genetic clusters of the genogroup I and II viruses, as determined by Ando et al. (1).
Thus, the present study clarified the relationship of the human NLVs and the BoCVs by showing that BoCVs formed a distinct third genogroup in the Norovirus genus, the first genogroup in this genus composed, so far, solely of enteric caliciviruses from animals. The data supported the suggestion of Ando et al. (1) but suggested an evolutionary relationship of BoCVs to genogroup I NLVs, supporting the comments and conclusions of Dastjerdi et al. (6), Liu et al. (28), and van der Poel et al. (45) that BoCVs were closer to NLV genogroup I than genogroup II. The identity of the Bo/Newbury2/76/UK to the human genogroups I and II NLV polymerase sequences agreed with that published previously by Dastjerdi et al. (6), but the amino acid identities of 57 to 73% reported by Dastjerdi et al. for the capsid protein sequences were considerably higher than those reported for the whole capsid protein in the present study. This is explained, since Dastjerdi et al. (6) examined a region of 116 amino acids from the relatively conserved S domain of the Bo/Newbury2/76/UK capsid protein, which, in the present study, had a similar level of identity to that found by Dastjerdi et al. No evidence for strains circulating in both humans and cattle was found, suggesting that at least the BoCVs identified in the present study are not a risk to human health. It was noteworthy that the nucleotide and amino acid sequence identities between the bovine and human NLVs were considerably lower than those reported between the swine vesicular exanthema caliciviruses and the San Miguel sea lion caliciviruses (34), supporting the opinion that the BoCVs are unlikely to infect humans. Whether occasional cross-species infectivity events occur between cattle and humans requires more sequence data of the caliciviruses circulating in cattle from throughout the world.
The choice of primer pairs used in the present study was deliberate in order to identify the BoCVs most closely related to human NLVs. The three sets of primer pairs are currently used successfully for diagnostic RT-PCR on human diarrheic fecal samples at the Central Public Health Laboratory by the authors. We may have detected a subset of BoCVs, but it seems unlikely that the use of additional primer pairs would identify BoCVs with a closer relationship to human NLVs. We have not detected all BoCVs circulating in the United Kingdom cattle population, as we have failed to amplify Newbury agent 1 (4, 7; personal observations). Other primer pairs will be needed to identify the complete range of BoCVs circulating in cattle, but we predict that these yet undefined BoCVs are unlikely to be closely related to the human NLVs.
The close relationship of the BoCVs identified in 1976, 1994, and between 1998 and 2000 in the United Kingdom and their close relationship to the German (28) and Dutch BoCVs (45) identified in 1980 and 1998 contrasted with the diversity identified with human NLVs over a similar time period (reviewed in references 1 and 13). However, there have been instances of closely related human NLVs identified many years apart from different continents (14, 19). We have no evidence to indicate that laboratory contamination accounted for the close relationship between the UK BoCVs. All RTs and PCRs were strictly controlled with negative samples, and no cross-contamination occurred. Although little heterogeneity was identified between the amino acid sequences, it should be noted that considerable heterogeneity was identified between some UK BoCVs at the nucleotide level. Ando et al. (1) suggested that BoCVs might be separated into two clusters represented by viruses similar to Bo/Newbury2/76/UK and Bo/Jena/80/DE. The GLSPG1/YGDD1 primer pair, which was used by Liu et al. (28) to identify Bo/Jena/80/DE, detected several UK BoCVs, but we failed to amplify viruses closely related to Bo/Jena/80/DE. The 68% amino acid identity of the capsid protein of Bo/Jena/80/DE to that of the UK BoCVs was comparable to the relationship of human NLVs within the two human genogroups (Table 3), confirming that the Bo/Jena/80/DE virus should be classified in the bovine genogroup III. BoCVs caused enteric disease in experimental calves (4, 48), caused pathology in the small intestine (18), and were found in association with outbreaks of calf diarrhea in the United Kingdom (37). However, van der Poel et al. (45) found BoCVs commonly in veal calves, highlighting the need for further studies to determine their role in bovine diarrhea outbreaks.
Acknowledgments
We thank members of the Veterinary Laboratory Agency (Graham David, Paul Duff, David Kirby and Gavin Watkins) for efficiently organizing and supplying the fecal samples.
We thank the Biotechnology and Biological Sciences Research Council (BBSRC) for studentship support to S.L.O.
REFERENCES
- 1.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 2.Berry, E. S., D. E. Skilling, J. E. Barlough, N. A. Vedros, L. J. Gage, and A. W. Smith. 1990. New marine calicivirus serotype infective for swine. Am. J. Vet. Res. 51:1184-1187. [PubMed] [Google Scholar]
- 3.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridger, J. C., G. A. Hall, and J. F. Brown. 1984. Characterisation of a calici-like virus (Newbury Agent) found in association with astrovirus in bovine diarrhea. Infect. Immun. 43:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cauchi, M. R., J. C. Doultree, J. A. Marshall, and P. J. Wright. 1996. Molecular characterization of Camberwell virus and sequence variation in ORF3 of small round-structured (Norwalk-like) viruses. J. Med. Virol. 49:70-76. [DOI] [PubMed] [Google Scholar]
- 6.Dastjerdi, A. M., J. Green, C. I. Gallimore, D. W. G. Brown, and J. C. Bridger. 1999. The bovine Newbury agent-2 is genetically more closely related to human SRSVs than to animal caliciviruses. Virology 254:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Dastjerdi, A. M., D. R. Snodgrass, and J. C. Bridger. 2000. Characterisation of the bovine enteric calici-like virus, Newbury agent 1. FEMS Microbiol. Lett. 192:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingle, K. E., P. R. Lambden, E. O. Caul, and I. N. Clarke. 1995. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J. Gen. Virol. 76:2349-2355. [DOI] [PubMed] [Google Scholar]
- 9.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 10.Green, J., C. I. Gallimore, J. P. Norcott, D. Lewis, and D. W. G. Brown. 1995. Broadly reactive reverse transcriptase polymerase chain reaction for the diagnosis of SRSV-associated gastroenteritis. J. Med. Virol. 47:392-398. [DOI] [PubMed] [Google Scholar]
- 11.Green, J., J. Vinje, C. I. Gallimore, M. Koopmans, A. Hale, D. W. Brown, J. C. Clegg, and J. Chamberlain. 2000. Capsid protein diversity among Norwalk-like viruses. Virus Genes 20:227-236. [DOI] [PubMed] [Google Scholar]
- 12.Green, K. Y. 2000.. Summary of the first international workshop on human caliciviruses. J. Infect. Dis. 181(Suppl. 2):S252-S253. [DOI] [PubMed] [Google Scholar]
- 13.Green, K. Y., R. M. Chanock, and A. Z. Kapikian. 2001. Human caliciviruses, p. 841-874. In Fields virology, 4th ed. Lippincott, Williams & Wilkins, Baltimore, Md.
- 14.Green, S. M., P. R. Lambden, E. O. Caul, C. R. Ashley, and I. N. Clarke. 1995. Capsid diversity in small round-structured viruses: molecular characterization of an antigenically distinct human enteric calicivirus. Virus Res. 37:271-283. [DOI] [PubMed] [Google Scholar]
- 15.Green, S. M., P. R. Lamden, Y. Deng, A. J. Lowes, S. Lineham, J. Bushell, J. E. Rogers, O. E. Caul, C. R. Ashley, and I. N. Clarke. 1995. Polymerase chain reaction detection of small round-structured viruses from two related hospital outbreaks of gastroenteritis using inosine containing primers. J. Med. Virol. 45:197-202. [DOI] [PubMed] [Google Scholar]
- 16.Günther, H., and P. Otto. 1987. Studies into diarrhoea of young calves. Seventh communication: “Zackenvirus” (Jena-Agens 117/80), a new diarrhoea pathogen to calf. Arch. Exp. Vet. Med. (Leipzig) 41:934-938. [PubMed] [Google Scholar]
- 17.Hale, A. D., S. E. Crawford, M. Ciarlet, J. Green, C. Gallimore, D. W. Brown, X. Jiang, and M. K. Estes. 1999. Expression and self-assembly of Grimsby virus: antigenic distinction from Norwalk and Mexico viruses. Clin. Diagn. Lab. Immunol. 6:142-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, G. A., J. C. Bridger, B. E. Brooker, K. R. Parsons, and E. Ormerod. 1984. Gnotobiotic calves experimentally infected with a calicivirus-like (Newbury) agent. Vet. Pathol. 21:208-215. [DOI] [PubMed] [Google Scholar]
- 19.Hardy, M. E., S. F. Kramer, J. J. Treanor, and M. K. Estes. 1997. Human calicivirus genogroup II capsid sequence diversity revealed by analyses of the prototype Snow Mountain agent. Arch. Virol. 142:1469-1479. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, X., C. Espul, W. M. Zhong, H. Cuello, and D. O. Matson. 1999. Characterization of a novel human calicivirus that may be a naturally occurring recombinant. Arch. Virol. 144:2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, X., D. O. Matson, F. R. Velazquez, J. J. Calva, W. M. Zhong, J. Hu, G. M. Ruiz-Palacios, and L. K. Pickering. 1995. Study of Norwalk-related viruses in Mexican children. J. Med. Virol. 47:309-316. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, S., K. Sakae, Y. Suzuki, K. Shinozaki, M. Okada, H. Ishiko, K. Kamata, K. Suzuki, K. Natori, T. Miyamura, and N. Takeda. 2000. Molecular cloning, expression, and antigenicity of Seto virus belonging to genogroup I Norwalk-like viruses. J. Clin. Microbiol. 38:3492-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 25.Lew, J. F., A. Z. Kapikian, X. Jiang, M. K. Estes, and K. Y. Green. 1994. Molecular characterization and expression of the capsid protein of a Norwalk-like virus recovered from a Desert Shield troop with gastroenteritis. Virology 200:319-325. [DOI] [PubMed] [Google Scholar]
- 26.Lew, J. F., A. Z. Kapikian, J. Valdesuso, and K. Y. Green. 1994. Molecular characterization of Hawaii virus and other Norwalk-like viruses: evidence for genetic polymorphism among human caliciviruses. J. Infect. Dis. 170:535-542. [DOI] [PubMed] [Google Scholar]
- 27.Lew, J. F., M. Petric, A. Z. Kapikian, X. Jiang, M. K. Estes, and K. Y. Green. 1994. Identification of minireovirus as a Norwalk-like virus in pediatric patients with gastroenteritis. J. Virol. 68:3391-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, B. L., P. R. Lambden, H. H. Günther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattick, K. L., J. Green, P. Punia, F. J. Belda, C. I. Gallimore, and D. W. G. Brown. 2000. The heteroduplex mobility assay (HMA) as a pre-sequencing screen for Norwalk-like viruses. J. Virol. Methods 87:161-169. [DOI] [PubMed] [Google Scholar]
- 31.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerging Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy, F. A., E. P. J. Gibbs, M. C. Horzinek, and M. J. Studdert. 1999. Veterinary virology, p. 533-541. Academic Press, San Diego, Calif.
- 33.Neill, J. D., R. F. Meyer, and B. S. Seal. 1995. Genetic relatedness of the caliciviruses: San Miguel sea lion and vesicular exanthema of swine viruses constitute a single genotype within the Caliciviridae. J. Virol. 69:4484-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neill, J. D., R. F. Meyer, and B. S. Seal. 1998. The capsid protein of vesicular exanthema of swine virus serotype A48: relationship to the capsid protein of other animal caliciviruses. Virus Res. 54:39-50. [DOI] [PubMed] [Google Scholar]
- 35.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 36.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds, D. J., J. H. Morgan, N. Chanter, P. W. Jones, J. C. Bridger, T. G. Debney, and K. J. Bunch. 1986. Microbiology of calf diarrhoea in southern Britain. Vet. Rec. 119:34-39. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 39.Schreier, E., F. Doring, and U. Kunkel. 2000. Molecular epidemiology of outbreaks of gastroenteritis associated with small round structured viruses in Germany in 1997/98. Arch. Virol. 145:443-453. [DOI] [PubMed] [Google Scholar]
- 40.Smith, A. W. 1987. San Miguel sea lion virus, p. 481-489. In M. J. Appel (ed.), Virus infections of carnivores. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 41.Smith, A. W., T. G. Akers, S. H. Madin, and N. A. Vedros. 1973. San Miguel sea lion virus isolation, preliminary characterization and relationship to vesicular exanthema of swine virus. Nature 244:108-110. [DOI] [PubMed] [Google Scholar]
- 42.Someya, Y., N. Takeda, and T. Miyamura. 2000. Complete nucleotide sequence of the chiba virus genome and functional expression of the 3C-like protease in Escherichia coli. Virology 278:490-500. [DOI] [PubMed] [Google Scholar]
- 43.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Poel, W. H. M., J. Vinjé, R. van der Heide, M. Herrera, A. Vivi, and M. P. G. Koopmans. 2000. Norwalk-like calicivirus genes in farm animals. Emerg. Infect. Dis. 6:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Corstens, M. K. Estes, S. M. Lemon, J. Manitoff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: classification and nomenclature of viruses, p. 725-735. In Seventh report of the ICTV. Academic Press, San Diego, Calif.
- 47.Vinje, J., and M. P. Koopmans. 2000. Simultaneous detection and genotyping of “Norwalk-like viruses” by oligonucleotide array in a reverse line blot hybridization format. J. Clin. Microbiol. 38:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woode, G. N., and J. C. Bridger. 1978. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J. Med. Microbiol. 11:441-452. [DOI] [PubMed] [Google Scholar]