Abstract
We have identified an HLA-A2-restricted CD8+ T-cell epitope, FLYALALLL, in the Epstein-Barr virus (EBV) latent membrane protein 2 (LMP2), an important target antigen in the context of EBV-associated malignancies. This epitope is TAP independent, like other hydrophobic LMP2-derived epitopes, but uniquely is dependent upon the immunoproteasome for its generation.
Immune intervention against virus-associated malignancies requires detailed knowledge of not only the immune response to the virus involved, but also the components of the antigen processing machinery required for efficient presentation of viral T-cell epitopes. Epstein-Barr virus (EBV), a B-lymphotropic gammaherpesvirus, is associated with a number of human malignancies, including posttransplant lymphoproliferative disease (PTLD), Hodgkin's disease (HD), and a tumor common in Southern Chinese populations, nasopharyngeal carcinoma (NPC) (21). In healthy individuals, EBV infection is kept under control by a strong virus-specific T-cell response, principally consisting of HLA class I-restricted CD8+ cytotoxic T lymphocytes (CTLs) (22). The dominant targets for these EBV-specific CTLs are the Epstein-Barr nuclear antigen 3 (EBNA3) family of proteins (12, 18, 22). Importantly, these antigens are expressed in most cases of EBV-positive PTLD, and the success of adoptive T-cell therapy in this context has become the paradigm for immune intervention against a tumor (24). However, in EBV-positive HD and NPC, virus gene expression, and therefore the available targets for T-cell recognition, is restricted to EBNA1 and the latent membrane proteins 1 and 2 (LMP1 and -2, respectively) (2, 20, 21). Of these, EBNA1 is protected from processing and presentation via the conventional HLA class I pathway, due to the presence of an internal glycine/alanine domain (1, 16), and LMP1 has thus far proven to elicit CD8+ T-cell responses only very rarely (13). Attention has therefore focused on LMP2 as a therapeutic target (15). Here, we report the identification of a novel HLA A*0201-restricted CD8+ T-cell epitope in LMP2, which is presented in a TAP-independent manner, but requires the immunoproteasome for its generation.
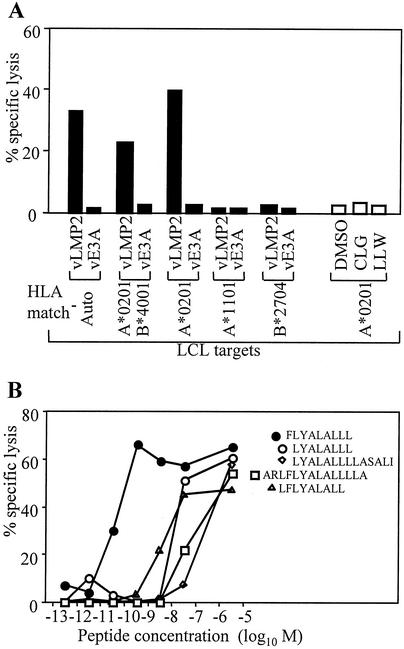
Stimulation of peripheral blood mononuclear cells (PBMCs) with cells of the autologous EBV-transformed lymphoblastoid cell line (LCL) has already identified a total of 11 CD8+ T-cell epitopes in LMP2, presented in the context of a range of HLA alleles, including HLA-A2, -A11, -A24, -B27, -B40, and -B63 (14, 22). In this study, a similar reactivation of PBMCs from an EBV-seropositive individual (donor A) carrying the HLA-A2, -A11, -B27, and -B40 alleles generated a number of CTL clones that did not recognize any of the previously identified CD8+ epitopes. As shown in the chromium release assay in Fig. 1A by using one such CTL clone, there was clear recognition of A*0201-positive LCLs overexpressing LMP2 from a vaccinia virus vector, but no recognition of either of the previously characterized A*0201-restricted LMP2 epitopes CLGGLLTMV and LLWTLVVLL (henceforth designated by the first three letters of their amino acid sequence). To identify the new epitope, such clones were screened in a T-cell-T-cell killing assay (3) against a panel of overlapping 14- and 15-mer LMP2 peptides, and their reactivity was mapped to the overlapping 14-mers, LMP2 amino acid positions 353 to 366 and 357 to 370. As shown in Fig. 1B, titration of these 14-mers and smaller peptides from within this region identified the minimal epitope as FLYALALLL (FLY; LMP2 amino acids 356 to 364); interestingly this sequence does not lie entirely within the original overlap so that the recognition of the position 357-to-370 14-mer was possible, even though it lacked the phenylalanine residue that forms position 1 of the optimal recognition sequence. Importantly, this epitope sequence is conserved in the LMP2 gene of all Caucasian and Chinese EBV isolates so far sequenced, including the viruses present in the tumor cells of EBV-positive HD and NPC (data not shown). Furthermore, analysis by enzyme-linked immunospot (ELISPOT) assay for peptide-induced gamma interferon (IFN-γ) release revealed that CD8+ T cells specific for the FLY epitope are present in a high proportion of EBV-seropositive, HLA-A*0201-positive donors at levels (up to 168/106 PBMCs) that were generally as strong as the response to CLG and stronger than the response to LLW in the same individuals (Table 1) (5). Clearly therefore the FLY epitope is immunogenic in vivo, and FLY-specific responses are likely to play a significant role in the physiologic control of EBV infection in A*0201-positive individuals. It is worth pointing out that the FLY T-cell clone illustrated in Fig. 1, like CD8+ T-cell clones expanded in vitro against several different EBV epitopes, including CLG and LLW, shows little if any baseline killing of autologous EBV-transformed LCL targets in conventional 5-h chromium release assays; only cells overexpressing the target antigen from a vaccinia virus vector elicit strong killing (5, 10). However, again, like clones against other epitopes, good recognition of the unmanipulated LCLs was observed when using IFN-γ release as the more sensitive ELISPOT assay (data not shown).
FIG. 1.
Identification and mapping of an LMP2-specific CTL clone directed against a novel HLA-A*0201-restricted epitope, FLYALALLL. (A) CTL clone donor A c30, derived by stimulation of PBMCs with irradiated autologous LCLs, was tested in a chromium release assay against autologous (auto) or partially HLA-matched LCL target cells infected with recombinant vaccinia viruses expressing either LMP2 (vLMP2) or, as a control, EBNA3A (vE3A), and A*0201-positive targets preexposed to the known LMP2-derived A*0201-restricted epitope peptides CLGGLLTMV (CLG) or LLWTLVVLL (LLW), or to dimethyl sulfoxide (DMSO) solvent control. (B) donor A c30 was tested in a chromium release assay against autologous LCL target cells preincubated with the synthetic peptides shown, representing LMP2 amino acids 353 to 370, at the indicated molar concentration. The minimal epitope was defined as FLYALALLL (FLY; LMP2 356 to 364). Results are expressed as the percentage of specific lysis observed in a standard 5-h chromium release assay at an effector/target ratio of 5:1.
TABLE 1.
Frequency of CD8+ T cells specific for HLA-A2-restricted epitopes in EBV LMP2a
| Donor no. | EBV statusb | HLA type | No. of SFCs/106 PBMCsc
|
||
|---|---|---|---|---|---|
| FLY | CLG | LLW | |||
| 1 | + | A1, A2, B8, B39 | 80 | 65 | 5 |
| 2 | + | A2, A11, B44, B35 | 45 | 30 | 35 |
| 3 | + | A2, A24, B40, B44 | 158 | 7 | 13 |
| 4 | + | A2, A11, B35, B44 | 10 | 15 | NTd |
| 5 | + | A2, A24, B27, B35 | 88 | 219 | 46 |
| 6 | + | A2, A32, B44 | 88 | 65 | 13 |
| 7 | + | A2, A11, B8, B44 | 52 | 50 | 8 |
| 8 | + | A1, A2, B8 | 168 | 68 | 53 |
| 9 | + | A2, A11, B8, B18 | 38 | 60 | 5 |
| 10 | + | A2, A31, B49, B60 | 18 | 33 | 8 |
| 11 | + | A2, A11, B18, B44 | 87 | 23 | 40 |
| 12 | + | A11, A24, B35, B52 | 0 | 0 | 0 |
| 13 | − | A2, B40, B44 | 0 | 0 | 0 |
The epitopes tested were FLYALALLL (FLY), CLGGLLTMV (CLG), and LLWTLVVLL (LLW).
EBV status was determined serologically by staining for viral capsid antigen. +, positive; −, negative.
Results are expressed as the number of spot-forming cells (SFCs)/106 PBMCs (after subtracting spots in control wells with no peptide).
NT, not tested.
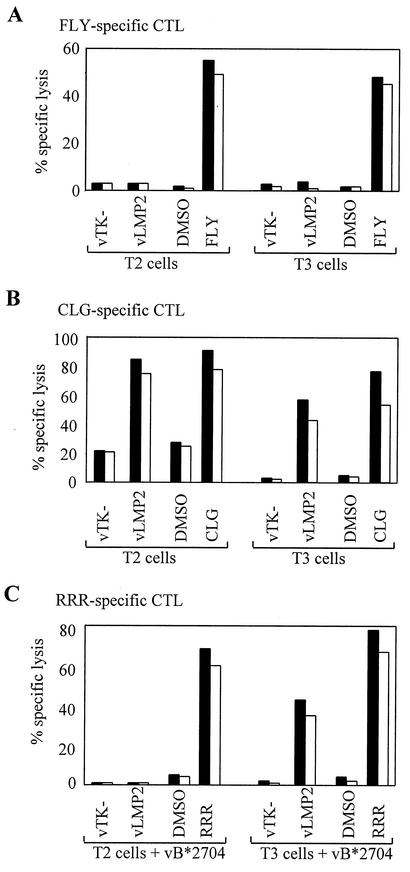
In a previous study, we identified a number of LMP2-derived epitopes that unexpectedly were presented in the background of TAP-negative T2 cells (14). This revealed a clear correlation between hydrophobicity of the epitope sequence and TAP-independent presentation. Thus, the A*0201-restricted epitopes LLW and CLG, with hydrophobicities of 8.14 and 4.14, respectively, were TAP independent, whereas epitopes RRR and IED restricted through B*2704 and B*4001 and with hydrophobicities of −0.60 and −0.59, respectively, were TAP dependent (14). Calculation of the hydrophobicity of the FLY epitope as 7.64 (29) predicted that this epitope would also be TAP independent. To investigate this, we carried out chromium release assays by using as targets either T2 cells or a rat TAP1/TAP2 transfectant of T2 (designated T3 cells), in both cases expressing LMP2 via a recombinant vaccinia virus (vLMP2). Figure 2A shows the results of such assays with FLY-specific effector CTLs. Interestingly LMP2 was not processed to generate the FLY epitope in either T2 or T3 cells, yet both target lines pulsed with the cognate peptide epitope were efficiently recognized. The inability of FLY-specific effectors to recognize endogenously expressed LMP2 in either T2 or T3 cell backgrounds was confirmed by using IFN-γ release as the detection assay (data not shown). Figure 2B and C show the different cytotoxicity assay results obtained when the same set of target cells were tested against reference effector CTL clones specific for TAP-independent (CLG) and TAP-dependent (RRR) epitopes from LMP2. As expected, the CLG epitope was presented from endogenously expressed LMP2 in both T2 and T3 cells, whereas the B27-restricted RRR epitope was only presented in the T3 cells, with, in this case, the restriction element being provided by a vaccinia virus expressing HLA-B*2704.
FIG. 2.
The LMP2 FLY epitope is not presented to CTLs in the T2 cell background. (A) Chromium release assays carried out with CTLs specific for the FLY/A*0201 epitope as effectors. Targets include T2 cells infected with a recombinant vaccinia virus expressing LMP2 (vLMP2) or control virus vTK− and T3 cells (rat TAP1/TAP2-transfectants of T2) infected with either vLMP2 or control virus vTK−. Both target cell lines were also preexposed to either the cognate epitope peptide or to dimethyl sulfoxide (DMSO) solvent control. (B) Assays conducted with effector CTLs specific for the TAP-independent CLG/A*0201 epitope on the same targets as those described above. (C) Assays conducted using as effectors CTLs specific for the TAP-dependent RRR/B*2704 epitope on the same targets described above, but in this case also infected with a vaccinia virus, vB*2704, expressing the HLA-B*2704 allele. Results are expressed as the percentage of specific lysis observed in a standard 5-h chromium release assay at effector/target ratios of 5:1 (black bars) and 2:1 (white bars).
Given the unique pattern of results obtained with FLY effectors in the assays described above, we sought to determine whether the requirements for processing of LMP2 to generate the FLY epitope were in some way different. In this context, we had earlier shown that all other LMP2-derived epitopes (both TAP dependent and TAP independent) appeared to be generated by proteasomal degradation, since specific inhibitors of the proteasome, such as lactacystin and epoxomicin, blocked their presentation in LMP2-expressing cells (14); this is indeed the pathway by which most CD8+ epitopes are produced from endogenously expressed antigens (4, 6, 23). The catalytic core unit of the proteasome, the 20S proteasome, has a cylindrical structure composed of four stacked rings made up of 14α and 14β subunits (30). Importantly, the catalytic activity of this constitutively expressed complex has been mapped to three β subunits, β1, β2 and β5. In professional antigen-presenting cells, such as B cells and certain other cell types following IFN-γ induction, the subunit composition of the proteasome alters such that the active site β subunits are replaced by three IFN-γ-inducible subunits, low-molecular-weight protein 2, low-molecular-weight protein 7, and MECL-1, resulting in the formation of the immunoproteasome (11, 19). We noted that the major histocompatibility complex (MHC) region deletion that rendered T2 cells TAP negative has also removed the genes encoding two of the IFN-γ inducible immunoproteasome subunits, low-molecular-weight proteins 2 and 7 (here referred to as ip-lmp2 and ip-lmp7, respectively) (25). Recent studies have shown that these subunits, either individually or collectively, are required for efficient generation of particular CD8+ epitope sequences from native antigen (8, 26, 27). In an attempt to study this further, we took advantage of a series of TAP and ip-lmp transfectants that had been generated on the background of cell line .174, which was equivalent to T2 in processing function and was in fact the line from which the T2 clone was originally derived. However, these experiments showed that coexpression of the human TAP1 and TAP2 proteins plus either ip-lmp2 or ip-lmp7 was insufficient to rescue presentation of the FLY epitope (data not shown). Because there was no .174 transfectant in which TAP1/TAP2 and both ip-lmp subunits had been restored, we moved to the use of epithelial or fibroblast targets (i.e., to cells in which the immunoproteasome subunits could be induced by IFN-γ).
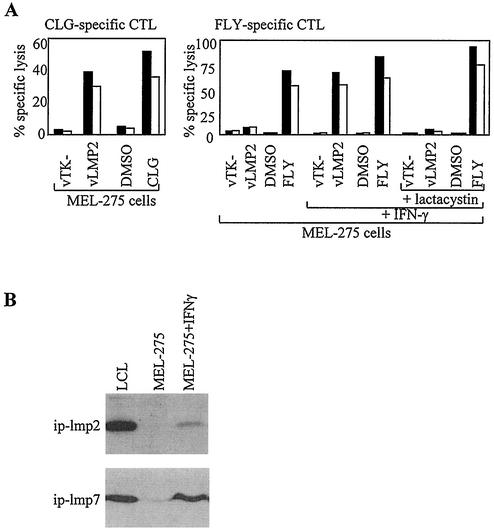
The first experiments of this type involved the TAP-positive, HLA-A*0201-positive melanoma cell line MEL-275, which preliminary work had identified as negative for the immunoproteasome subunits. As shown in Fig. 3A, assays with CLG-specific effectors first confirmed that after infection by vLMP2, these cells can efficiently present the CLG epitope to levels almost equivalent to those of peptide-pulsed targets. In contrast, the same target cells were not recognized by FLY-specific effectors; however, good recognition was achieved if the cells were first treated with IFN-γ (Fig. 3A), and such treatment was associated with the induction of significant levels of the ip-lmp2 and ip-lmp7 subunits (Fig. 3B). This presentation of FLY by IFN-γ-induced cells was completely blocked by pretreatment with the proteasomal inhibitor lactacystin (Fig. 3A) and by a second specific inhibitor, epoxomicin (17) (data not shown), while such IFN-γ-induced and drug-treated cells retained the ability to present exogenously added FLY peptide. This strongly suggests that the IFN-γ-induced immunoproteasome is necessary for generation of the FLY epitope.
FIG. 3.
Presentation of the LMP2 FLY epitope requires IFN-γ induction of the immunoproteasome. (A) The melanoma cell line MEL-275 was infected with a recombinant vaccinia virus expressing LMP2 (vLMP2) or a control virus, vTK−, or preexposed to the cognate epitope peptide or to dimethyl sulfoxide (DMSO) solvent control and then used as targets in chromium release assays with CLG-specific CTL effectors (left panel). MEL-275 cells were either untreated or pretreated with IFN-γ for 48 h prior to exposure to the vaccinia viruses or peptide described above and then used as targets for FLY-specific CTL effectors (right panel). IFN-γ-induced MEL-275 cells were also incubated with lactacystin for 1 h prior exposure to the vaccinia viruses or peptide described above and then used as targets for FLY-specific CTL effectors. Results of chromium release assays are shown as the percentage of specific lysis observed in a standard 5-h chromium release assay at effector/target ratios of 5:1 (black bars) and 2:1 (white bars). (B) Western blot analysis of MEL-275, both before and after IFN-γ induction, for expression of immunoproteasome subunits ip-lmp2 and ip-lmp7. A standard LCL was used as a positive control.
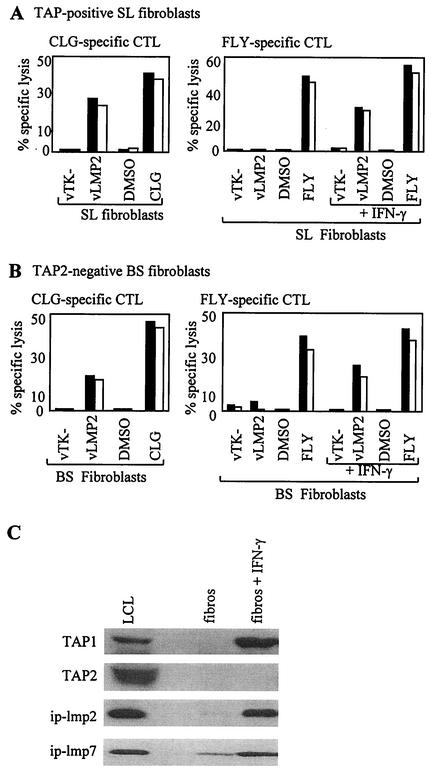
We subsequently showed that, like MEL-275, normal fibroblasts from an HLA-A*0201-positive donor (donor B) were able to present the CLG epitope but not the FLY epitope from vaccinia-expressed LMP2, but acquired the ability to present following IFN-γ induction (Fig. 4A). This proved important in view of the availability of fibroblasts from an HLA-A*0201-positive patient (patient C) with a homozygous TAP2 gene mutation that abrogated TAP function (S.G. and V.C., unpublished data). When the experiment was repeated in this TAP-defective fibroblast background, uninduced cells again presented the TAP-independent CLG epitope from LMP2 but not the FLY epitope; importantly, however, IFN-γ induction did allow FLY to be presented (Fig. 4B). Figure 4C confirms that the immunoproteasome subunits ip-lmp2 and ip-lmp7 are indeed induced by IFN-γ in patient C fibroblasts, as is TAP1, but these cells are incapable of making TAP2. There is one report on a study using mouse RMA/S cells, also expressing only TAP1, that raises the possibility that a TAP1 homodimer is capable of transporting peptides (7). However, from work with T2 cells, human TAP1 has an absolute requirement for TAP2 to transport peptides (9), and so we consider it very unlikely that in patient C fibroblasts a homodimer of TAP1 could be responsible for the ability of the FLY epitope to access the HLA class I presentation pathway. The data are therefore consistent with FLY being a TAP-independent-epitope. This is in accord with our proposed hypothesis that cytosolically generated peptides, if sufficiently hydrophobic, can access the endoplasmic reticulum by a novel TAP-independent pathway (14).
FIG. 4.
Presentation of the LMP2 FLY epitope is TAP-independent. (A) TAP-positive HLA-A*0201-positive donor B fibroblasts were infected with a recombinant vaccinia virus expressing LMP2 (vLMP2) or a control virus vTK− or preexposed to the cognate epitope peptide or to dimethyl sulfoxide (DMSO) solvent control and then used as targets in chromium release assays with CLG-specific CTL effectors (left panel). The same cells were either untreated or pretreated with IFN-γ for 48 h prior to exposure to the vaccinia viruses or peptide described above and then used as targets for FLY-specific CTL effectors (right panel). (B) The TAP2-negative HLA-A*0201-positive patient C fibroblast cells were infected with a recombinant vaccinia virus expressing LMP2 (vLMP2) or a control virus vTK− or preexposed to the cognate epitope peptide or to dimethyl sulfoxide solvent control and then used as targets in chromium release assays with CLG-specific CTL effectors (left panel). Patient C fibroblasts were either untreated or pretreated with IFN-γ for 48 h prior to exposure to the vaccinia viruses or peptide described above and then used as targets for FLY-specific CTL effectors (right panel). Results of chromium release assays are shown as the percentage of specific lysis observed in a standard 5-h chromium release assay at an effector/target ratio of 5:1 (black bars) and 2:1 (white bars). (C) Western blot analysis of patient C fibroblasts (fibros), both before and after IFN-γ induction, for expression of immunoproteasome subunits ip-lmp2 and ip-lmp7 and the TAP1 and TAP2 subunits. A standard LCL line was used as a positive control.
As to the route whereby the FLY epitope is generated from endogenously expressed LMP2, our data from both epithelial and fibroblast systems clearly show that the processing of FLY (i) is dependent upon IFN-γ induction of the target cells, (ii) is blocked by specific inhibitors of the proteasome, and (iii) is coincident with appearance of the IFN-γ-inducible immunoproteasome components ip-lmp2 and ip-lmp7 (Fig. 3 and 4) (data not shown). Besides the ip-lmp subunits, it is of course possible that some other IFN-γ-induced proteasomal activity is responsible for the change in processing capacity. In this context, the most likely candidate would be the proteasome modulator complex, PA28, which has been shown to influence the generation of antigenic peptides in a manner independent of the presence of immunoproteasome subunits (28). However, we found that PA28 is present at significant levels in donor B and patient C fibroblasts even before IFN-γ induction, in addition to being present in .174 and T2 cells (data not shown). This would suggest that PA28 is not the limiting factor in the generation of the FLY epitope. We therefore infer that FLY is indeed dependent upon the immunoproteasome components ip-lmp2 and ip-lmp7 for its generation.
In summary, we have identified a new EBV-encoded CD8+ T-cell epitope which could prove a therapeutically useful target in that it is restricted through a relatively common HLA allele, HLA-A*0201, and is derived from LMP2, one of the few viral proteins expressed in EBV-positive malignancies such as HD and NPC. Interestingly, though the epitope displays the expected TAP-independent phenotype, it also represents the first EBV epitope whose generation has been shown to be immunoproteasome dependent. These data emphasize a more general lesson: that as more “tumor-associated” epitopes are described, one needs to study the requirements for their generation within cells and to determine whether these requirements are met in the tumor cells themselves. In the particular context of the FLY epitope, it will be important to determine whether EBV-positive HD and NPC cells, for which there are very few if any truly representative cell lines in culture, have the capacity to present this epitope from endogenously expressed LMP2 in vivo.
Acknowledgments
This work was supported by Cancer Research UK and by the Medical Research Council, UK.
We thank I. Correa (Imperial College, London, United Kingdom), P. Cresswell (Yale University, New Haven, Conn.), A. Kelly (Cambridge University, Cambridge, United Kingdom), S. Powis (University of Dundee, Dundee, United Kingdom), M. Rowe (University of Wales College of Medicine, Cardiff, United Kingdom), and J. Trowsdale (Cambridge University) for kind gifts of cell lines and antibodies.
REFERENCES
- 1.Blake, N., S. Lee, I. Redchenko, W. Thomas, N. Steven, A. Leese, P. Steigerwald-Mullen, M. G. Kurilla, L. Frappier, and A. Rickinson. 1997. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity 7:791-802. [DOI] [PubMed] [Google Scholar]
- 2.Brooks, L., Q. Y. Yao, A. B. Rickinson, and L. S. Young. 1992. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J. Virol. 66:2689-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrows, S. R., A. Suhrbier, R. Khanna, and D. J. Moss. 1992. Rapid visual assay of cytotoxic T cell specificity utilizing synthetic peptide induced T-cell-T-cell killing. Immunology 76:174-175. [PMC free article] [PubMed] [Google Scholar]
- 4.Cerundolo, V., A. Benham, V. Braud, S. Mukherjee, K. Gould, B. Macino, J. Neefjes, and A. Townsend. 1997. The proteasome-specific inhibitor lactacystin blocks presentation of cytotoxic T lymphocyte epitopes in human and mouse cells. Eur. J. Immunol. 27:336-341. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, A. L. N., A. B. Rickinson, W. A. Thomas, R. J. Jarrett, J. Crocker, and S. P. Lee. 2001. Epstein-Barr virus-specific cytotoxic T lymphocyte responses in the blood and tumor site of Hodgkin's disease patients: implications for a T-cell based therapy. Cancer Res. 61:6219-6226. [PubMed] [Google Scholar]
- 6.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 7.Gabathuler, R., G. Ried, G. Kolaitis, J. Driscoll, and W. A. Jeffries. 1994. Comparison of cell lines deficient in antigen presentation reveals a functional role for TAP-1 alone in antigen processing. J. Exp. Med. 180:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gileadi, U., H. T. Moins-Teisserenc, I. Correa, B. L. Booth, P. R. Dunbar, A. K. Sewell, J. Trowsdale, R. E. Phillips, and V. Cerundolo. 1999. Generation of an immunodominant CTL epitope is affected by proteasome subunit composition and stability of the antigenic protein. J. Immunol. 163:6045-6052. [PubMed] [Google Scholar]
- 9.Heemels, M. T., and H. Ploegh. 1995. Generation, translocation, and presentation of MHC class I-restricted peptides. Annu. Rev. Biochem. 64:463-491. [DOI] [PubMed] [Google Scholar]
- 10.Hill, A. B., S. P. Lee, J. S. Haurum, N. Murray, Q. Y. Yao, M. Rowe, N. Signoret, A. B. Rickinson, and A. J. McMichael. 1995. Class I major histocompatibility complex-restricted T lymphocytes specific for Epstein-Barr virus (EBV) nuclear antigens fail to lyse the EBV-transformed B lymphoblastoid lines against which they were raised. J. Exp. Med. 181:2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hisamatsu, H., N. Shimbara, Y. Saito, P. Kristensen, K. B. Hendil, T. Fujiwara, E. Takahashi, N. Tanahashi, T. Tamura, A. Ichihara, and K. Tanaka. 1996. Newly identified pair of proteasomal subunits regulated reciprocally by interferon γ. J. Exp. Med. 183:1807-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khanna, R., S. R. Burrows, M. G. Kurilla, C. A. Jacob, I. S. Misko, T. B. Sculley, E. Kieff, and D. J. Moss. 1992. Localization of Epstein-Barr virus cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development. J. Exp. Med. 176:169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khanna, R., S. R. Burrows, J. M. Nicholls, and L. M. Poulsen. 1998. Identification of cytotoxic T cell epitopes within Epstein-Barr virus (EBV) oncogene latent membrane protein 1 (LMP1): evidence for HLA A2 supertype-restricted immune recognition of EBV-infected cells by LMP1-specific cytotoxic T lymphocytes. Eur. J. Immunol. 28:451-458. [DOI] [PubMed] [Google Scholar]
- 14.Lautscham, G., S. Mayrhofer, G. Taylor, T. Haigh, A. Leese, A. Rickinson, and N. Blake. 2001. Processing of a multiple membrane spanning Epstein-Barr virus protein for CD8+ T cell recognition reveals a proteasome-dependent, transporter associated with antigen processing-independent pathway. J. Exp. Med. 194:1053-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, S. P., R. Tierney, W. A. Thomas, J. M. Brooks, and A. B. Rickinson. 1997. Conserved CTL epitopes within EBV latent membrane protein 2: a potential target for CTL-based tumor therapy. J. Immunol. 158:3325-3334. [PubMed] [Google Scholar]
- 16.Levitskaya, J., M. Coram, V. Levitsky, S. Imreh, P. M. Steigerwald-Mullen, G. Klein, M. G. Kurilla, and M. G. Masucci. 1995. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 375:685-688. [DOI] [PubMed] [Google Scholar]
- 17.Meng, L., R. Mohan, B. H. Kwok, M. Elofsson, N. Sin, and C. M. Crews. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96:10403-10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray, R. J., M. G. Kurilla, J. M. Brooks, W. A. Thomas, M. Rowe, E. Kieff, and A. B. Rickinson. 1992. Identification of target antigens for the human cytotoxic T cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J. Exp. Med. 176:157-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nandi, D., H. Jiang, and J. J. Monaco. 1996. Identification of MECL-1 (LMP-10) as the third IFN-γ-inducible proteasome subunit. J. Immunol. 156:2361-2364. [PubMed] [Google Scholar]
- 20.Niedobitek, G. E., E. Kremmer, H. Herbest, L. Whitehead, C. W. Dawson, E. Niedobitek, C. vonOstau, N. Rooney, F. A. Grasser, and L. S. Young. 1997. Immunohistochemical detection of the Epstein-Barr virus encoded latent membrane protein 2A in Hodgkin's disease and infectious mononucleosis. Blood 90:1664-1672. [PubMed] [Google Scholar]
- 21.Rickinson, A. B., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 22.Rickinson, A. B., and D. J. Moss. 1997. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 15:405-431. [DOI] [PubMed] [Google Scholar]
- 23.Rock, K. L., C. Gramm, L. Rothstein, K. Clark, R. Steijn, L. Dick, D. Hwang, and A. L. Goldberg. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78:761-771. [DOI] [PubMed] [Google Scholar]
- 24.Rooney, C. M., C. A. Smith, C. Y. Ng, S. K. Loftin, J. W. Sixbey, Y. Gan, D. K. Srivastava, L. C. Bowman, R. A. Krance, M. K. Brenner, and H. E. Heslop. 1998. Infusion of cytotoxic T cells for the prevention and treatment of Epstein-Barr virus-induced lymphoma in allogeneic transplant recipients. Blood 92:1549-1555. [PubMed] [Google Scholar]
- 25.Salter, R. D., and P. Cresswell. 1986. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 5:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sijts, A. J., T. Ruppert, B. Rehermann, M. Schmidt, U. H. Koszinowski, and P. M. Kloetzel. 2000. Efficient generation of a hepatitis B virus cytotoxic T lymphocyte epitope requires the structural features of immunoproteasomes. J. Exp. Med. 191:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sijts, A. J., S. Standera, R. E. M. Toes, T. Ruppert, N. J. C. M. Beekman, P. A. Van Veelen, F. A. Ossendorp, C. J. M. Melief, and P. M. Kloetzel. 2000. MHC class I antigen processing of an adenovirus CTL epitope is linked to the levels of immunoproteasome in infected cells. J. Immunol. 164:4500-4506. [DOI] [PubMed] [Google Scholar]
- 28.Sijts, A., Y. Yuancheng, K. Janek, S. Kral, A. Paschen, D. Schadenorf, and P. M. Kloetzel. 2002. The role of the proteasome activator PA28 in MHC class I antigen processing. Mol. Immunol. 39:165-169. [DOI] [PubMed] [Google Scholar]
- 29.Sweet, R. M., and D. Eisenberg. 1983. Correlation of sequence hydrophobicities measures similarity in the three-dimensional protein structure. J. Mol. Biol. 171:479-488. [DOI] [PubMed] [Google Scholar]
- 30.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]