Abstract
Previous studies have shown that infection of G0-synchronized human fibroblasts by human cytomegalovirus (HCMV) results in a block to cellular DNA synthesis. In this study, we have examined the effect of viral infection on the formation of the host cell DNA prereplication complex (pre-RC). We found that the Cdc6 protein level was significantly upregulated in the virus-infected cells and that there was a delay in the expression of the Mcm family of proteins. The loading of the Mcm proteins onto the DNA pre-RC complex also appeared to be defective in the virus-infected cells. This inhibition of DNA replication licensing was associated with the accumulation of geminin, a replication inhibitor. Cdt1, which participates in the loading of the Mcm proteins, was also downregulated and modified differentially in the infected cells. Early viral gene expression was sufficient for the virus-induced alteration of the pre-RC, and the immediate-early protein IE1 was not required. These studies show that the inhibition of replication licensing in HCMV-infected cells is one of the multiple pathways by which the virus dysregulates the host cell cycle.
Human cytomegalovirus (HCMV), an ubiquitous betaherpesvirus, is the leading viral cause of birth defects and poses a serious health threat to immunocompromised individuals (40). The development of strategies to prevent HCMV infection requires an understanding of the initial interactions between the virus and the host cell that promote the progression of the viral replication cycle and subsequent pathogenesis. As is the case with mitogens, HCMV infection of quiescent cells results in the rapid activation of the cellular proto-oncogenes c-fos, c-jun, and c-myc as well as an increased expression of ornithine decarboxylase, thymidine kinase, DNA polymerase alpha, and dihydrofolate reductase (1, 8, 19, 22, 52). In addition, increased levels of p53 and hyperphosphorylated Rb are observed in the virus-infected cells (23). HCMV also induces elevated levels of cyclin E and cyclin B and their associated kinase activities (23). In contrast, the expression of cyclin A and its associated kinase activity is inhibited (23). These combined effects suggest that HCMV adopts a strategy of early cellular activation that facilitates viral replication but simultaneously inhibits host cell DNA synthesis by an undefined mechanism.
Work from our laboratory and others has shown that the HCMV infection blocks cell cycle progression in primary human fibroblasts. In these studies, cells that were synchronized by serum starvation, contact inhibition, or both conditions, as well as asynchronous, proliferating cells, were used (2, 7, 23, 29, 46). The arrest occurs primarily at G1/S, but blockage at other points in the cell cycle also has been observed. It has been proposed that the immediate-early proteins (IE1 and IE2) and the virion constituent UL69 of HCMV contribute to the virus-mediated alteration in cell growth control (3, 30, 36, 55, 56).
Previously, it was demonstrated that the cell cycle phase at the time of the infection influences the virus-induced cell cycle dysregulation (11, 46). Cells that are infected on release from G0 arrest and most cells in G1 do not initiate cellular DNA synthesis at a time corresponding to S phase in the mock-infected cells. In contrast, nearly 50% of the cells infected in S phase are able to pass through G2/M before they arrest (11). Although the failure to induce cyclin A in the virus-infected cells probably plays a role in the blockage of cellular DNA synthesis, we were interested in determining whether the virus might affect key steps in cellular DNA replication prior to the requirement for cyclin A.
DNA replication in eukaryotic cells is precisely regulated such that the genomic DNA is replicated completely and only once during a single cell cycle (6, 12, 28). The first step involves the assembly of prereplication complexes (pre-RC) at the replication origins. This happens in a stepwise manner. The origin recognition complex (Orc) (44, 50, 51), a multisubunit complex, binds to the origins of DNA replication and remains bound during most of the cell cycle (see the model in Fig. 6A). Cdc6 then binds to the complex and facilitates the loading of the family of Mcm (Mcm2-7) proteins. Pre-RC formation, also referred to as “licensing,” occurs during the interval between the end of mitosis and the middle of the G1 phase (35, 39). Recently, it has been found that another protein, Cdt1, is recruited to the pre-RC independently of Cdc6 and is also required for the loading of the Mcm2-7 complex (32, 37, 38, 45). Cdt1 itself is regulated by a protein called geminin, which has been implicated as an inhibitor of DNA replication. The evidence suggests that it interacts with Cdt1 and thus blocks the binding of the Mcm complex to the pre-RC (31, 34, 48, 58). The activation of the pre-RC occurs at the G1/S boundary after licensing and is mediated by the action of S-phase cyclin-dependent kinases (Cdks), mainly cyclin A/Cdk2, cyclin E/Cdk2, and Cdc7/Dbf4 (24, 54). These protein kinases trigger a chain of reactions that lead to the binding of Cdc45 to the origin and the phosphorylation of Cdc6 and the Mcms. As a result, the DNA duplex unwinds, facilitating the loading of the DNA polymerase machinery (53). The hexameric Mcm complex possesses helicase activity and is thought to be involved in both the initiation and elongation steps of DNA synthesis (26, 27). The phosphorylation of key components of this process by the Cdks facilitates the initiation of replication and at the same time helps to prevent rereplication during the G2 and M phases of the cell cycle (10, 15). The ubiquitin-proteasome degradation pathway also contributes to the process of DNA replication by regulating the timely disappearance of certain proteins (9, 34, 43).
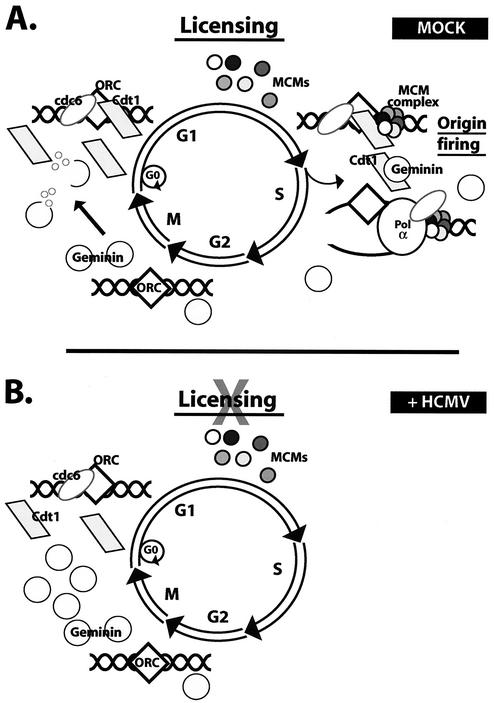
FIG. 6.
Model for the inhibition of replication licensing in HCMV-infected cells. (A) Formation of the pre-RC occurs in G1 by sequential loading of Cdc6, Cdt1, and Mcm proteins on an origin of DNA replication. Activation of the pre-RC or origin firing occurs in S phase under the influence of S-phase Cdks. Formation of the pre-RC is prevented during the remainder of the S, G2, and M phases mainly by the presence of the replication inhibitor geminin. On exit from M phase, geminin is degraded, thus allowing subsequent association of replication initiator proteins onto the origin in G1. (B) In HCMV-infected cells, accumulation of geminin inhibits the loading of Mcm proteins onto the origin, thereby blocking cellular DNA replication at the step of licensing.
In this work, we have extended our studies of the virus-host cell interactions by examining the effect of the HCMV infection on the cellular replication machinery. We find that HCMV induces elevated levels of Cdc6 and geminin but delays the expression of Mcm proteins. In addition, we present evidence that replication licensing is defective in the infected cells at the step of Mcm loading. These results suggest that the virus has evolved multiple and redundant pathways to inhibit the cell cycle. A possible mechanism underlying these effects and its implications are discussed.
MATERIALS AND METHODS
Cells and virus.
Primary human foreskin fibroblasts were obtained from the University of California, San Diego, Medical Center, and cultured in minimum essential medium with Earle's salts (Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum, l-glutamine (2 mM), penicillin (200 U/ml), streptomycin (200 μg/ml), and amphotericin B (1.5 μg/ml). The Towne strain of HCMV was obtained from the American Type Culture Collection and propagated as described previously (49). The recombinant Towne strain of HCMV that contains a deletion in exon 4 of the IE1 gene (CR208) (16) was a generous gift from Edward S. Mocarski (Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, Calif.).
Synchronization and infection.
All experiments were performed under G0 synchronization conditions as previously described (46). The cells were allowed to become confluent, and after 3 days of confluence, they were trypsinized and replated at a lower density to induce progression into the cell cycle. At the time of replating, they were infected with HCMV at a multiplicity of infection of 5 or mock infected with tissue culture supernatants as described previously (46). At different times post infection (p.i.), the cells were harvested, washed with phosphate-buffered saline, counted, and processed as described for each experiment. All experiments were performed at least twice.
Western blot analysis.
The cells were lysed in Laemmli reducing sample buffer (62.5 mM Tris [pH 6.8], 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 8 μg each of aprotinin and pepstatin A per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM benzamidine, 1 mM sodium metabisulfite, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 5 mM β-glycerophosphate) at 104 cells/μl of buffer. The cells were sonicated, boiled for 5 min, and centrifuged for 5 min at 16,000 × g. Equal amounts of lysate (105 cells/lane) were loaded onto an SDS-8% polyacrylamide gel for SDS-polyacrylamide gel electrophoresis (PAGE) unless otherwise stated. Following electrophoresis, the proteins were transferred to nitrocellulose (Schleicher & Schuell) and Western blot analysis was performed using appropriate mouse or rabbit antibody followed by appropriate horseradish peroxidase-linked secondary antibody. The Pierce Supersignal West pico and West femto chemiluminescent detection methods were used to visualize the proteins as specified by the manufacturer.
Subcellular fractionation.
At appropriate times p.i., the cells were harvested, washed with cold phosphate-buffered saline, and lysed in CSK buffer (10 mM HEPES-KOH, [pH 7.4], 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 0.5% NP-40), containing 8 μg each of aprotinin and pepstatin A per ml, 1 mM PMSF, 1 mM benzamidine, 1 mM sodium metabisulfite, 50 mM sodium fluoride, 1 mM sodium orthovanadate, and 5 mM β-glycerophosphate, at 2 × 104 cells per μl. The cells (3 × 106) were kept on ice for 15 min with occasional mild vortexing and then centrifuged at low speed (325 × g for 3 min). The supernatant was collected, and the pellet was washed twice; the supernatant was retained after each wash. The three resulting supernatants were centrifuged at high speed (16,000 × g for 5 min) and designated as the soluble fractions—fraction 1 (F1), fraction 2 (F2), and fraction 3 (F3). The residual pellet (F4) remaining after three washes was resuspended in lysis buffer and sonicated. An equal volume of 2× SDS-PAGE loading buffer was added to each of the fractions, and the samples were boiled for 5 min and analyzed by SDS-PAGE and Western blotting.
Phosphatase treatment.
The cells were lysed in buffer A (50 mM Tris-HCl [pH 7.5], 10 mM KCl, 1 mM MgCl2, 10% glycerol, 300 mM NaCl, 0.1% NP-40, 8 μg each of aprotinin and pepstatin A per ml, 1 mM PMSF, 1 mM benzamidine) or in buffer B (buffer A plus 50 mM sodium fluoride, 1 mM sodium orthovanadate, and 5 mM β-glycerophosphate). After incubation on ice for 5 min, the cells were subjected to three cycles of freezing and thawing (5 min at −80°C and then 5 min at 37°C). The lysate was then centrifuged at 16,000 × g for 10 min, the supernatant was collected, and the protein concentration was determined using the Bio-Rad protein assay. For λ-phosphatase treatment, the lysate (200 μg) was incubated with 1× λ-phosphatase buffer (New England Biolabs), 2 mM MnCl2, and 1,000 U of λ-phosphatase (New England Biolabs) for 30 min at 30°C. The controls (cell lysate in the presence of phosphatase inhibitors) were incubated in parallel. The reactions were terminated by adding SDS-PAGE loading buffer, and the samples were then boiled and analyzed by SDS-PAGE and Western blotting.
Antibodies.
The sources of the antibodies used were as follows: Orc1 (MS-649-P0; Neomarkers), Cdc6 (SC-9964; Santa Cruz Biotechnology), Mcm2 (68676E; BD Pharmingen), Mcm3 (68686E; BD Pharmingen), Mcm4 (68696E; BD Pharmingen), Mcm5 (68706E; BD Pharmingen), Mcm6 (68716E; BD Pharmingen), Mcm7 (MS-862-P0; NeoMarkers), lamins A/C (SC-7292; Santa Cruz Biotechnology), proliferating-cell nuclear antigen (PCNA) (SC-56; Santa Cruz Biotechnology), Dbf4 (SC-11354; Santa Cruz Biotechnology), and acetylated histone H3 (06-599; Upstate Biotech). The antibody against Cdt1 was a kind gift from Hideo Nishitani (Graduate School of Medical Science, Kyushu University, Kyushu, Japan), and the antibody against geminin was a kind gift from Anindya Dutta (Department of Pathology, Brigham and Women's Hospital and Harvard Medical School, Boston, Mass.). The antibody against Cdc7 was a gift from Hisao Masai (Department of Molecular and Developmental Biology, Institute of Medical Science, University of Tokyo, Tokyo, Japan). The CH16.0 antibody against the HCMV IE1 and IE2 proteins was obtained from the Goodwin Institute.
RESULTS
The expression of proteins associated with cellular DNA replication is altered by the HCMV infection.
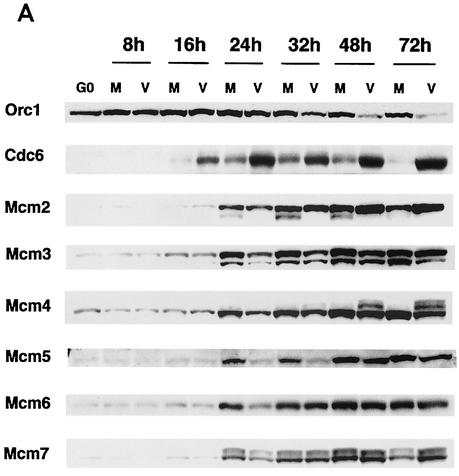
We and others have demonstrated that infection of G0-synchronized human fibroblasts with HCMV results in dysregulation of the expression of various cellular proteins and arrest of the cell cycle (2, 7, 29, 46). Cellular DNA synthesis was blocked in cells infected at the time of release from G0, and this was associated with the absence of cyclin A mRNA and protein in these cells. To examine more closely the effects of the HCMV infection on earlier events involved in the initiation of cellular DNA synthesis that occur before a requirement for cyclin A, we used Western blot analysis to measure the steady-state level of proteins associated with the formation of the prereplication complex during the course of the infection. As shown in Fig. 1, the expression of the respective proteins in the mock and viral samples was noticeably different between 16 and 32 h p.i.
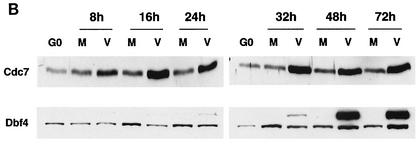
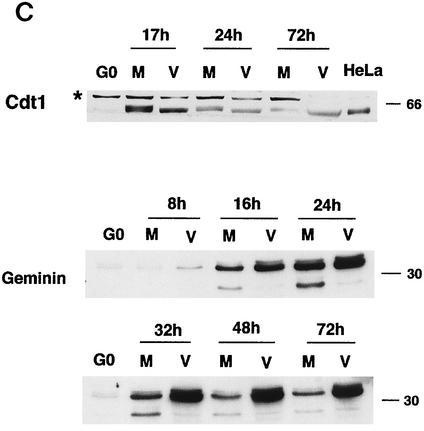
FIG. 1.
Effects of the HCMV infection on the accumulation of cellular replication proteins. Human fibroblasts were synchronized at G0 by contact inhibition. The cells were then stimulated to enter the cell cycle by replating at lower density and were simultaneously infected with either HCMV or tissue culture supernatant (mock) in 10% fetal bovine serum. At various time points, the cells were harvested, lysed in SDS loading buffer, and analyzed by Western blotting. Lysates from an equal number of cells (105) were loaded in each lane for the mock-infected (lanes M) and virus-infected (lanes V) samples. The proteins shown are Orc1, Cdc6, and Mcm proteins (A), Cdc7 and Dbf4 (B), and geminin and Cdt1 (C). HeLa in panel C is a HeLa cell lysate obtained from Transduction Laboratories and was used to identify the band that corresponds to Cdt1. The asterisk in panel C indicates reactivity with a nonspecific protein.
The levels of Orc1 were slightly higher in both the mock- and virus-infected cells than in the G0 cells after 8 h p.i. and remained at these levels up to the 24-h time point (Fig. 1A). However, after 32 h the Orc1 level declined in the infected cells while remaining the same in the mock-infected cells.
The Cdc6 level was low in G0 and then began to increase between 16 and 24 h p.i. in both the mock- and virus-infected cells, with the level in the infected cells being considerably higher than that in the controls (Fig. 1A). In the mock-infected cells, the Cdc6 level remained high up to 32 h and then declined gradually, reflecting the cycling of this protein during the cell cycle. In contrast, Cdc6 protein levels in the infected cells remained high throughout the infection. Analysis of the mRNA showed that this increase was not due to effects at the transcriptional level (data not shown).
The level of Mcm proteins was low in G0, began to increase at 16 to 24 h p.i. in both the mock- and virus-infected cells, and remained relatively constant thereafter. In general, the accumulation of Mcm proteins lagged in the infected cells, with Mcm5 and Mcm6 showing the greatest difference between virus- and mock-infected samples (Fig. 1A).
Several changes in mobility of the Mcm proteins were also noticed. A faster-migrating form of Mcm2, which has been reported to be the hyperphosphorylated species, was present in the mock-infected but not in the virus-infected cells at the 24-, 32-, and 48-h time points. We also detected a slower-migrating form of Mcm4 in the infected cells at 48 and 72 h p.i. Overall, these observations indicate that the expression of the Mcm proteins is delayed in the infected cells during the first 24 h p.i. and that there is probably differential modification of some of the proteins.
Cdc7 was upregulated in the infected cells between 8 and 16 h p.i., and the level remained high throughout the infection (Fig. 1B). For Dbf4, the regulatory subunit of Cdc7, there was a modest induction in the mock-infected cells at around 16 h p.i. (Fig. 1B). Although the level of Dbf4 was somewhat lower in the infected cells between 16 to 32 h p.i., it increased at the later time points. The most striking change was the accumulation of a higher-molecular-weight form of Dbf4 in the infected cells at late times in the infection. The mobility of this slower-migrating protein did not change after phosphatase treatment (data not shown), indicating that the infected-cell specific protein does not represent a highly phosphorylated form of Dbf4. Whether this protein is an alternative form of Dbf4 or simply a virus-induced cross-reacting species remains to be determined.
We also measured the expression of two other proteins involved in the initiation of replication, Cdt1 and geminin (Fig. 1C). A 65-kDa protein band corresponding to Cdt1 appeared in the mock-infected cells at 16 to 18 h p.i., after which its level diminished steadily. The asterisk next to the band migrating slower than the 66-kDa marker indicates reactivity with a nonspecific protein. The induction of Cdt1 also occurred in the virus-infected cells, but the protein was detected as a faster-migrating form throughout the infection. At 17 h p.i., the Cdt1 protein level was slightly lower in the virus-infected cells than in the mock-infected control. In contrast, the expression of geminin was upregulated in the infected cells as early as 8 h p.i. and its level increased significantly as the infection progressed. There also appeared to be accumulation of a slightly slower-migrating form in the infected cells. A faster-migrating band was observed in the mock-infected samples between 24 and 32 h p.i., and its level decreased at later times. This band was also seen in the infected cells, but the level remained very low throughout the infection (Fig. 1C).
HCMV infection affects the formation of the pre-RC.
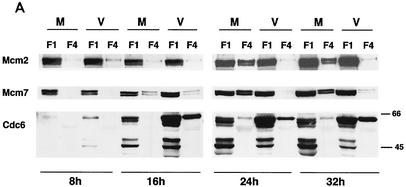
It has been reported that the assembly of the pre-RC complex or RC on a DNA replication origin is associated with a nuclear matrix and/or chromatin structure that can not be extracted by nonionic detergents (13, 25, 35). To analyze the components of the pre-RC in the virus- and mock-infected cells more closely, a subcellular fractionation was carried out as described in Materials and Methods. The cells were extracted with nonionic detergent to remove all of the soluble proteins, and the fractions containing the soluble proteins (F1) and chromatin-nuclear matrix-bound proteins (F4) were then analyzed (Fig. 2A). It should be noted that the Western blot shown has been overexposed so that the F4 lanes can be visualized. Thus, the exposure for most of the F1 lanes is out of the linear range.
FIG. 2.
HCMV infection leads to the alteration of pre-RC formation. The cells were lysed at 8, 16, 24, and 32 h p.i. in CSK buffer containing 0.5% NP-40, and the soluble (F1, F2, and F3) and chromatin-matrix (F4) fractions were prepared as described in Materials and Methods. (A) Only the soluble (F1) and chromatin-matrix (F4) fractions are shown. The wash fractions, F2 and F3, contained only traces of the proteins. Lysates from an equal number of cells (3 × 105) were loaded in each lane. The Western blot shown has been overexposed so that the F4 lanes can be visualized. Thus, the exposure for most of the F1 lanes is out of the linear range. (B) As controls, the fractionated samples from the 24-h time point were assayed for lamins and acetylated histone H3 (chromatin-matrix fraction) and PCNA (soluble fraction).
The level of Cdc6 in both F1 and F4 was higher in the virus-infected cells than in the mock-infected cells at all time points. Lower exposure revealed the presence of two or three Cdc6 bands in the soluble fraction (F1), whereas the chromatin-matrix fraction (F4) contained primarily the slower migrating form. The soluble fractions of both the mock-infected and virus-infected samples also showed several bands of approximately 45 kDa, which may be degradation products.
The amount of Mcm2 and Mcm7 present in the soluble fraction (F1) was comparable for the mock-infected and virus-infected samples throughout the time course. Although the chromatin-matrix fraction (F4) from the virus-infected cells had traces of these two proteins, the mock-infected sample showed significant amounts of Mcm2 and Mcm7 at the 24- and 32-h p.i. time points. Acetylated histone H3, lamins A and C, and PCNA served as controls for the fractionation, with the acetylated histone H3 and lamins A and C representing the chromatin-matrix fraction and PCNA representing the soluble fraction (Fig. 2B).
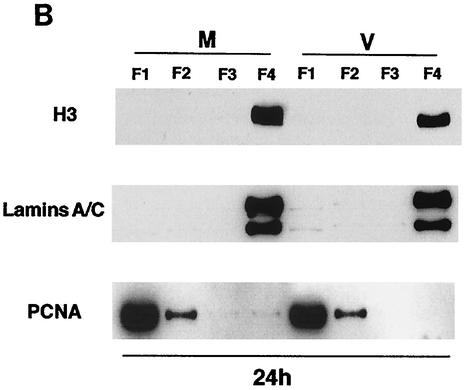
Cells infected in G1 show partial Mcm loading.
Previously, we showed that the pattern of HCMV-mediated cell cycle arrest is related to the phase of the cell cycle at the time of the infection (46). To determine whether the observed effects of the infection on the Mcm proteins and Cdc6 showed a similar dependence on the phase of the cell cycle, the cells were synchronized at G0 as described previously and plated at lower density and, at 12 h postplating (corresponding to the G1 phase), either mock infected or infected with the virus. The cells were then harvested at 12 h p.i., and the proteins present in the soluble (F1 and F2) and insoluble (F4) fractions were analyzed (Fig. 3). As observed for the G0 infection, the amount of Cdc6 present in F1 and F4 from the virus-infected cells was larger than that in the respective fractions from the mock-infected cells. However, in contrast to what was observed for the G0 infection, there was some accumulation of Mcm2 and Mcm7 in F4 from the virus-infected samples during the G1 infection.
FIG. 3.
Cells infected in G1 do not show a complete absence of licensing. The human foreskin fibroblasts were synchronized at G0 by contact inhibition and stimulated to enter the cell cycle by replating at lower density. At 12 h postplating, the cells were infected with HCMV or mock infected. After an additional 12 h, the cells were harvested and subjected to fractionation as described in Materials and Methods. Lanes in the Western blot contain the soluble (F1 and F2) and chromatin-matrix (F4) fractions from an equal number of cells (3 × 105). The Western blot has been overexposed so that the F4 lanes can be visualized.
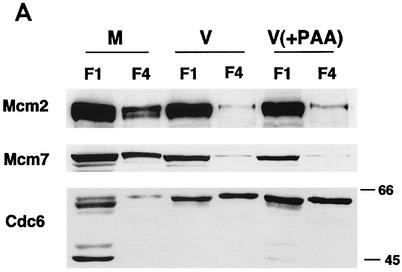
Early viral gene expression is sufficient for the virus-induced alteration of the pre-RC and the HCMV IE1-72 protein is not required.
To examine whether viral DNA replication is required for the effect on pre-RC formation, the cells were infected with HCMV in the presence and absence of phosphonoacetic acid (PAA), an inhibitor of viral DNA replication. The cells were harvested at 24 h p.i., and the soluble (F1) and chromatin-matrix (F4) fractions were analyzed (Fig. 4A). In the presence of PAA, there was little detectable accumulation of Mcm2 and Mcm7 in the chromatin-matrix fraction, similar to the control viral infection. The pattern of accumulation of Cdc6 in the infected cells also was unaffected by the presence of PAA.
FIG. 4.
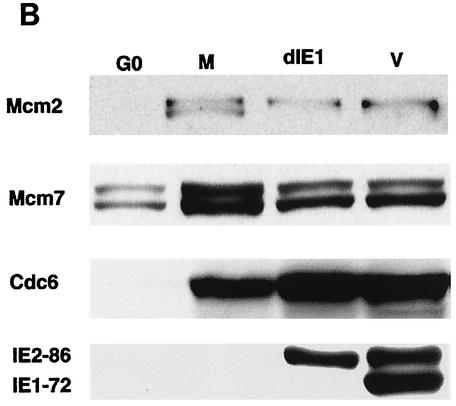
The effects on the pre-RC in infected cells do not require expression of the IE1-72 gene product or late proteins. (A) The cells were G0 synchronized and either mock infected (M) or infected with HCMV (V) in the absence or presence of 0.36 mM PAA (V+PAA). The cells were harvested after 24 h p.i. and subjected to fractionation, and the soluble (F1) and chromatin-matrix (F4) fractions from an equal number of cells (3 × 105) were analyzed by SDS-PAGE and Western blotting. The Western blot has been overexposed so that the F4 lanes can be visualized. Thus, the exposure for the F1 lanes is out of the linear range. (B) The cells were G0 synchronized by contact inhibition and mock infected (M), infected with CR208 (dIE1), or infected with Towne strain (V). The cell lysates were prepared at 24 h p.i. and analyzed as in panel A.
HCMV encodes two immediate-early proteins, IE1-72 and IE2-86, which play a role in the expression of viral early genes. To address whether IE1-72 is required for the virus-mediated alteration in the expression of Mcm proteins or Cdc6, an IE1 deletion mutant virus (CR208) (16) was used. The cells were synchronized in G0 as before and then mock infected or infected with wild-type HCMV or CR208. The cells were harvested for Western blot analysis at 24 h p.i., and the expression of Mcm2, Mcm7 and Cdc6 in the mutant virus-infected cells was compared to that in the cells infected with wild-type virus. As shown in Fig. 4B, similar effects were observed in the HCMV wild-type- and CR208-infected cells, indicating that IE1-72 is not required for the virus-mediated changes in Mcm and Cdc6 expression.
Modification of Mcm2, Mcm4, and Cdt1 in infected cells.
Protein phosphorylation plays a critical role in the regulation of various proteins. Western blot analysis revealed protein bands with altered mobility for Mcm2, Mcm4, and Cdt1 in the virus-infected cells. All of these proteins have multiple Cdk phosphorylation sites, and their phosphorylated forms have been described (15, 38, 42). Previously, we found that cyclinE/Cdk2 and cyclinB/Cdk1 activity in the infected cells was upregulated while cyclinA/Cdk2 activity was downregulated. Thus, we were interested in determining whether the modified forms of the proteins correlated with the Cdk activities present in the infected cells. Extracts from nocodazole-arrested uninfected cells served as a control for the phosphorylated forms of the respective proteins.
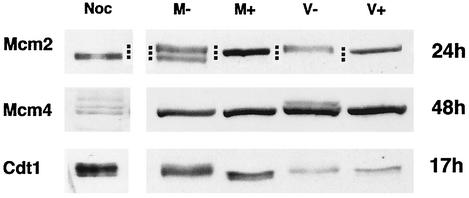
In uninfected cells, Mcm2 appears as a doublet, with the lowest band representing the hyperphosphorylated protein and the upper band a partially phosphorylated form. When the proteins that were harvested at 24 h p.i. from the mock-infected cells were treated with phosphatase, the unphosphorylated Mcm2 migrated at an intermediate position (Fig. 5). In the virus-infected cells, only the slowest-migrating partially phosphorylated protein was observed, and after phosphatase treatment it comigrated with the phosphatase-treated form from the mock-infected samples.
FIG. 5.
Alteration of phosphorylation of Mcm2 and Mcm4 in infected cells. Human fibroblasts were confluence synchronized at G0 and mock infected or infected with HCMV. The cells were harvested at 24 h p.i. for detection of Mcm2, at 48 h p.i. for detection of Mcm4, and at 17 h p.i. for detection of Cdt1. Cell lysates were treated with lambda phosphatase as described in Materials and Methods. The treated (M+ and V+) and untreated (M− and V−) samples were assayed for Mcm2, Mcm4, and Cdt1 by Western blotting. Noc corresponds to the nocodazole-arrested cell extract. The dots represent the mobility of three respective bands.
Mcm4 from the nocodazole-arrested cell extract appeared as several slower-migrating forms (Fig. 5). At 48 h p.i., Mcm4 from the infected cells was moderately phosphorylated, and after phosphatase treatment, it ran as a faster-migrating band, which comigrated with Mcm4 (plus or minus phosphatase) from the mock-infected cells.
Cdt1 from the viral samples migrated slightly faster than the protein from the mock-infected cells but slower than the phosphatase-treated mock-infected samples (Fig. 5). There was a small difference in the migration of Cdt1 from the virus-infected samples after phosphatase treatment, but it still migrated slightly slower than did the phosphatase-treated protein from the mock-infected cells. Although the mobility differences are small, they were consistently observed. These results suggest that the altered mobility of the viral species is not due entirely to phosphorylation.
DISCUSSION
In this paper, we have focused on the effect of HCMV infection on the early steps of cellular DNA synthesis. Our findings can be summarized as follows: (i) the expression of the Mcm proteins is delayed in the infected cells; (ii) the loading of the Mcm proteins onto the pre-RC is defective, as evidenced by the absence of Mcm2 and Mcm7 in the chromatin-matrix fraction of the infected cells; (iii) the expression of a second licensing factor, Cdt1, is downregulated, and it is present as a faster-migrating form in the virus-infected cells; and (iv) the expression of geminin, an inhibitor of cellular DNA replication, is upregulated in the infected cells. Taken together, these results suggest that the licensing of host cell DNA replication is impaired in the HCMV-infected cells. Thus, DNA replication is blocked before a requirement for active cyclin A kinase.
According to the current view of mammalian cell DNA replication (a model is given in Fig. 6), formation of the pre-RC occurs after mitosis during G1. In actively cycling cells, the levels of Orc1 and Mcm proteins remain relatively constant throughout the cell cycle while the levels of some of the replication initiation proteins (i.e., Cdc6, Cdt1, Dbf4, and geminin) fluctuate. Since the studies reported here used G0-synchronized cells, progression of the cell cycle requires the synthesis and/or induction of the majority of the replication proteins. We observed the induction of Cdc6, Mcm proteins, Cdc7, Dbf4, and Cdt1 in both the mock- and virus-infected cells (Fig. 1). The induction of several Mcm RNAs has been described in the HCMV-infected cells (47), and the expression of these proteins in the infected cells may be delayed at the translational level.
In uninfected cells, the induction of Cdc6 during G1 occurs at the transcriptional level and is regulated by the E2F transcription factors (17, 57). To ensure that only a single round of DNA replication occurs during each cell cycle, the level of mammalian Cdc6 is highly regulated at both the transcriptional and posttranslational levels. One of the two forms of Cdc6 can be extracted with nonionic detergent and is present in variable amounts (Fig. 2A). The level of the other insoluble form remains constant during the cell cycle (4, 14). Biochemical studies suggest that Cdc6 undergoes phosphorylation at the G1/S transition and is then exported to the cytoplasm (5, 25, 42). The mobility of Cdc6 in the mock- and virus-infected cells remained identical (Fig. 1A), indicating that the virus did not significantly affect the phosphorylation of Cdc6. In early G1, the ubiquitin-mediated proteolytic breakdown of Cdc6 is regulated by the anaphase-promoting complex, an E3 ubiquitin ligase (43). Our finding that there are high levels of Cdc6 in the virus-infected cells without a concomitant increase in the mRNA level suggests that this degradative pathway may be affected by the infection (Fig. 1A and 2A), and this possibility is being investigated in our laboratory.
Previously, we reported that HCMV infection induces hyperphosphorylation of Rb (23). In this study, we have also observed virus-mediated changes in the phosphorylation of Mcm2 and Mcm4, as evidenced by their mobility differences and sensitivity to phosphatase treatment (Fig. 1A and 5). Both Mcm2 and Mcm4 exhibit different phosphorylation states that depend on the cell cycle phase and their association with chromatin (15). Mcm2 is a key physiological substrate for Cdc7/Dbf4 kinase and undergoes phosphorylation at the time of pre-RC activation (24, 33). Mcm2 also can be phosphorylated by Cdks, and the hyperphosphorylated form is present in the G2/M phase of the cell cycle and contributes to the suppression of rereplication. The reason for the lack of the hyperphosphorylated form of Mcm2 in infected cells is unknown, particularly since the Cdc7/Dbf4 kinase activity does not seem to be significantly affected by the infection (data not shown). Nevertheless, this result is consistent with the absence of chromatin-bound Mcm2 in the infected cells. In contrast to Mcm2, the hyperphosphorylated form of Mcm4 is present at later times during the infection (Fig. 1A). In uninfected cells, Mcm4 is phosphorylated by cyclinA/Cdk2 and cyclinB/Cdk1, primarily during mitosis (15). The protein is then dephosphorylated as the cells progress to G1 to allow for association of the hexameric Mcm complex with the chromatin (41). Phosphorylation of Mcm4 by Cdks leads to the loss of Mcm4, Mcm6, and Mcm7 helicase activity (18, 20, 21). Thus, the presence of hyperphosphorylated Mcm4 in the virus-infected cells might inhibit the activity of the hexameric Mcm complex as the infection progresses (Fig. 5). Cdt1 also undergoes different modifications in S phase and mitosis (38). Our results suggest that Cdt1 in the virus-infected cells is modified, but only partially, by phosphorylation (Fig. 1C and 5). At present, it is not known whether phosphorylation is required for its activity or for its interaction with geminin (see below).
To study the composition of the pre-RC, we used subcellular fractionation. The accumulation of Cdc6 in the chromatin-matrix fraction in the mock- and virus-infected cells suggests that pre-RC formation proceeded normally through Cdc6 loading (Fig. 2A and 6). The presence of the slower-migrating form of Cdc6 in the chromatin-matrix fraction is consistent with observations from other laboratories (14). After origin firing, the rebinding of Mcm proteins is prevented by the presence of high Cdk activities in the G2 and M phases of the cell cycle. Although the elevated levels of cyclinE/Cdk2 and cyclinB/Cdk1 kinase activities in the infected cells might suggest a reason for the absence of Mcm loading (Fig. 2A), we would then expect that the Mcm proteins should be present in their hyperphosphorylated forms. As noted above, only Mcm4 was significantly phosphorylated in the infected cells. We did not observe any mobility differences for Mcm3, Mcm5, Mcm6, and Mcm7 up to 32 h p.i. (Fig. 1A), and the hypophosphorylated form of Mcm2 predominated in the infected cells (Fig. 1A and 5).
We hypothesize that the high level of geminin in the infected cells may play a key role in blocking Mcm loading (Fig. 1C and 6). Geminin is a known inhibitor of DNA replication, possibly through its interaction with Cdt1 (31, 48, 58). However, the mode of association of these two proteins has not yet been well defined. The presence of Cdt1 in the immunoprecipitate of geminin from both the mock- and virus-infected cells at 17 h p.i. suggests that the complex formation between these proteins is not affected (results not shown). Thus, the high ratio of geminin to Cdt1 (Fig. 1C) could be responsible for preventing the loading of the Mcm proteins onto the chromatin in the virus-infected cells. The degradation of geminin, which occurs at the metaphase-anaphase transition, is thought to be mediated by the anaphase-promoting complex (APC) in uninfected cells, and this degradation continues through early G1. Hence, the low geminin concentration in G1 allows Cdt1 to promote the association of Mcm proteins onto the pre-RC. The finding that in the infected cells there is concomitant accumulation of geminin and cdc6, which are both normally regulated by the APC E3 ubiquitin ligase, provides additional support for the hypothesis that there might be virus-induced alteration of the APC pathway.
Previously, it has been shown that a small population of cells infected in the G1 phase enter the S phase rather than arrest (11, 46). In support of this, we observed some accumulation of Mcm proteins in the chromatin-matrix fraction of G1-infected cells (compare F4 of virus-infected cells at 24 h p.i. [Fig. 2A] and F4 of virus-infected cells [Fig. 3]). Thus, during the first 12 h postplating, some synthesis of replication proteins as well as licensing can take place before the addition of virus. Since we have observed the inhibition of replication licensing in G0-infected cells treated with PAA, which blocks viral DNA replication (Fig. 4A), it appears that neither viral DNA synthesis nor late-gene expression is required for the virus-induced inhibition of replication licensing.
Taken together, the studies presented here suggest that HCMV has adopted multiple redundant pathways to halt the cell cycle. Inhibition of the replication licensing thus provides another mechanism to block cell DNA replication in the infected cells.
Acknowledgments
We thank Elizabeth White for careful reading of the manuscript and members of the Spector laboratory for suggestions and help. We also are grateful for the generous gifts of antibodies from Hideo Nishitani, Anindya Dutta, and Hisao Masai. We are grateful for the gift of the IE1 mutant virus from Edward S. Mocarski.
This work was supported by NIH grants CA73490 and CA34729.
REFERENCES
- 1.Boldogh, I., S. AbuBakar, C. Z. Deng, and T. Albrecht. 1991. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J. Virol. 65:1568-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:156-160. [DOI] [PubMed] [Google Scholar]
- 3.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coverley, D., C. Pelizon, S. Trewick, and R. A. Laskey. 2000. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 113:1929-1938. [DOI] [PubMed] [Google Scholar]
- 5.Delmolino, L. M., P. Saha, and A. Dutta. 2001. Multiple mechanisms regulate subcellular localization of human CDC6. J. Biol. Chem. 276:26947-26954. [DOI] [PubMed] [Google Scholar]
- 6.Diffley, J. F. 2001. DNA replication: building the perfect switch. Curr. Biol. 11:R367-370. [DOI] [PubMed] [Google Scholar]
- 7.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estes, J. E., and E. S. Huang. 1977. Stimulation of cellular thymidine kinases by human cytomegalovirus. J. Virol. 24:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferreira, M. F., C. Santocanale, L. S. Drury, and J. F. Diffley. 2000. Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol. Cell. Biol. 20:242-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Findeisen, M., M. El-Denary, T. Kapitza, R. Graf, and U. Strausfeld. 1999. Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6, and MCM in egg extracts of Xenopus laevis. Eur. J. Biochem. 264:415-426. [DOI] [PubMed] [Google Scholar]
- 11.Fortunato, E. A., V. Sanchez, J. Y. Yen, and D. H. Spector. 2002. Infection of cells with human cytomegalovirus during S phase results in a blockade to immediate-early gene expression that can be overcome by inhibition of the proteasome. J. Virol. 76:5369-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita, M. 1999. Cell cycle regulation of DNA replication initiation proteins in mammalian cells. Front. Biosci. 4:D816-D823. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, M., T. Kiyono, Y. Hayashi, and M. Ishibashi. 1997. In vivo interaction of human MCM heterohexameric complexes with chromatin. Possible involvement of ATP. J. Biol. Chem. 272:10928-10935. [DOI] [PubMed] [Google Scholar]
- 14.Fujita, M., C. Yamada, H. Goto, N. Yokoyama, K. Kuzushima, M. Inagaki, and T. Tsurumi. 1999. Cell cycle regulation of human CDC6 protein. Intracellular localization, interaction with the human MCM complex, and CDC2 kinase-mediated hyperphosphorylation. J. Biol. Chem. 274:25927-25932. [DOI] [PubMed] [Google Scholar]
- 15.Fujita, M., C. Yamada, T. Tsurumi, F. Hanaoka, K. Matsuzawa, and M. Inagaki. 1998. Cell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinase. J. Biol. Chem. 273:17095-17101. [DOI] [PubMed] [Google Scholar]
- 16.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hateboer, G., A. Wobst, B. O. Petersen, L. Le Cam, E. Vigo, C. Sardet, and K. Helin. 1998. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol. 18:6679-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrickson, M., M. Madine, S. Dalton, and J. Gautier. 1996. Phosphorylation of MCM4 by cdc2 protein kinase inhibits the activity of the minichromosome maintenance complex. Proc. Natl. Acad. Sci. USA 93:12223-12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirai, K., and Y. Watanabe. 1976. Induction of alpha type DNA polymerases in human cytomegalovirus-infected WI-38 cells. Biochim. Biophys. Acta 447:328-339. [DOI] [PubMed] [Google Scholar]
- 20.Ishimi, Y., and Y. Komamura-Kohno. 2001. Phosphorylation of Mcm4 at specific sites by cyclin-dependent kinase leads to loss of Mcm4,6,7 helicase activity. J. Biol. Chem. 276:34428-34433. [DOI] [PubMed] [Google Scholar]
- 21.Ishimi, Y., Y. Komamura-Kohno, Z. You, A. Omori, and M. Kitagawa. 2000. Inhibition of Mcm4,6,7 helicase activity by phosphorylation with cyclin A/Cdk2. J. Biol. Chem. 275:16235-16241. [DOI] [PubMed] [Google Scholar]
- 22.Isom, H. C. 1979. Stimulation of ornithine decarboxylase by human cytomegalovirus. J. Gen. Virol. 42:265-278. [DOI] [PubMed] [Google Scholar]
- 23.Jault, F. M., J.-M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang, W., D. McDonald, T. J. Hope, and T. Hunter. 1999. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 18:5703-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, W., N. J. Wells, and T. Hunter. 1999. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl. Acad. Sci. USA 96:6193-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Labib, K., and J. F. Diffley. 2001. Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev. 11:64-70. [DOI] [PubMed] [Google Scholar]
- 27.Labib, K., S. E. Kearsey, and J. F. Diffley. 2001. MCM2-7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol. Biol. Cell 12:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei, M., and B. K. Tye. 2001. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell. Sci. 114:1447-1454. [DOI] [PubMed] [Google Scholar]
- 29.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madine, M., and R. Laskey. 2001. Geminin bans replication licence. Nat. Cell Biol. 3:E49-E50. [DOI] [PubMed] [Google Scholar]
- 32.Maiorano, D., J. Moreau, and M. Mechali. 2000. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404:622-625. [DOI] [PubMed] [Google Scholar]
- 33.Masai, H., E. Matsui, Z. You, Y. Ishimi, K. Tamai, and K. Arai. 2000. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 by Cdks. J. Biol. Chem. 275:29042-29052. [DOI] [PubMed] [Google Scholar]
- 34.McGarry, T. J., and M. W. Kirschner. 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93:1043-1053. [DOI] [PubMed] [Google Scholar]
- 35.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishitani, H., Z. Lygerou, T. Nishimoto, and P. Nurse. 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404:625-628. [DOI] [PubMed] [Google Scholar]
- 38.Nishitani, H., S. Taraviras, Z. Lygerou, and T. Nishimoto. 2001. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 276:44905-44911. [DOI] [PubMed] [Google Scholar]
- 39.Okuno, Y., A. J. McNairn, N. den Elzen, J. Pines, and D. M. Gilbert. 2001. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 20:4263-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 41.Pereverzeva, I., E. Whitmire, B. Khan, and M. Coue. 2000. Distinct phosphoisoforms of the Xenopus Mcm4 protein regulate the function of the Mcm complex. Mol. Cell. Biol. 20:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen, B. O., J. Lukas, C. S. Sorensen, J. Bartek, and K. Helin. 1999. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 18:396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen, B. O., C. Wagener, F. Marinoni, E. R. Kramer, M. Melixetian, E. L. Denchi, C. Gieffers, C. Matteucci, J. M. Peters, and K. Helin. 2000. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14:2330-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quintana, D. G., and A. Dutta. 1999. The metazoan origin recognition complex. Front. Biosci. 4:D805-D815. [DOI] [PubMed] [Google Scholar]
- 45.Rialland, M., F. Sola, and C. Santocanale. 2002. Essential role of human CDT1 in DNA replication and chromatin licensing. J. Cell Sci. 115:1435-1440. [DOI] [PubMed] [Google Scholar]
- 46.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: Influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song, Y. J., and M. F. Stinski. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. USA 99:2836-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tada, S., A. Li, D. Maiorano, M. Mechali, and J. J. Blow. 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamashiro, J. C., L. J. Hock, and D. H. Spector. 1982. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169). J. Virol. 42:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatsumi, Y., T. Tsurimoto, K. Shirahige, H. Yoshikawa, and C. Obuse. 2000. Association of human origin recognition complex 1 with chromatin DNA and nuclease-resistant nuclear structures. J. Biol. Chem. 275:5904-5910. [DOI] [PubMed] [Google Scholar]
- 51.Vashee, S., P. Simancek, M. D. Challberg, and T. J. Kelly. 2001. Assembly of the human origin recognition complex. J. Biol. Chem. 276:26666-26673. [DOI] [PubMed] [Google Scholar]
- 52.Wade, M., T. F. Kowalik, M. Mudryj, E. S. Huang, and J. C. Azizkhan. 1992. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol. Cell. Biol. 12:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 54.Walter, J. C. 2000. Evidence for sequential action of cdc7 and cdk2 protein kinases during initiation of DNA replication in Xenopus egg extracts. J. Biol. Chem. 275:39773-39778. [DOI] [PubMed] [Google Scholar]
- 55.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiebusch, L., and C. Hagemeier. 2001. The human cytomegalovirus immediate early 2 protein dissociates cellular DNA synthesis from cyclin-dependent kinase activation. EMBO J. 20:1086-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams, R. S., R. V. Shohet, and B. Stillman. 1997. A human protein related to yeast Cdc6p. Proc. Natl. Acad. Sci. USA 94:142-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wohlschlegel, J. A., B. T. Dwyer, S. K. Dhar, C. Cvetic, J. C. Walter, and A. Dutta. 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290:2309-2312. [DOI] [PubMed] [Google Scholar]