Abstract
Infectious bursal disease virus (IBDV), a member of the Birnaviridae family, is the causative agent of one of the most important infectious poultry diseases. Major aspects of the molecular biology of IBDV, such as assembly and replication, are as yet poorly understood. We have previously shown that encapsidation of the putative virus-encoded RNA-dependent RNA polymerase VP1 is mediated by its interaction with the inner capsid protein VP3. Here, we report the characterization of the VP1-VP3 interaction. RNase A treatment of VP1- and VP3-containing extracts does not affect the formation of VP1-VP3 complexes, indicating that formation of the complex requires the establishment of protein-protein interactions. The use of a set of VP3 deletion mutants allowed the mapping of the VP1 binding motif of VP3 within a highly charged 16-amino-acid stretch on the C terminus of VP3. This region of VP3 is sufficient to confer VP1 binding activity when fused to an unrelated protein. Furthermore, a peptide corresponding to the VP1 binding region of VP3 specifically inhibits the formation of VP1-VP3 complexes. The presence of Trojan peptides containing the VP1 binding motif in IBDV-infected cells specifically reduces infective virus production, thus showing that formation of VP1-VP3 complexes plays a critical role in IBDV replication.
Infectious bursal disease virus (IBDV) is a double-stranded nonenveloped RNA virus belonging to the Birnaviridae family (15). IBDV replicates in B-lymphocyte precursors developing within the Fabricius bursa of young chickens, leading to a severe immunodeficiency (20, 23).
The genome of IBDV is formed by two segments of double-stranded RNA (dsRNA). Segment A (3.2 kb) contains two partially overlapping open reading frames (ORFs) encoding the viral polyprotein and the nonstructural protein VP5. Segment B (2.8 kb) encodes VP1, the putative RNA-dependent RNA polymerase (RdRp) (18). VP5 (17 kDa) is dispensable for virus replication in vitro but important for pathogenicity (22, 27). By using both recombinant vaccinia viruses (rVVs) and IBDV VP5 knockout mutants, it has been recently shown that VP5 is a cytolytic protein that accumulates within the plasma membrane of infected cells and promotes the egress of the viral progeny (17, 28). The major capsid proteins are synthesized as a 109-kDa polyprotein that is autoproteolytically processed, rendering three proteins, VPX (48 kDa), VP3 (32 kDa), and VP4 (28 kDa). The C-terminal region of VPX is further processed to form VP2 (37 kDa) (20), (15). VPX-VP4, VP4-VP3, and VPX-VP2 cleavage sites have been described previously (6, 24). VPX, VP2, and VP3 are the major structural proteins. VP4 is responsible for the proteolytic maturation of the polyprotein, and it is not incorporated into the virion (2, 9, 12). VP3 contains a highly hydrophilic C-terminal region rich in prolines and charged amino acids (see Fig. 2A). It has been proposed that this region might bind genomic dsRNA. Indeed, it has recently been shown that VP3 is an RNA-binding protein (13). VP1 (97 kDa) shares a number of primary sequence features with RNA polymerases from diverse origins (4). However, direct evidence demonstrating its RNA polymerase activity is, as yet, lacking. VP1, also encapsidated into the IBDV particle, is linked to both ends of the dsRNA genome as well as to the internal surface of the capsid (16, 21). The VP1 counterpart of the infectious pancreatic necrosis virus, the prototype member of the Birnaviridae family, has been shown to be guanylated in vitro, and although it lacks a capping activity, this guanylation might be associated with a putative primase activity (8, 14, 19).
FIG. 2.
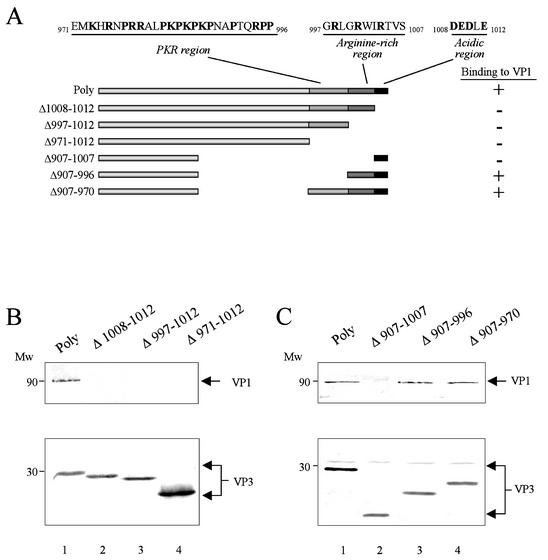
The 16 C-terminal residues of VP3 are necessary for the VP1-VP3 interaction. (A) The diagram depicts the set of VP3 deletion mutants used for the mapping of the VP1BM. Indicated by boxes are the three arbitrary VP3 C-terminal regions: (i) PKR; (ii) arginine rich; and (iii) acidic. COS cells were transfected with plasmids expressing the VP3 deletion mutants. The ability (+) or inability (−) of each mutant to bind to VP1 is also shown. (B and C) At 48 h p.t., cells were harvested and used to prepare protein extracts for in vitro binding assays. The extracts were mixed with an extract obtained from VT7/VP1-infected COS cells. After incubation, samples were subjected to IP with anti-VP3 serum. The resulting samples were subjected to SDS-PAGE and electroblotted onto nitrocellulose. To determine the position of the VP1 polypeptide, the upper part of the filter was incubated with anti-VP1 serum and the lower part was incubated with anti-VP3 serum. After washing, filters were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin. Signals were developed by enhanced chemiluminescence. The positions of molecular weight markers (Mw) and the VP1 and VP3 polypeptides (arrowheads) are indicated. (B) Effect of the VP3 carboxy-terminal deletions. (C) Effect of the VP3 internal deletions.
IBDV virions consist of an icosahedral single-shelled capsid of 65 to 70 nm with a T = 13 symmetry (3, 11). This is an exception among dsRNA viruses (i.e., reoviruses, rotaviruses, and orbiviruses) that usually contain two or three concentric protein shells (1). The external surface of the IBDV capsid is formed by trimeric subunits of VPX/2, and the inner surface is formed by trimeric subunits of VP3 (3, 5). In addition, VP1 interacts with VP3, leading to its encapsidation into virus-like particles (VLPs) in the absence of the IBDV genome (16, 25), supporting the idea that VP3 functions as a connector between VP1 and VPX/2 during the assembly of IBDV virions. However, neither the sequences responsible for VP1-VP3 interaction nor the biological role of the VP1-VP3 complexes has been analyzed.
In spite of the great impact for the poultry industry, little is known about key aspects of the molecular biology of IBDV. In this report we describe the characterization of the VP1-VP3 interaction that plays a crucial role during virus morphogenesis.
MATERIALS AND METHODS
Cells and viruses.
The VT7/VP1 and VT7/VP3 rVVs have been previously described (10, 16). Expression experiments were carried out with either BSC-1 or COS cells (American Type Culture Collection). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% newborn calf serum. The SalI IBDV strain adapted to grow in BSC-1 cells was grown and titrated as previously described (17).
Construction of plasmids expressing polyprotein deletion mutants.
The previously described plasmid pcDNA3/Poly (24) was digested with EcoRI and NotI, and the fragment corresponding to the complete ORF of the IBDV polyprotein was purified and cloned into the expression vector pCINeo (Promega Corporation, Madison, Wis.), previously digested with the same enzymes, giving rise to the plasmid pCINeo-Poly, which contains the IBDV polyprotein gene under the control of the cytomegalovirus early promoter.
The oligonucleotides used for the generation of polyprotein mutants are described in Table 1. The plasmids pPoly-Δ907-1012, pPoly-Δ907-1007, and pPoly-Δ907-996 were constructed by annealing the corresponding oligonucleotides and cloning the resulting DNA fragments into pCINeo-Poly linearized with XbaI and NotI.
TABLE 1.
Synthetic oligonucleotides used for the generation of constructs
| Name | Nucleotide sequence (5′-3′) |
|---|---|
| pPoly-Δ907-1012 | CTAGACTAATAAGC |
| pPoly-Δ907-1012c | GGCCGCTTATTAGT |
| pPoly-Δ907-996 | CTAGACGGTCGGCTGGGCCGCTGGATCAGGACCGTCTCTGATGAGGACCTTGAGTGAGCTGAGC |
| pPoly-Δ907-996c | GGCCGCTCACTCAAGGTCCTCATCAGAGACGGTCCTGATCCAGCGGCCCAGCCGACCGT |
| pPoly-Δ907-1007 | CTAGACGATGAGGACCTTGAGTGAGC |
| pPoly-Δ907-1007c | GGCCGCTCACTCAAGGTCCTCATCGT |
| pPoly-Δ100-1012 | GCGCGCGGCCGCTTAAGAGACGGTCCTGATCCAGCG |
| pPoly-Δ997-1012 | GCGCGCGGCCGCTTAAGGGGGTCTCTGTGTTGGAGC |
| pPoly-Δ971-1012 | GCGCGCGGCCGCTTACATCGCAGTCAAGAGCAGATC |
| Δ 3′ | GCGCGAGCTCCGTTTCCCTCACAATCCACGCGAC |
| pPoly-Δ907-970 | CGCGTCTAGACGAGATGAAGCATCGCAATCCC |
| pPoly-Δ907-970c | GCGCGCGGCCGCTCACTCAAGGTCCTCATCAG |
| pGFP-997-1012 | AATTCTGGTCGGCTGGGCCGCTGGATCAGGACCGTCTCTGATGAGGAGCTTGAGTGA |
| pGFP-997-1012c | GATCTCACTCAAGGTCCTCATCAGAGACGGTCCTGATCCAGCGGCCCAGCCGACCAG |
| pGFP-R-A | AATTCTGGTCGGCTGGGCGCCTGGATCAGGACCGTCTCTGATGAGGACCTTGAGTGA |
| pGFP-R-Ac | GATCTCACTCAAGGTCCTCATCAGAGAAGGTCCTGATCCAGGCGCCCAGCCGACCAG |
| pGFP-2R-A | AATTCTGGTGCCCTGGGCCGCTGGATCGCCACCGTCTCTGATGAGGACCTTGAGTGA |
| pGFP-2R-Ac | GATCTCACTCAAGGTCCTCATCAGAGACGGTGGCGATCCAGCGGCCCAGGGCACCAG |
| pGFP-3R-A | AATTCTGGTGCCCTGGGCGCCTGGATCGCCACCGTCTCTGATGAGGACCTTGAGTGA |
| pGFP-3R-Ac | GATCTCACTCAAGGTCCTCATCAGAGACGGTGGCGATCCAGGCGCCCAGGGCACCAG |
| pGFP-3R-K | AATTCTGGTAAGCTGGGCAAGTGGATCAAGACCGTCTCTGATGAGGACCTTGAGTGA |
| pGFP-3R-Kc | GATCTCACTCAAGGTCCTCATCAGAGACGGTCTTGATCCACTTGCCCAGCTTACCAG |
| pGFP-RXR | AATTCTGGTCTGCGGGGCCGCTGGAGGATCACCGTCTCTGATGAGGACCTTGAGTGA |
| pGFP-RXRc | GATCTCACTCAAGGTCCTCATCAGAGAAGGTCCTGATCCAGGCGCCCAGCCGACCAG |
| pGFP-DEDLE-A | AATTCTGGTCGGCTGGGCCGCTGGATCAGGACCGTCTCTGCCGCCGCCCTTGCCTGA |
| pGFP-DEDLE-Ac | GATCTCAGGCAAGGGCGGCGGCAGAGACGGTCCTGATCCAGCGGCCCAGCCGACCAG |
| pGFP-poly-A | AATTCTGCCCGGGCCGCCCGCGCCGCCAGGGCCGCCGCCGATGAGGACGCCGAGTGA |
| pGFP-poly-Ac | GATCTCACTCGGCGTCCTCATCGGCGGCGGCCCTGGCGGCGCGGGCGGCCCGGGCAG |
For the construction of plasmids pPoly-Δ1008-1012, pPoly-Δ997-1012, and pPoly-Δ971-1012, a set of DNA fragments was generated by PCR by using the coupled oligonucleotides described in Table 1 as primers and a DNA fragment containing the complete ORF of the IBDV polyprotein described above as a template. After purification, these PCR-derived fragments were digested with XbaI and NotI and ligated to the plasmid pPoly-Δ907-1012 previously linearized with the same enzymes.
The plasmid pPoly-Δ907-970 was constructed by PCR by using the coupled oligonucleotides described in Table 1 as primers and a DNA fragment containing the complete ORF of the IBDV polyprotein as a template. After purification, these PCR-derived DNA fragments were digested with XbaI and NotI and ligated to the plasmid pPoly previously linearized with the same enzymes.
Construction of plasmids for the expression of green fluorescent protein (GFP)-VP3 fusion proteins.
These constructs were generated by annealing the corresponding coupled oligonucleotides (Table 1). The resulting DNA fragments were cloned into pEGFPct (Clontech Laboratories, Inc., Palo Alto, Calif.) linearized with EcoRI and BamHI.
Formation and detection of VP3-VP1 complexes.
Protein extracts containing either VP3 or VP1 were obtained by infecting confluent monolayers of COS cells with VT7/VP3 or VT7/VP1 at a multiplicity of infection (MOI) of 5 PFU/cell. After infection, cultures were maintained in the presence of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM). At 20 h postinfection (p.i.), cells were harvested and protein extracts were prepared as previously described (16). When noted at 4 h p.i., infected cells were washed twice with methionine-free DMEM and metabolically labeled by adding methionine-free DMEM containing 100 μCi of [35S]methionine (Amersham International, Little Chalfont, United Kingdom)/ml and IPTG. Labeled cells were harvested at 20 h p.i., and processed as previously described (16). Extracts containing the polyprotein deletion mutants and GFP fusion proteins were obtained by transfecting monolayers of COS cells with the corresponding expression plasmids as previously described (24). Transfected cells were harvested at 48 h posttransfection (p.t.), and protein extracts were prepared. In vitro binding assays were performed by mixing a constant amount of the extract containing VP1 and the volume of extract, containing polyprotein deletion mutants or GFP fusion proteins, required to reach equivalent amounts of the proteins under study. The samples were mixed and incubated overnight (ON) at 4°C. Then, immunoprecipitation (IP) was carried out as previously described (16), with either anti-VP3 (10) or anti-GFP (Boehringer Mannheim GmbH, Mannheim, Germany) sera. After IP, samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. After blotting, membranes were cut in half. The upper halves were developed with anti-VP1 antibodies (16), and the lower ones were developed with either anti-VP3 or anti-GFP antibodies. Immunoreactive bands were detected as described above. When noted, radioactive signals were detected with a Storm gel imaging system (Molecular Dynamics, Sunnyvale, Calif.). Results were analyzed with ImageQuant software (Molecular Dynamics). For the analysis of GFP mutants, the anti-GFP chemiluminescent signal was detected by using a GEL DOC 2000 system (Bio-Rad Laboratories, Hercules, Calif.) and analyzed with Quantity One software (Bio-Rad Laboratories).
RNase treatment.
Lysates from cells infected with VT7/VP1 or VT7/VP3 were treated with either 0.2 or 1 mg of RNase A/ml for 15 min at 37°C. RNase-treated samples were then used for in vitro binding assays as described above.
Competition assays with synthetic peptides.
The synthetic peptides used in our study were H2N-GRLGRWIRTVSDEDLE-COOH (GRL) and H2N-GALGAWIATVSDEDLE-COOH (GAL). GRL contains the 16 C-terminal residues of the VP3 protein. The sequence of GAL, used as a control for the competition assays, is identical to that of GRL, with the exception that the arginine residues have been replaced by alanine residues. Both peptides were purified by high-pressure liquid chromatography and analyzed for concentration and composition to verify their identity. For the competition assays, increasing amounts of each peptide were added to the VP1-VP3 complex in the in vitro binding assays. Thereafter, assays were continued as described above.
Inhibition of virus replication with synthetic peptides.
The synthetic peptides used in our study were Biot-H2N-RQILIWFQNRRMKWKKGRLGRWIRTVSDEDLE-COOH (GRL-biot) and Biot-H2N-RQILIWFQNRRMKWKKGALGAWIATVSDEDLE-COOH (GAL-biot). The 16 N-terminal residues of both peptides contain the penetratin peptide sequence corresponding to the cell internalization motif of the antennapedia protein from Drosophila melanogaster (7). This sequence facilitates the internalization of the peptides into IBDV-susceptible cells. The 16 C-terminal residues of GRL-biot and GAL-biot are identical to those described for the oligopeptides GRL and GAL, respectively. Both peptides were biotinylated to allow their detection by immunofluorescence with a rhodamine-streptavidin conjugate. Peptides were purified by high-pressure liquid chromatography and analyzed for concentration and composition to verify their identity. Peptides were resuspended at a concentration of 5 mM in dimethyl sulfoxide. BSC-1 cell monolayers, grown in 60-mm-diameter dishes, were washed twice with phosphate-buffered saline, and then 1 ml of DMEM containing various peptide concentrations was added. As a control for these experiments, samples containing the same amount of dimethyl sulfoxide as those present in the different peptide dilutions were prepared and used to treat cell cultures. Cultures were then maintained for 1 h at 37°C to allow the internalization of the peptides. Thereafter, monolayers were washed three times with DMEM and maintained at 37°C. At 24 h posttreatment, cells were fixed and the extent of induced apoptosis was detected as described before (17). Monolayers treated with a 10 μM concentration of either peptide were infected with IBDV at an MOI of 0.5 PFU/cell. After 1 h of adsorption, cells were washed three times with DMEM and then left on the incubator with DMEM supplemented with 2% newborn calf serum. At different times p.i., supernatants were harvested and, after removal of cell debris by low speed centrifugation, used for virus titration with BSC-1 cells.
RESULTS
RNA does not mediate interaction between VP1 and VP3.
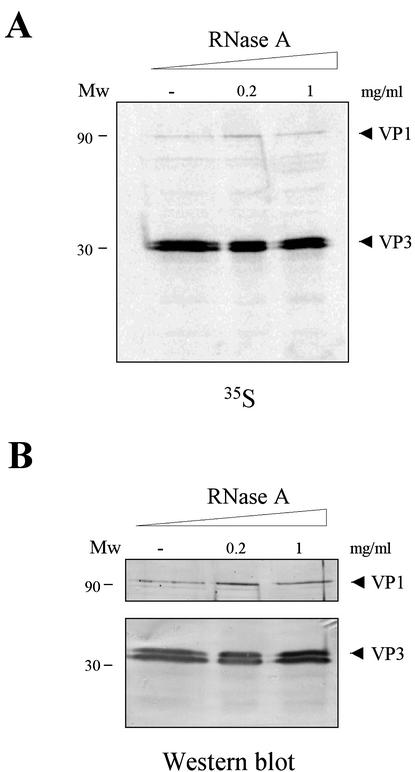
The formation of VP1-VP3 complexes leading to the encapsidation of VP1 into IBDV VLPs has been previously described (16). We were interested in identifying the sequence(s) and mechanisms employed by VP3 to bind VP1. VP1, which is considered to be the IBDV RdRp, binds RNA (21). Moreover, results from our laboratory have shown that VP3 is an RNA-binding protein capable of binding both single-stranded RNA and dsRNA (13). Hence, it seemed feasible that RNA might mediate the interaction of both proteins. For this, we generated independent VP1- and VP3-containing extracts and subjected them to RNase treatment before allowing the interaction to take place. We took advantage of two previously described rVVs, VT7/VP1 and VT7/VP3, expressing VP1 and VP3, respectively (10, 16). BSC-1 cells were independently infected with these rVVs. After infection, cells were maintained in the presence of the specific inducer IPTG. At 4 h p.i., cells were metabolically labeled with [35S]methionine. Cells were collected at 20 h p.i. and used to obtain protein extracts for the in vitro binding assays. Cell extracts were treated with RNase A. Thereafter, VP1- and VP3-containing extracts were mixed, incubated at 4°C ON, and immunoprecipitated with a specific anti-VP3 serum. After IP, samples were subjected to SDS-PAGE, and the formation of VP1-VP3 complexes was analyzed by either autoradiography or Western blot analysis with anti-VP1 or anti-VP3 sera. It has previously been shown that anti-VP3 serum does not immunoprecipitate VP1 (16). As shown in Fig. 1, the formation of VP1-VP3 complexes is not affected by the RNase treatment, thus indicating that the interaction does not require the presence of RNA in the protein extracts. This result strongly suggests that VP1-VP3 complexes are formed by a direct protein-protein interaction.
FIG. 1.
Characterization of the role of RNA for the VP1-VP3 interaction. (A) Cultures of BSC-1 cells were infected with VT7/VP1 or VT7/VP3, metabolically labeled with [35S]methionine, harvested at 20 h p.i., and used to prepare protein extracts. The extracts were either untreated (−) or treated with 0.2 or 1 mg of RNase A/ml. Thereafter, samples were mixed, and the VP1-VP3 interaction was allowed to proceed ON at 4°C. After incubation, samples were subjected to IP with anti-VP3 rabbit antiserum. The resulting samples were subjected to SDS-PAGE and electroblotted onto nitrocellulose. The filters were subjected to autoradiography. (B) Western blot analysis of the immunoprecipitated products. After autoradiography, filters were excised in half. The upper and lower halves were incubated with anti-VP1 and anti-VP3 sera, respectively. After incubation with the corresponding antisera, filters were developed by the addition of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin. In both cases, the signal was detected by enhanced chemiluminescence. In each panel, the positions of standard protein molecular size (in kilodaltons) markers are indicated on the left (Mw). Arrowheads in both panels indicate the positions of bands corresponding to the VP1 and VP3 polypeptides.
It is interesting that two VP3 bands were observed in VT7/VP3-infected cell extracts. One of these bands presents at the expected size while the other seems to be smaller. The nature of the smaller VP3 species remains unclear. It could be generated by internal initiation of translation at an AUG codon located 15 residues downstream from the bona fide initiator codon. Alternatively, the protein might undergo a proteolytic trimming.
The last 16 aa of VP3 are necessary for formation of VP1-VP3 complexes.
VP3, a 363-amino-acid (aa)-long polypeptide, contains a highly hydrophilic C-terminal region rich in charged amino acids and proline residues. Due to its electrostatic and hydrophilic features, this region is predicted to be well exposed, and hence, it has the potential to bind proteins and nucleic acids. In fact, it has been proposed that the C-terminal region of VP3 could bind viral dsRNA (15). Considering this, we focused on studying the possible role of this region for the VP1 interaction. Taking into consideration both the amino acid composition and the net charge, we arbitrarily divided the VP3 C-terminal tail into three independent regions: (i) an acidic region, formed by the sequence 1008DEDLE1012; (ii) an arginine-rich region, formed by the sequence 997GRLGRWIRTVS1007; and (iii) a proline-lysine-arginine-rich region (PKR region), formed by the sequence 971EMKHRNPRRALPKPKPKPNAPTQRPP996 (Fig. 2A). In order to analyze the possible role of these regions, three VP3 mutants containing C-terminal deletions of increasing lengths were generated. Deletions present in Δ1008-1012, Δ997-1012, and Δ971-1012 cover one, two, or three of the described regions, respectively. To determine the level of expression of these proteins, the constructs were transfected into COS cells, and the VP3 expression was analyzed by Western blotting. The polyproteins encoded by the different constructs were cotranslationally processed, and VP3 accumulated to a similar level to that observed after expression of the wild-type (wt) one, indicating that the deletions do not lead to protein instability or degradation (data not shown).
To characterize the ability of these VP3 mutants to interact with VP1, in vitro binding assays were carried out. As a source of VP1, COS cells were infected with VT7/VP1. Cell extracts were prepared at 20 h p.i. As a source of VP3, plasmids expressing the deletion mutants were individually transfected into COS-1 cells. At 48 h p.t., cells were harvested and processed for IP. Prior to the IP assays, the amount of VP3 in each sample was analyzed by Western blotting and then normalized. Then, extracts from cells infected with VT7/VP1 were incubated with extracts from cells transfected with the different deletion mutants. After IP with anti-VP3, samples were subjected to SDS-PAGE and Western blotting with anti-VP1 or anti-VP3 serum.
In contrast to wt VP3, which efficiently coimmunoprecipitated a band of 97 kDa, corresponding to VP1 (Fig. 2B, lane 1), incubation with the C-terminal deletion mutants did not lead to VP1 coimmunoprecipitation (Fig. 2B, lanes 2 to 4). Remarkably, a 5-aa deletion (Δ1008-1012) completely abolishes the VP1-VP3 interaction, showing that the acidic region is essential for VP1 binding (Fig. 2B, lane 2).
Since the VP3 mutant containing the smallest deletion was completely unable to bind VP1, the possibility that the PKR and/or arginine-rich regions might also play a role in the interaction with VP1 could not be ruled out. To determine the role of these two regions in VP1 binding, three internal deletion mutants of increasing sizes were generated. For this, one, two, or three of the VP3 C-terminal regions were inserted on the C terminus of the mutant polyprotein Δ907-1012. Δ907-1012 contains a C-terminal deletion of 105 aa and encodes a mutant VP3 unable to bind VP1 (data not shown). In these internal deletion mutants, the acidic region, known to be critical for the interaction, was preserved. Mutant Δ907-970 contains an internal deletion that does not affect the hydrophilic region (Fig. 2A). Mutants Δ907-996 and Δ907-1007 present deletions spanning the PKR region alone or both the PKR and the arginine-rich regions, respectively (Fig. 2A). The ability of these VP3 mutants to form complexes with VP1 was analyzed. As shown in Fig. 2C (lane 4), deletion of aa 907 to 970 (Δ907-970) does not affect the VP1 interaction, showing that deletions out of the VP3 hydrophilic tail do not alter the formation of VP1-VP3 complexes. Interestingly, deletion of residues 971 to 996, covering the PKR region (Δ907-996), does not affect the binding ability (Fig. 2C, compare lanes 3 and 4), showing that this region is not involved in the formation of VP1-VP3 complexes. However, the further deletion of residues 997 to 1007 (Δ907-1007), containing the arginine-rich region, completely abolishes it, even though the acidic region is present (Fig. 2C, lane 2). These results demonstrate that both the acidic and the arginine-rich regions are required for the interaction. Taken together, these results clearly show that the VP3 sequence, 997GRLGRWIRTVSDEDLE1012, is essential for the interaction with VP1.
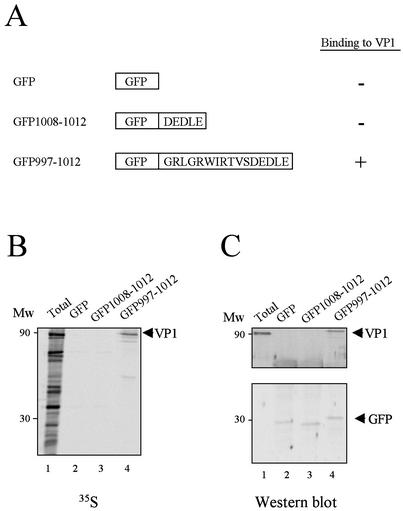
The VP3 sequence of aa 997 to 1012 confers VP1 binding activity to a heterologous protein.
As described above, the 16 C-terminal VP3 residues are necessary for the formation of VP1-VP3 complexes. It was therefore important to determine whether this sequence was sufficient to confer VP1 binding activity to a heterologous protein. For this, sequences DEDLE and GRLGRWIRTVSDEDLE were inserted in frame at the C-terminal region of GFP (Fig. 3A). To determine the ability of these GFP-VP3 fusion proteins to form complexes with VP1, an in vitro binding assay was carried out. COS cells were transfected with GFP or GFP-VP3 fusion-expressing plasmids. At 48 h p.t., cells were harvested and processed to generate extracts for the binding assays. To normalize the amount of the wt GFP and GFP-VP3 used for the assays, the concentration of the protein in each extract was analyzed by Western blotting. [35S]methionine metabolically labeled extracts from cells infected with VT7/VP1 were incubated with extracts from cells transfected with either wt GFP or GFP-VP3 fusion proteins. IP was performed with a specific anti-GFP antibody. After IP, samples were subjected to SDS-PAGE, transferred to nitrocellulose, and analyzed by either autoradiography or Western blotting with anti-VP1 or anti-GFP serum.
FIG. 3.
The 16 C-terminal residues of VP3 are sufficient to confer the ability to interact with VP1 to the GFP. To further analyze the role of the C-terminal region of VP3, two plasmids were generated expressing GFP fused to the VP3 C-terminal 5 and 16 residues, respectively. COS cells were infected with VT7/VP1 and metabolically labeled with [35S]methionine. At 20 h p.i., cells were harvested and the corresponding extracts were used to carry out in vitro binding assays with extracts obtained from cells transfected with the different GFP-VP3 fusion proteins. After incubation, samples were subjected to IP with anti-GFP mouse antisera, analyzed by SDS-PAGE, and electroblotted onto nitrocellulose. (A) The diagram shows the structure of the GFP-VP3 fusion proteins used for the assays. The ability (+) or inability (−) of each protein to bind to VP1 is also shown. (B) Autoradiography of the nitrocellulose filter obtained after the electroblotting of the SDS-PAGE of IP samples. The positions of molecular weight markers (Mw) and the VP1 polypeptide (arrowhead) are indicated. (C) Western blot analysis of the IP products. After blotting, the upper part of the filter was incubated with rabbit anti-VP1, and the lower one was incubated with mouse anti-GFP antisera. After washing, filters were incubated with horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse immunoglobulin, respectively. Signals were developed by enhanced chemiluminescence.
As expected, both fusion proteins were efficiently immunoprecipitated by the anti-GFP antiserum (Fig. 3C). As shown in Fig. 3B and C (lanes 2), wt GFP does not coprecipitate VP1. Interestingly, fusion of both the VP3 arginine-rich and acidic regions (GFP997-1012) precipitated a polypeptide of 97 kDa (Fig. 3B and C, lanes 4) that was specifically recognized by the anti-VP1 antiserum (Fig. 3C, lane 4), demonstrating that VP1 was efficiently bound by this GFP-VP3 fusion polypeptide. However, the fusion of the acidic region (GFP1008-1012) does not confer VP1 binding properties to GFP (Fig. 3B, lane 3). This along with the results presented above shows that the acidic region is necessary but not sufficient to bind VP1. By contrast, our results clearly demonstrate that the sequence 997GRLGRWIRTVSDEDLE1012 is sufficient to allow interaction with VP1.
GFP997-1012 showed the expected electrophoretic mobility, and GFP1008-1012 produced a band migrating faster in SDS-PAGE than that of wt GFP (Fig. 3C, lanes 3 to 4). Since the sequence of the plasmid that encoded this protein was confirmed by nucleotide sequence analysis, the aberrant mobility of this fusion protein is likely due to the presence of the highly acidic DEDLE at the C terminus of GFP.
The results described so far lead us to the conclusion that the ability of VP3 to interact with VP1 relies on the sequence from aa 997 to 1012 containing the arginine-rich and acidic regions. Accordingly, this region is hereafter referred to as the VP1 binding motif (VP1BM) of VP3.
Mutational analysis of the VP1BM.
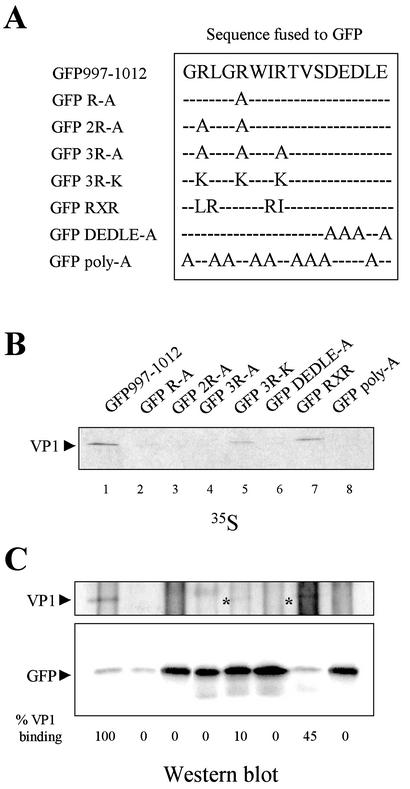
To further define the VP1BM, we sought to characterize the effect of specific point mutations within the VP1BM for the formation of complexes. To that purpose, a series of mutations were introduced within the VP1BM of the GFP997-1012 fusion protein (Fig. 4A). The ability of the corresponding mutant polypeptides to interact with VP1 was determined as described above.
FIG. 4.
Mutational analysis of the VP3 VP1BM. To characterize the VP1BM of VP3, a series of GFP-VP3 fusion mutants were generated by site-directed mutagenesis. (A) The diagram shows the mutations introduced within the GFP-VP3 fusion protein. (B) COS cells were infected with VT7/VP1 and metabolically labeled with [35S]methionine. At 20 h p.i., cell extracts were prepared and mixed with extracts from COS cells transfected with the different GFP-VP3 constructs. After incubation, reaction mixtures were subjected to IP with anti-GFP mouse antiserum. IP samples were subjected to SDS-PAGE and electroblotted onto nitrocellulose. The filter was subjected to autoradiography. The VP1 polypeptide (arrowhead) is indicated. (C) Western blot analysis of the IP products. To determine the position of the VP1 polypeptide, the upper part of the filter was incubated with anti-VP1 rabbit antiserum followed by the addition of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin. To determine the position of the GFP fusion polypeptides, the lower part of the filter was incubated with anti-GFP mouse antiserum followed by addition of horseradish peroxidase-conjugated goat anti-mouse immunoglobulin. The signal was detected by enhanced chemiluminescence. Asterisks indicate fainter VP1 bands. The amounts of VP1 coimmunoprecipitated by the different GFP constructs (B) were determined as described in Material and Methods. VP1 coimmunoprecipitation efficiencies (%VP1 binding) were calculated by using the value obtained with the GFP997-1012 construct as 100%.
To analyze the role of arginines 998, 1001, and 1004, GFP997-1012 fusion proteins containing the replacement of one (GFP-R-A), two (GFP-2R-A), or three (GFP-3R-A) arginines by alanine residues were generated (Fig. 4A). As shown in Fig. 4B (lanes 2 to 4), none of these mutants is able to bind VP1. To further confirm the importance of these arginine residues, a GFP997-1012 fusion protein harboring the conservative replacement of the three arginines by lysine residues was generated (GFP-3R-K) (Fig. 4A). Somewhat unexpectedly, this mutation dramatically reduces the ability of the fusion protein to bind VP1. A densitometric analysis demonstrated that this mutant polypeptide coimmunoprecipitated only 10% of the VP1 bound by GFP997-1012 (Fig. 4B, lane 5). This indicates that the identity of the charged residues (arginine rather than lysine) is critical for the establishment of the VP1-VP3 interaction.
Two hydrophobic residues separate the arginine residues within the VP1BM (R-X-X-R; X = uncharged residues). Thus, we wondered whether this spacing might be also important for VP1 binding. To examine this possibility, we generated a GFP997-1012 fusion protein, GFP-RXR (Fig. 4A), in which the spacing had been modified (R-X-R instead of R-X-X-R). To this end, we rearranged the positions of the arginines by exchanging residues R998 and R1004 for G999 and L1003, respectively. As shown in Fig. 4B, lane 7, even though the net basic charge is not altered, the rearrangement significantly reduces the VP1 binding (up to 45%) with respect to that observed for GFP997-1012. This indicates that the sequence necessary for VP1 binding is very sensitive to internal rearrangement of the basic residues. In summary, the results presented demonstrate that the number, identity, and periodicity of arginines 998, 1001, and 1004 play a key role in the VP1-VP3 interaction.
The relevance of the acidic region was also explored by using a mutant containing four alanines replacing the acidic residues (GFP DEDLE-A) (Fig. 4A). This mutation completely abolishes the interaction with VP1 (Fig. 4B, lane 6), thus confirming our previous data obtained with VP3 mutant proteins containing C-terminal deletions (Fig. 2B). This result shows that the acidic region is, by itself, essential for the VP1BM activity and that the inhibition induced by the deletion of this sequence in the context of the VP3 protein, described above, is not due to conformational changes affecting the adjacent arginine-rich region. Therefore, we conclude that, to confer VP1 binding capacity, both the arginine-rich and the acidic regions are required.
The requirement of the VP1BM containing arginines positioned two residues apart from each other prompted us to investigate the hypothesis that the VP1BM of VP3 might function in a precise conformation. That being the case, uncharged residues might play a role in presenting the charged residues under a precise conformation. Consequently, to test whether a simple cluster of charged residues is able to bind VP1, we generated a construct in which all of the amino acids, except the charged ones, were replaced by alanines (GFP-Poly-A) (Fig. 4A). As shown in Fig. 4B, lane 8, a complete inhibition of the interaction was observed with this mutant, indicating that the uncharged residues of the VP1BM might play a role in the VP1-VP3 interaction. This suggests that uncharged and hydrophobic residues also contribute to the proper VP1BM configuration.
A synthetic peptide corresponding to the VP1BM specifically inhibits binding of VP3 to VP1.
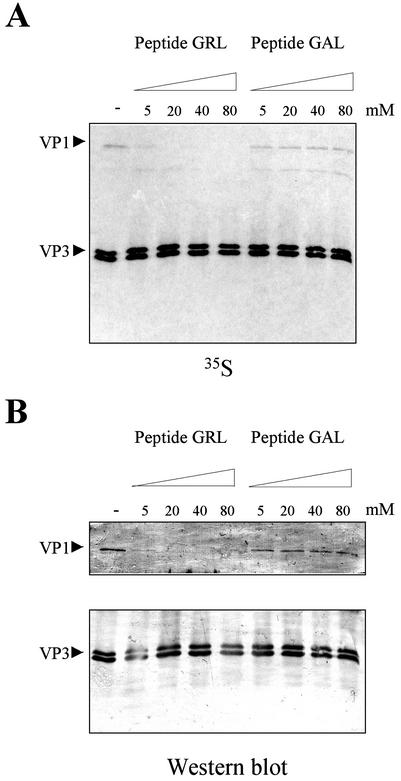
To further confirm that the sequence of aa 997 to 1012 contains the VP1BM, we analyzed the formation of VP1-VP3 complexes in the presence of increasing concentrations of a synthetic peptide, GRL (GRLGRWIRTVSDEDLE), containing the VP1BM sequence. As a negative control, experiments were performed in parallel with the peptide GAL (GALGAWIATVSDEDLE), which contains a sequence similar to that of the VP1BM but with the substitution of alanine residues for the three essential arginines.
[35S]methionine metabolically labeled extracts, obtained from BSC-1 cells infected with either VT7/VP1 or VT7/VP3, were mixed and incubated in the presence of increasing concentrations of the peptide GRL or GAL. After incubation, samples were subjected to IP with anti-VP3 antiserum, separated by SDS-PAGE, and analyzed by either autoradiography or Western blotting with specific anti-VP1 and anti-VP3 sera. The VP1-VP3 interaction is strongly reduced by the presence of GRL at a concentration of 5 μM and completely reduced at a concentration of 20 μM (Fig. 5). However, no binding inhibition was observed when the GAL peptide was used, even at a concentration of 80 μM, showing the specificity of the inhibitory effect of GRL (Fig. 5). This result confirms the key role of the VP1BM for the formation of complexes. Additionally, the total absence of inhibitory effect by the GAL peptide supports our previous finding that arginine residues 998, 1001, and 1004 are required for VP1-VP3 complex formation.
FIG. 5.
VP1/VP3 peptide competition assays. (A) Cultures of BSC-1 cells were infected with VT7/VP1 or VT7/VP3, metabolically labeled with [35S]methionine, harvested at 20 h p.i., and used to prepare protein extracts. Then, in vitro binding assays were carried out in the presence of increasing amounts of the synthetic peptide GRL corresponding to the VP3 VP1BM. The peptide GAL, containing a mutated version of the VP3 VP1BM was used as a control for the experiment. After incubation, the samples were subjected to IP with anti-VP3 rabbit antiserum. Then samples were analyzed by SDS-PAGE and electroblotted onto nitrocellulose. The filter was subjected to autoradiography. Arrowheads indicate the positions of the VP1 and VP3 polypeptides. (B) Western blot analysis of the IP products. The filter was cut in half, and the upper part was incubated with anti-VP1, and the lower one was incubated was incubated with anti-VP3 rabbit serum. Thereafter, filters were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin. The signal was detected by enhanced chemiluminescence.
A synthetic peptide containing the VP1BM sequence inhibits virus replication.
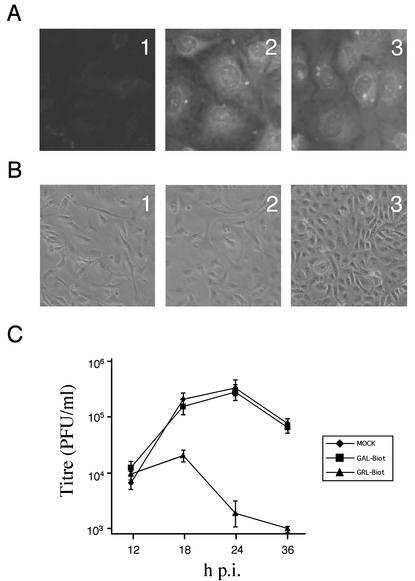
The results described above conclusively showed that in vitro formation of VP1-VP3 complexes is efficiently prevented by the presence of a peptide containing the VP1BM sequence. It was therefore interesting to determine the effect of a peptide containing this sequence within the cytoplasm of IBDV-infected cells. To analyze this, a couple of oligopeptides, GRL-biot and GAL-biot, were synthesized containing either the wt or a nonfunctional version of the VP1BM sequence, respectively. In order to allow their internalization into the cell cytoplasm, the penetratin peptide from the homeodomain of the antennapedia transcription factor was incorporated at the N terminus of both oligopeptides. An analysis of the potential proapoptotic effect of the peptides was tested. For this, twofold dilutions of the peptides were added to BSC-1 monolayers. Twenty-four hours after the treatment, cells were fixed and analyzed by tunnel assay. No significant apoptotic effects were detected in cultures treated with peptide concentrations below 20 μM (data not shown). Accordingly, a concentration of 10 μM was selected for the further experiments. As shown in Fig. 6A, at the selected concentration, GRL-biot and GAL-biot were efficiently internalized in BSC-1 cells. Peptide-treated cells were infected with IBDV, and the extent of cytopathic effect (CPE) and virus production were monitored. As shown in Fig. 6B, treatment with GRL-biot prevents IBDV-induced CPE and causes a significant reduction in extracellular virus production (Fig. 6C). No effect on virus replication was detected in either GAL-biot- or mock-treated cells. These results demonstrate that the presence of a peptide containing the VP1BM sequence causes a clear reduction in virus replication and strongly suggests that in vivo formation of VP1-VP3 complexes plays a major role in virus replication.
FIG. 6.
Analysis of the effect of Trojan peptides containing the VP3 VP1BM on IBDV replication. Two peptides, GRL-biot and GAL-biot, containing the wt and a nonfunctional version of the VP3 VP1BM, respectively, were used for the assays. (A) Detection of the intracellular accumulation of GAL-biot (2) and GRL-biot (3) in BSC-1 cells. The cultures were maintained for 1 h in the presence of either peptide at a concentration of 10 μM in DMEM. Thereafter, cultures were washed three times and maintained at 37°C for 20 h. After this period, monolayers were fixed and incubated with a rhodamine-streptavidin conjugate. The peptides were visualized by immunofluorescence. Panel 1 shows an image obtained under the same conditions from a mock-treated cell culture. To analyze the effect of both peptides on virus replication, peptide- or mock-treated cells were infected with IBDV at an MOI of 0.5 PFU/cell. (B) The extent of IBDV-induced CPE was analyzed at 36 h p.i. by phase-contrast microscopy. The panels show images from mock-treated cells (1), GAL-biot-treated cells (2), and GRL-biot-treated cells (3). (C) Supernatants from the infected cultures, collected at the indicated times p.i., were used for virus titration on fresh BSC-1 monolayers. The results shown in the graph correspond to the titration of three independent samples for each treatment.
DISCUSSION
Encapsidation of the viral polymerase into the virion during the viral morphogenesis is a major step in the replicative cycle of dsRNA viruses. It was previously described that VP1, the putative RdRp of IBDV, is incorporated into IBDV VLPs through the interaction with VP3 (16). In this study, we have studied the molecular mechanism regulating the interaction between VP1 and VP3. Our results show that VP1-VP3 complexes are formed in vitro after treatment with RNase A (Fig. 1). This indicates that the formation of VP1-VP3 complexes does not require the presence of RNA. The competition experiments carried out with the RNA-free synthetic peptides discussed below also show that the VP1BM of VP3 does not require the presence of RNA molecules in order to be functional. However, the role of small RNA molecules, protected from RNase and bound to VP1, that might be important for the precise folding of this protein cannot be completely ruled out.
The C-terminal region of VP3 is characterized by the presence of three contiguous sequences rich in proline and charged amino acid residues: (i) the acidic region (1008DEDLE1012), (ii) the arginine-rich region (997GRLGRWIRTVS1007), and (iii) the PKR region (971EMKHRNPRRALPKPKPKPNAPTQRPP996). In view of its hydrophilic nature, the VP3 C-terminal tail constitutes an excellent candidate for the interaction with VP1. Three complementary approaches lead us to the conclusion that the VP3 sequence 997GRLGRWIRTVSDEDLE1012 is necessary and sufficient for the formation of the VP1-VP3 complexes. First, deletions of either the VP3 arginine-rich or the VP3 acidic region abolishes VP1-VP3 interaction. Second, the fusion of the sequence 997GRLGRWIRTVSDEDLE1012 to GFP confers VP1-binding properties to the fusion protein. Finally, the formation of VP1-VP3 complexes is specifically inhibited by a synthetic peptide corresponding to residues 997 to 1012 of VP3. Accordingly, this region has been named the VP1BM.
The molecular features of VP1BM were dissected by mutational analysis. Our results from both deletion mutants on the C terminus of VP3 and point mutants on GFP-VP3 fusion proteins show that neither the acidic nor the arginine-rich regions are independently able to support VP1 binding. Interestingly, the results presented here suggest that VP1BM activity requires a precise conformation. Since internal deletions preserving the arginine-rich and acidic regions allow VP1-VP3 interaction, VP1BM is independent of the overall VP3 conformation. Additionally, VP1BM functions when fused to GFP, a heterologous protein. Noteworthy, VP1BM activity is extremely sensitive to point mutations. Mutations affecting uncharged residues completely abolish VP1 interaction, suggesting that uncharged residues might be presenting charged residues in a proper conformation. Replacement of arginines R998 and R1004 by G999 and L1003, respectively, reduces VP1 binding up to a 55%, indicating that the native positioning of the arginine residues is essential for the formation of VP1-VP3 complexes. In order to get information about the possible VP1BM conformation, an extensive search of protein data banks was carried out. Unfortunately, no significant homologies were obtained. Considering that basic residues are positioned every three residues, it is tempting to speculate that VP1BM might fold into an α-helix, with arginine residues exposed to the same face. Interestingly, in the sequence alignment of VP3s from different IBDV strains, the composition of the VP1BM is strictly conserved (data not shown). This strongly suggests an extreme importance of this sequence for VP3 activity.
VP3 forms trimeric subunits located at the inner surface of the virion shell (3, 5). Given that only a few VP1 molecules are encapsidated, a small fraction of the total VP3 molecules might interact with VP1. This indicates that some level of regulation exists and raises the intriguing question of how VP1 binding activity of VP3 is regulated. It seems feasible that most VP1BMs are not exposed when VP3 trimers are formed. According to this hypothesis, a heterologous protein containing the VP1BM would show a higher VP1 binding efficiency than that of VP3. A comparison of the IP results obtained after incubating VP1 with GFP997-1012 and VP3, respectively, suggests that this might be the case.
It should be noted that VP3 acts as an anchor, interacting also with VPX/2 and the genomic dsRNA. Given this, it is exciting to speculate on the possibility that binding to VPX/2 and/or genomic dsRNA might modulate the formation of VP1-VP3 complexes. Alternatively, posttranslational modifications may also play a role in VP3 folding and, hence, on VP1BM activity. Additionally, it could be argued that the VP3 C-terminal region interacts with genomic RNA, disturbing the VP1-VP3 interaction. However, results from our laboratory have shown that the VP3 RNA-binding motif maps near the N-terminal region of the protein. This implies that VP1BM does not bind the genomic dsRNA and, hence, that a direct interaction with RNA is not modulating VP1BM activity. Indeed, a mutant lacking the VP3 VP1BM binds RNA in vitro as efficiently as the wt protein (13).
According to the results of our mutational analysis, the existence of additional regions implicated in interacting with VP1 could not be ruled out since no N-terminal deletions were generated. Nevertheless, the specific inhibition of the VP1-VP3 complex binding produced by the presence of the synthetic GRL peptide undoubtedly supports the notion that this sequence displays the main VP1 binding activity of VP3. It should be noted that a peptide containing alanine residues instead of arginines fails to inhibit the VP1-VP3 interaction. This result, consistent with our mutational analysis of GFP fusion proteins, demonstrates that in the context of the complete VP3 polypeptide, arginines 998, 1001, and 1004 mediate VP1 binding. In this respect, the strong inhibition of VP1 binding resulting from the replacement of these arginines by lysine residues was somewhat surprising. Although the reason for this is unclear, it is feasible that the bigger size of the side chain in lysine residues might alter the VP1BM conformation or produce a steric hindrance for the interaction.
The VP3 VP1BM sequence is extremely well conserved among IBDV strains. Indeed, our mutational analysis shows that VP1BM functionality is strongly affected even by conservative point mutations. On the other hand, removal of short sequences from the C-terminal end of VP3 completely abolishes the formation of virus particles (A. Maraver, A. Oña, F. Abaitua, D. González, R. Clemente, A. Díaz, J. R. Castón, F. Pazos, and J. F. Rodriguez, submitted for publication). Accordingly, it is expected that VP3 VP1BM mutations will severely impair the replicative capacity of IBDV, preventing the use of reverse genetics to further characterize, in vivo, the role of this motif. Consequently, an alternative strategy was undertaken. The effect of Trojan peptides containing the VP3 VP1BM on virus replication was analyzed. These peptides might intracellularly compete the VP1-VP3 interaction and thus affect the formation of VP1-VP3 complexes in vivo. Trojan peptides, containing the penetrating sequence of the antennapedia transcription factor from Drosophila, have been successfully used to introduce peptides into different cell systems in order to characterize the functional role of small protein motifs in different biological processes (7, 26). The observation that the presence of a peptide containing the VP3 VP1BM in IBDV-susceptible cells causes a specific inhibition of the production of virus progeny strongly reinforces the notion that the formation of VP1-VP3 complexes plays a crucial role during virus morphogenesis. In addition, this finding opens the possibility of designing new strategies for controlling IBDV.
Acknowledgments
This work was supported by grant no. BIO2000-0905 from the Comisión Interministerial de Ciencia y Tecnología and by grant no. 07B/0032/1998 from the Subdirección General de Investigación of the Comunidad Autónoma de Madrid. E.L. and A.M. were recipients of fellowships from Comunidad Autónoma de Madrid and Ministerio de Educación, respectively.
We thank Amelia Nieto, Jose R. Castón, Grazyna Kochan, and Juan Carlos Ramírez for helpful discussions. We are indebted to Bernard Moss for the VOTE expression system. We also thank Inés Poveda for excellent photography work.
REFERENCES
- 1.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruenn, J. A. 1991. Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 19:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castón, J. R., J. L. Martínez-Torrecuadrada, A. Maraver, E. Lombardo, J. F. Rodríguez, J. I. Casal, and J. L. Carrascosa. 2001. C terminus of infectious bursal disease virus major capsid protein vp2 is involved in definition of the T number for capsid assembly. J. Virol. 75:10815-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Costa, B., C. Chevalier, C. Henry, J. C. Huet, S. Petit, J. Lepault, H. Boot, and B. Delmas. 2002. The capsid of infectious bursal disease virus contains several small peptides arising from the maturation process of pVP2. J. Virol. 76:2393-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derossi, D., G. Chassaing, and A. Prochiantz. 1998. Trojan peptides: the penetratin system for intracellular delivery. Trends Cell Biol. 8:84-87. [PubMed] [Google Scholar]
- 8.Dobos, P. 1993. In vitro guanylylation of infectious pancreatic necrosis virus polypeptide VP1. Virology 193:403-413. [DOI] [PubMed] [Google Scholar]
- 9.Fahey, K. J., I. J. O'Donnell, and A. A. Azad. 1985. Characterization by Western blotting of the immunogens of infectious bursal disease virus. J. Gen. Virol. 66:1479-1488. [DOI] [PubMed] [Google Scholar]
- 10.Fernández-Arias, A., S. Martínez, and J. F. Rodríguez. 1997. The major antigenic protein of infectious bursal disease virus, VP2, is an apoptotic inducer. J. Virol. 71:8014-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirai, K., and S. Shimakura. 1974. Structure of infectious bursal disease virus. J. Virol. 14:957-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kibenge, F. S., B. Qian, J. R. Cleghorn, and C. K. Martin. 1997. Infectious bursal disease virus polyprotein processing does not involve cellular proteases. Arch. Virol. 142:2401-2419. [DOI] [PubMed] [Google Scholar]
- 13.Kochan, G., D. González, and J. F. Rodríguez. Characterization of the RNA binding activity of VP3, a major structural protein of IBDV. Arch. Virol., in press. [DOI] [PMC free article] [PubMed]
- 14.Kordyban, S., G. Magyar, H. K. Chung, and P. Dobos. 1997. Incomplete dsRNA genomes in purified infectious pancreatic necrosis virus. Virology 239:62-70. [DOI] [PubMed] [Google Scholar]
- 15.Leong, J. C., D. Brown, P. Dobos, F. S. B. Kibenge, J. E. Ludert, H. Muller, and B. Nicholson. 2000. Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 16.Lombardo, E., A. Maraver, J. R. Castón, J. Rivera, A. Fernández-Arias, A. Serrano, J. L. Carrascosa, and J. F. Rodríguez. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol. 73:6973-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lombardo, E., A. Maraver, I. Espinosa, A. Fernández-Arias, and J. F. Rodríguez. 2000. VP5, the nonstructural polypeptide of infectious bursal disease virus, accumulates within the host plasma membrane and induces cell lysis. Virology 277:345-357. [DOI] [PubMed] [Google Scholar]
- 18.Macreadie, I. G., and A. A. Azad. 1993. Expression and RNA dependent RNA polymerase activity of birnavirus VP1 protein in bacteria and yeast. Biochem. Mol. Biol. Int. 30:1169-1178. [PubMed] [Google Scholar]
- 19.Magyar, G., H. K. Chung, and P. Dobos. 1998. Conversion of VP1 to VPg in cells infected by infectious pancreatic necrosis virus. Virology 245:142-150. [DOI] [PubMed] [Google Scholar]
- 20.Muller, H., and H. Becht. 1982. Biosynthesis of virus-specific proteins in cells infected with infectious bursal disease virus and their significance as structural elements for infectious virus and incomplete particles. J. Virol. 44:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller, H., and R. Nitschke. 1987. The two segments of the infectious bursal disease virus genome are circularized by a 90,000-Da protein. Virology 159:174-177. [DOI] [PubMed] [Google Scholar]
- 22.Mundt, E., B. Kollner, and D. Kretzschmar. 1997. VP5 of infectious bursal disease virus is not essential for viral replication in cell culture. J. Virol. 71:5647-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saif, Y. M. 1998. Infectious bursal disease and hemorrhagic enteritis. Poult. Sci. 77:1186-1189. [DOI] [PubMed] [Google Scholar]
- 24.Sánchez, A. B., and J. F. Rodríguez. 1999. Proteolytic processing in infectious bursal disease virus: identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology 262:190-199. [DOI] [PubMed] [Google Scholar]
- 25.Tacken, M. G., P. J. Rottier, A. L. Gielkens, and B. P. Peeters. 2000. Interactions in vivo between the proteins of infectious bursal disease virus: capsid protein VP3 interacts with the RNA-dependent RNA polymerase, VP1. J. Gen. Virol. 81:209-218. [DOI] [PubMed] [Google Scholar]
- 26.Williams, E. J., D. J. Dunican, P. J. Green, F. V. Howell, D. Derossi, F. S. Walsh, and P. Doherty. 1997. Selective inhibition of growth factor-stimulated mitogenesis by a cell-permeable Grb2-binding peptide. J. Biol. Chem. 272:22349-22354. [DOI] [PubMed] [Google Scholar]
- 27.Yao, K., M. A. Goodwin, and V. N. Vakharia. 1998. Generation of a mutant infectious bursal disease virus that does not cause bursal lesions. J. Virol. 72:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao, K., and V. N. Vakharia. 2001. Induction of apoptosis in vitro by the 17-kDa nonstructural protein of infectious bursal disease virus: possible role in viral pathogenesis. Virology 285:50-58. [DOI] [PubMed] [Google Scholar]