Abstract
The filamentous virion of the closterovirus Beet yellows virus (BYV) consists of a long body formed by the major capsid protein (CP) and a short tail composed of the minor capsid protein (CPm) and the virus-encoded Hsp70 homolog. By using nano-liquid chromatography-tandem mass spectrometry and biochemical analyses, we show here that the BYV 64-kDa protein (p64) is the fourth integral component of BYV virions. The N-terminal domain of p64 is exposed at the virion surface and is accessible to antibodies and mild trypsin digestion. In contrast, the C-terminal domain is embedded in the virion and is inaccessible to antibodies or trypsin. The C-terminal domain of p64 is shown to be homologous to CP and CPm. Mutation of the signature motifs of capsid proteins of filamentous RNA viruses in p64 results in the formation of tailless virions, which are unable to move from cell to cell. These results reveal the dual function of p64 in tail assembly and BYV motility and support the concept of the virion tail as a specialized device for BYV cell-to-cell movement.
Despite the enormous variation in molecular architecture, virions of all viruses share the primary function of encapsidation and protection of the virus genome. In addition, virions have more specialized roles at various steps of virus infection. Virions of bacteriophages and animal viruses mediate interactions with the cell surface receptors and subsequent entry and activation of the virus genome (17). In particular, some bacteriophages have specialized molecular assemblies that puncture cells and/or eject the genome from the virion (20, 31, 48). Virions of animal viruses, such as poxviruses, herpesviruses, and lentiviruses, are capable of translocation within and between cells by hijacking cellular motility systems (19, 37).
For plant viruses, active cell-to-cell movement is one of the critical aspects of the life cycle. Therefore, all nondefective plant viruses evolved to encode dedicated movement proteins (MPs) that translocate the virus toward and through the plasmodesmata, the organelles responsible for intercellular communication in plants (9, 25, 27, 33). Some plant viruses move from cell to cell in a nonvirion form, whereas others require functional capsid proteins (CPs) and virion formation for their movement (8, 9, 25). In particular, CP-dependent movement has been described in members of the genera Comovirus and Caulimovirus. The icosahedral virions of these viruses are translocated intercellularly inside MP-induced tubules that traverse plasmodesmal channels of neighboring cells (22, 49). Among filamentous viruses, members of the Potexvirus and Potyvirus genera require functional CPs for cell-to-cell movement (10, 14). However, it remains unclear whether these viruses are actually translocated as virions (26, 40, 41).
A striking relationship between virion morphology and cell-to-cell movement has been revealed in Closteroviridae, a large family of plant positive-strand RNA viruses (5, 21). Unlike other filamentous viruses of plants, which have uniform structure, virions of closteroviruses consist of two distinct morphological units, a long body and a short tail (2, 50). The body and tail both show helical symmetry of the protein subunits, which is typical of all elongated viruses, and consist of the homologous major and minor CPs (CP and CPm), respectively. The gene for CPm most likely evolved via tandem gene duplication in an ancestral closterovirus (7). In addition to CP and CPm, minor amounts of two other closterovirus proteins copurify with virions and are thought to be involved in virion assembly or stabilization (43, 50). One of these proteins, an Hsp70 homolog (Hsp70h), has been recently shown to be an integral virion component (32) that is specifically required for tail assembly (5). The role of another protein of ∼60 kDa (p64), which is conserved among closteroviruses, has remained obscure.
At least five proteins encoded in the ∼16-kb genome of Beet yellows virus (BYV), the prototype closterovirus, are essential for cell-to-cell movement (Fig. 1A) (4, 36). One of these is a small hydrophobic protein of ∼6 kDa (p6). The others include the integral virion components CP, CPm, and Hsp70h, as well as p64. These results indicated that the unusually complex BYV virions have a critical role in virus movement. Further studies revealed a strict correlation between the assembly of the tailed virions and the ability of the virus to move from cell to cell (5). Suppression of tail assembly by mutation of CPm or Hsp70h resulted in the formation of tailless virions, which were competent for genome protection and infectivity assayed on single cells but were defective in cell-to-cell movement. These results prompted the hypothesis that the closterovirus tail was a specialized movement device powered by the ATPase activity of Hsp70h (5).
FIG. 1.
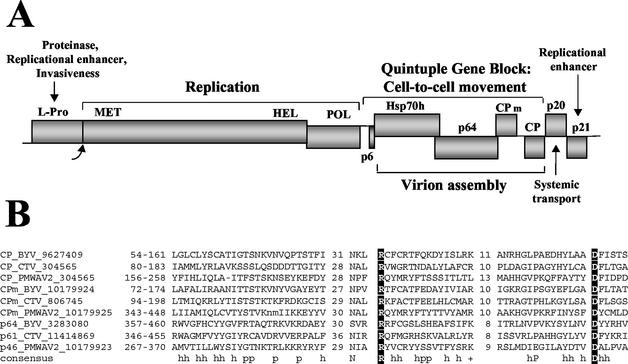
(A) Diagram of the BYV genome showing the ORFs that code for leader proteinase (L-Pro), replication-associated proteins possessing putative methyltransferase (MET), RNA helicase (HEL), and RNA polymerase (POL) domains, a 6-kDa protein (p6), Hsp70h, a 64-kDa protein (p64), CPm, CP, a 20-kDa protein (p20), and a 21-kDa protein (p21). (B) Multiple alignment of the CPs, CPm's, and CP-like C-terminal domains of the ∼60-kDa proteins of three closteroviruses. The alignment was constructed with the MACAW program, and only the three conserved blocks are shown (see text). The consensus line shows the amino acid residues that are present in the majority of the proteins in each of the three groups: h indicates a hydrophobic residue, p indicates a polar residue, and a plus sign indicates a positively charged residue. The two signature residues that are conserved in most of the coat proteins of elongated plant viruses are shown in white against a black background. The ranges of the aligned regions in the corresponding protein sequences are indicated in front of each sequence; the values between the blocks are the lengths of the variable spacers, which are not shown. Each protein is identified by its name, the virus name abbreviation, and the Gene Identifier (GI number) from the NR database. PMWAV2, Pineapple mealybug wilt-associated virus 2 (30).
Here we identify p64 as the fourth integral component of BYV virions and show that it is essential for tail assembly. We further show that p64 consists of a C-terminal domain that is homologous to CP and a unique N-terminal domain. Mutation of the signature motifs of the CPs in the C-terminal domain of p64 abrogates cell-to-cell movement of BYV. These results indicate that the bipartite morphology and the complex movement function of the closterovirus virions evolved via triplication of the gene for CP and acquisition of additional RNA sequences that encode Hsp70h and the N-terminal domain of p64.
MATERIALS AND METHODS
Protein sequence analysis.
The nonredundant (NR) protein sequence database at the National Center for Biotechnology Information (NCBI; National Institutes of Health, Bethesda, Md.) was searched by using the PSI-BLAST program (3). Additionally, the NCBI Conserved Domain Database was searched by using the RPS_BLAST program (28). Multiple alignments of protein sequences were constructed by using the MACAW program (46), with subsequent manual refinement. Protein secondary-structure prediction was performed by using the Jpred program (12).
Isolation, sedimentation, and stability of virions.
BYV virions were isolated from infected Nicotiana benthamiana plants and separated in 10 to 40% sucrose density gradients as described previously (32). Virion preparations were used to determine the stability of the p64-virion complexes in the presence of various concentrations of LiCl. Reaction mixtures containing 100 μl of BYV virions (1 mg/ml) were incubated on ice for 1 h in 0, 0.5, 0.8, 1, 1.5, and 2 M concentrations of LiCl (the final volume of reaction mixtures was 600 μl). Virions were pelleted by ultracentrifugation at 100,000 × g for 1 h at 4°C, and their protein composition was analyzed as described below.
Nano-LC-MS/MS.
Virion proteins were separated by sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel electrophoresis (PAGE); an ∼65-kDa protein band was visualized by staining with 300 mM CuCl2 for 5 min, cut out, and destained; and in-gel trypsin digestion was performed overnight at 37°C as previously described (47). The resulting tryptic peptides were separated by nano-liquid chromatography (nano-LC) with a 40-cm PicoFrit column (New Objective, Cambridge, Mass.) packed with 5 μm, 300-Å-pore-size, Luna C18 silica gel particles (Phenomenex, Torrance, Calif.) as previously described (23). Nano-LC was conducted by using exponential dilution to produce gradient separations (15). To sequence tryptic peptides, online nano-LC-electrospray ionization tandem mass spectrometry (MS/MS) was performed by using an LC-Q ion trap mass spectrometer (Finnigan, San Jose, Calif.). The instrument was operated in the full-scan mode with the spraying potential set to 2.5 kV (applied to the injector), the temperature of the heated inlet capillary at 180°C, the capillary potential at 46 V, and the tube lens offset potential at 30 V. The maximum injection time was 50 ms. The instrument was set to acquire a mass spectrum between 400 and 2,000 m/z, followed by an MS/MS scan. For operation in the MS/MS mode, the maximum injection time was increased to 500 ms, the isolation width was set to 2 Da, and the relative collision energy was set to 35% with a 30-ms activation time. By using the instrument's data-dependent algorithm, the most intense ion from the full scan spectrum was selected for MS/MS if its signal exceeded 2 × 104 counts. The peptide sequences were compared to those in the NR NCBI database by using the Sequest search engine (Finnigan).
Analyses of virion composition.
The PAGE and immunoblot analyses with anti-CP and anti-CPm sera were conducted as previously described (5, 32). The antisera to the N-terminal (amino acid residues 2 to 218) and C-terminal (residues 305 to 553) regions of p64 were raised by using histidine-tagged, glutathione S-transferase-fused recombinant proteins that were expressed in Escherichia coli strain BL21(DE3). To this end, corresponding coding regions were cloned as BamHI-NsiI fragments into the pGEX-2T plasmid (Pharmacia), which was modified to incorporate an NsiI site followed by six histidine codons and a stop codon between the BamHI and SmaI sites present in the original polylinker. The presence of the glutathione S-transferase and histidine tags permitted purification of the recombinant products with glutathione or metal affinity resin. However, because the products obtained were insoluble, each of them was purified under denaturing conditions with only the TALON metal affinity resin (Clontech) as recommended by the manufacturer. The proteins eluted from the resin were submitted to Cocalico Biologicals (Reamstown, Pa.) for generation of rabbit antisera. To characterize the specificities of the resulting antisera, the entire p64 open reading frame (ORF) was cloned into plasmid pTL7SN and used to generate the corresponding RNA transcripts as previously described (29). The resulting p64 mRNA was translated in wheat germ extract (Promega) to yield l-[35S]cysteine (Amersham)-labeled products that were used in immunoprecipitation assays (32). The immunoblot analyses of p64 were conducted with the ECL Western blotting system (Amersham) and anti-p64 sera at a 1:2,000 dilution. Goat anti-rabbit secondary antibodies were used at a 1:4,000 dilution.
Limited digestion of BYV virions with sequencing grade modified trypsin (Promega) was conducted with 20 mM sodium phosphate buffer, pH 7.4, for 1 h at room temperature. The reaction mixtures (total volume, 175 μl) contained 1 mg of BYV virions per ml and trypsin at a 1:50, 1:100, or 1:200 (wt/wt) ratio to the total virion protein. After digestion, reactions were stopped by adding 5 μl of 100 mM phenylmethylsulfonyl fluoride and diluted to 400 μl and virions were separated from digestion products by ultracentrifugation as indicated above.
Generation and characterization of BYV mutants.
Site-directed mutagenesis and plasmid pNB-4 were used to generate p64 mutants (35, 36). Mutagenic primers 5′-CCGTTTCTGTTCGTGCCAGATTTTGTGGCAG and 5′-GTGAAATACTCCTATCTAAACGTTGCCTATTACAGACACG (the mutated nucleotides are in bold) were used to replace the Arg-416 and Asp-455 codons in the p64 ORF with Ala codons to yield mutants R416A and D455A, respectively. The NdeI-BamHI fragments from the mutant pNB-4 variants were cloned into plasmid pBYV-4 to test for virion assembly in protoplast transfection experiments. Alternatively, these fragments were cloned into plasmid pBYV-GFP for assay of virus cell-to-cell movement in plant inoculation experiments (5).
RESULTS
The p64 protein of BYV contains a CP-Like domain.
In database searches performed with the PSI-BLAST program, the BYV p64 sequence showed statistically significant sequence similarity only to orthologs from several other closteroviruses, such as Beet yellow stunt virus and Citrus tristeza virus. However, when the orthologous sequences from all other sequenced closteroviruses (as determined by analysis of genome organization) that appeared in these searches with nonsignificant expectation (E) values were manually added to the position-specific scoring matrix, statistically significant similarity (E < 0.005) was detected between the C-terminal portion of p64 and closterovirus CPs and CPm's. When the sequences of these three groups of closterovirus proteins were aligned by the MACAW program, counterparts of the three prominent motifs that have been detected previously in the CPs of all filamentous plant viruses (13) and in closterovirus CPm's (7) were identified in p64 and its orthologs (Fig. 1B). The alignments of the two distal motifs centered around the invariant arginine and aspartate residues, respectively, were highly statistically significantly similar (E < 10−11). These motifs could be detected in all available sequences of closterovirus p64 orthologs (data not shown). Despite the low sequence similarity (note that the alignment in Fig. 1B contains only two invariant residues), these observations strongly suggested that the C-terminal domains of the ∼60-kDa proteins of closteroviruses are homologous to the closterovirus CPs and CPm's (Fig. 2) and may have similar functions.
FIG. 2.
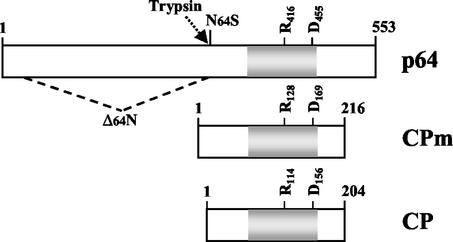
Diagrams of p64, CPm, and CP showing the homologous region (gray; same as in Fig. 1B), as well as the positions of the invariant arginine (R416) and aspartic acid (D455) residues. An approximate trypsin cleavage position (arrow) was used to arbitrarily delimit the N- and C-terminal domains of p64. The premature stop codon in the N64S mutant and the part of the p64 ORF that is deleted in the Δ64N mutant are also shown. The first and last amino acid residues of each protein are indicated.
Examination of the N-terminal domains of the ∼60-kDa proteins revealed a unique pattern of conservation that was not detected in any other known protein family despite an extensive search of the Conserved Domain Database. Secondary-structure prediction suggested that this is a unique, predominantly α-helical domain (data not shown).
p64 is associated with virions.
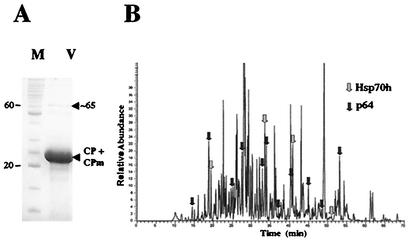
Identification of the CP-like domain of p64 prompted us to ask if this domain enables the incorporation of p64 into BYV virions. Virions were extensively purified and subjected to protein analysis via PAGE. As shown in Fig. 3A, in addition to a bulky band corresponding to CP and CPm, a distinct minor band of ∼65 kDa was present in the gel. To determine the identity of the protein(s) present in this band, it was excised and treated with trypsin and the resulting peptides were separated and sequenced by nano-LC-MS/MS. In total, 15 peptides, shown in Fig. 3B, were sequenced and compared to a database. Among those, 11 peptides were derived from BYV p64, whereas the remaining 4 originated from BYV Hsp70h (Table 1). The total coverage of the p64 and Hsp70h sequences was ∼21 and 8%, respectively. These results demonstrated that the original ∼65-kDa band contained a mixture of ∼64-kDa p64 and ∼65-kDa Hsp70h. The latter protein was previously found to be an integral component of BYV virions (5, 32).
FIG. 3.
(A) Analysis of BYV virion proteins by SDS-PAGE. M, protein markers; V, virions. The positions of the 20- and 60-kDa markers and an ∼65-kDa band (arrowhead) are indicated. (B) Separation of the tryptic peptides derived from the ∼65-kDa proteins present in virions by nano-LC. Arrows mark the positions and origins of the 15 sequenced peptides.
TABLE 1.
Tryptic peptides derived from ∼65-kDa virion proteins and sequenced by nano-LC-MS/MS
| Peptide sequencea | Identity | Positionb |
|---|---|---|
| ALISTACEAFK | Hsp70h | 133-143 |
| IDISFLK | Hsp70h | 258-264 |
| NDSPMLLVDCAAHNLSISSK | Hsp70h | 379-398 |
| VVADLHK | Hsp70h | 514-520 |
| FFGGR | p64 | 21-25 |
| NFSDSTGESFVR | p64 | 58-69 |
| EFSLLLTFPK | p64 | 70-78 |
| LSDYNVSELNVVDVK | p64 | 100-114 |
| FVSLIFK | p64 | 164-170 |
| ALYDEFLK | p64 | 211-218 |
| IPTINTHDSSTFLYK | p64 | 306-319 |
| DNPELK | p64 | 348-353 |
| FGVGFPPITRLNVPVKYSYLNVDYYR | p64 | 433-458 |
| EVALQUAR | p64 | 488-494 |
| NEVSPHAR | p64 | 511-518 |
The sequences are shown in single-letter code from the N terminus to the C terminus.
The positions within 598-residue-long Hsp70h and 553-residue-long p64 are shown.
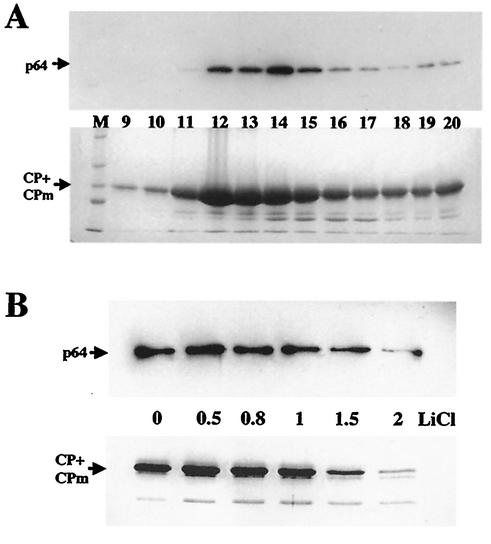
However, the possibility could not be ruled out that p64 was merely a contaminant that copurified with virions. To test whether p64 was specifically associated with virions, we separated virions in a sucrose density gradient and analyzed each fraction for the presence of CP and p64 with the corresponding antisera. As shown in Fig. 4A, peak fractions 12 to 15, which contained most of the CP and morphologically intact virions (32), also contained most of the p64. The comigration of p64 with virions is in agreement with their specific physical association.
FIG. 4.
(A) Comigration of p64 and virions in a sucrose density gradient. (Top) Immunoblot analysis of gradient fractions (numbered from the bottom up) with antiserum to the recombinant N-terminal domain of p64. (Bottom) Same gradient fractions separated by PAGE and stained with Coomassie brilliant blue. M, protein molecular size markers. (B) Stability of p64-virion complexes in the presence of LiCl. Virions incubated in the presence of the indicated molar concentrations of LiCl were precipitated by ultracentrifugation and analyzed by immunoblotting with anti-p64 serum (top) or SDS-PAGE and Coomassie staining (bottom) as in panel A. The minor band below the CP+CPm band likely represents a degradation product that occasionally appears in some virion preparations (compare to Fig. 5 in reference 32).
To determine if p64 could be selectively removed from virions, we treated virions with increasing concentrations of LiCl, collected them by centrifugation, and analyzed their protein composition. As expected, this treatment resulted in partial virion disassembly; the extent of disassembly correlated with the LiCl concentration (Fig. 4B). Importantly, there was also a correlation between the amounts of CP and p64 in virions that withstood disassembly (Fig. 4B). This suggested that p64 was tightly associated with virions and could not be readily dissociated without virion disassembly. Similar results were obtained when virions were treated for increasing time intervals with a low SDS concentration or with 2 M NaCl (data not shown). Collectively, these results parallel those previously obtained with Hsp70h (32) and indicate that p64 and Hsp70h alike are tightly associated with virions and are integral virion components.
Domain topology of p64 in BYV virions.
We next tested whether, as predicted by computational analysis, the C-terminal domain of p64 is functionally analogous to CP and is embedded in the virion. To assess the topology of p64 within the virion, we first tested the accessibility of p64 domains to antibodies specific to each domain. Figure 5A shows that each of these antibodies was able to specifically immunoprecipitate isotope-labeled p64 translated in a cell-free system. When these antibodies were incubated with BYV virions, it was found that the antibody specific for the N-terminal domain of p64, but not the one specific for the C-terminal domain, immunoprecipitated virions (Fig. 5B). This result confirmed the specific physical association of p64 with BYV virions and the prediction that the C-terminal domain of p64 is functionally analogous to CP.
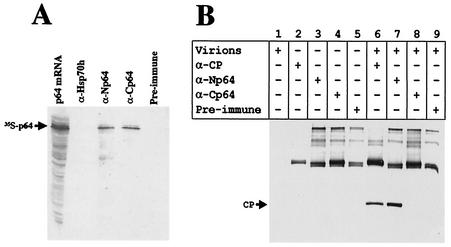
FIG. 5.
(A) Immunoprecipitation of 35S-labeled p64 with antisera raised against the recombinant N-terminal domain of p64 (lane α-Np64) or C-terminal domain of p64 (lane α-Cp64). Lane p64 mRNA, products of the in vitro translation reaction programmed with the p64 mRNA. Antiserum to Hsp70h (lane α-Hsp70h) and preimmune serum (lane Pre-immune) were used as negative controls. The proteins were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and subjected to autoradiography. (B) Immunoprecipitation of BYV virions with anti-Np64 (α-Np64) and anti-Cp64 (α-Cp64) sera. Anti-CP (α-CP) and preimmune sera were used as positive and negative controls, respectively. The products of immunoprecipitation reactions were analyzed by immunoblotting with anti-CP serum. The principal components of the reactions corresponding to each lane are shown in the table above the photograph. The bands in the upper portion of the membrane correspond to rabbit antibodies present in immunoprecipitated material and recognized by goat anti-rabbit serum conjugated to alkaline phosphatase.
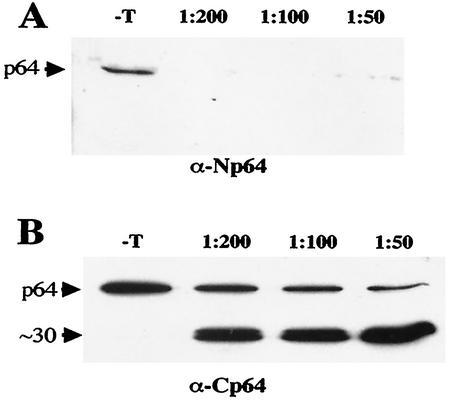
We also tested the accessibility of the p64 domains to limited trypsin digestion. Virions were treated with increasing concentrations of trypsin, precipitated by ultracentrifugation to remove the digestion products, and subjected to immunoblot analysis. As shown in Fig. 6A, even treatment with the highest trypsin dilution used resulted in virtually complete degradation of the N-terminal domain of p64. In contrast, the C-terminal domain of ∼30 kDa was resistant to trypsin digestion and remained associated with virions (Fig. 6B). On the basis of the molecular weight of this domain, Lys-325 was selected arbitrarily as a likely site of cleavage by trypsin (Fig. 2). The difference in the apparent amounts of the full-size p64 protein observed between Fig. 6A and B is likely due to difference in the relative sensitivities of the antisera used. No change in the electrophoretic mobility of CP was observed, indicating that the CP present in virions was protected from trypsin digestion under the experimental conditions used (data not shown). Thus, the results of immunoprecipitation and trypsin digestion were in agreement with the domain topology of p64 inferred on the basis of computational analysis.
FIG. 6.
(A and B) Relative resistance of the N- and C-terminal domains, respectively, of virion-associated p64 to limited trypsin digestion. −T, no trypsin added (negative control); 1:200, 1:100, and 1:50, trypsin dilutions. The type of antiserum used for immunoblot analysis of the treated and precipitated virions is shown below each panel. The positions of p64 and its ∼30-kDa degradation product are shown on the left.
p64 is required for virion tail assembly and cell-to-cell movement of BYV.
The presence of p64 in virions prompted us to ask if p64 is required for virion assembly and, if so, if there is a connection between the roles of p64 in assembly and cell-to-cell movement. Previously, we constructed two p64 mutants, each of which was defective in cell-to-cell movement (4). In the first of these mutants, Δ64N, most of the N-terminal domain was deleted, whereas the second mutant, N64S, expressed only the N-terminal domain (Fig. 2). Here, we designed mutants R416A and D455A, in which the invariant Arg-416 and Asp-455 residues in the CP-like domain (Fig. 1B) were replaced with Ala. As demonstrated previously for CPs of several filamentous viruses, mutations of these highly conserved amino acid residues completely abolished the assembly function of CP (5, 13, 14, 18).
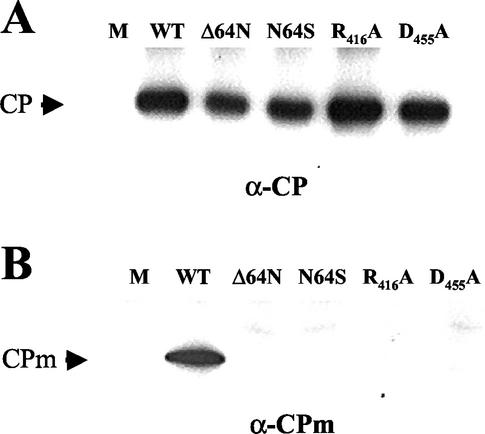
Each of the four BYV mutants was transfected into tobacco protoplasts, and virions were isolated 4 days posttransfection. To assess the presence of virion bodies and tails in the mutant virions, we used immunoblot analysis and antibodies specific for BYV CP and CPm, respectively (5). As shown in Fig. 7A, none of the four p64 mutations affected the formation of virion bodies: the amount of CP found in mutant virions was similar to that found in wild-type virions. However, in sharp contrast to the wild type, the mutant virions contained no detectable CPm, indicating that tail assembly was impaired (Fig. 7B). These results showed that p64 is required for virion tail assembly and supported the functional importance of the conserved Arg-416 and Asp-455 residues in this process. Moreover, the lack of tail formation in the Δ64N and N64S mutants suggested that each of the two domains of p64 is required for its function in tail formation.
FIG. 7.
Protein composition of virions assembled by four BYV variants possessing mutant p64. Virions were isolated from protoplasts transfected with the corresponding BYV variant and analyzed by immunoblotting with anti-CP (A) and anti-CPm (B) sera. M and WT, mock-transfected protoplasts and protoplasts transfected with wild-type BYV, respectively. The mutants' names are shown above each panel, and the type of antiserum used for immunoblot analysis is shown below each panel.
The cell-to-cell movement phenotypes of the R416A and D455A mutants was tested in plant inoculation experiments with a green fluorescent protein-tagged BYV variant (36). Screening of 12 leaves showed that, by 10 days postinoculation, the parental green fluorescent protein-tagged BYV formed 118 green fluorescent infection foci with a mean diameter of 4.4 ± 2.4 cells. The mutant variants produced similar numbers of infection foci (130 and 99, respectively). However, all of these foci were unicellular, demonstrating that the R416A and D455A mutations abolished the cell-to-cell movement of the virus. Thus, each of the four p64 mutants tested was defective in both the ability to form virion tails and the ability to move from cell to cell.
DISCUSSION
Among the members of plant positive-strand RNA virus families, members of the Closteroviridae family stand apart in having the largest genomes, at 15 to 20 kb, in contrast to the ∼6 kb genomes of their cousins, the members of the Tobamoviridae family. Our major goal in studying the biology of closteroviruses is to elucidate the unique functions and the underlying evolutionary mechanisms that account for the increase in the genetic capacity of closteroviruses. Some of the extra genes encode parts of the sophisticated RNA synthesis machinery of the closterovirus. In addition to the core replicase domains that are shared by all Sindbis virus-like RNA viruses of plants and animals (24), closteroviruses encode unique domains that could account for their ability to replicate large RNAs and recognize an array of subgenomic promoters (16, 21). Moreover, efficient replication of closterovirus RNA requires two types of replication enhancers, the leader proteinases (34) and the ∼20-kDa proteins (Fig. 1A) (35, 39, 42).
This work, along with previous results (2, 4, 5, 32, 43, 50), reveals another major function of the unique genes of closteroviruses by showing that proteins encoded by these genes contribute to the formation of morphologically and functionally complex virions. Closteroviruses have two gene blocks that are conserved within this family: the replication-related gene block and the so-called quintuple-gene block or QGB (Fig. 1A) (4, 21). As we show here, p64 is the fourth of the five QGB-encoded proteins that are incorporated into tailed BYV virions. Although each of the QGB proteins is essential for BYV cell-to-cell movement, only p6 is a BYV MP that is not a virion component (5).
How could this unique constellation of genes with dual functions in virion formation and cell-to-cell movement evolve? This work and previous analyses (7) showed that one of the underlying mechanisms was tandem gene duplication, which occurred at least twice to yield the coding regions for CPm and the CP-like domain of p64 (Fig. 2). Since the three CP-like domain-containing genes are present in all of the closteroviruses whose genomes have been sequenced so far, it appears that both duplications occurred prior to the divergence of these viruses from their common ancestor. Other important events in the evolution of closteroviruses apparently included acquisition of the coding regions for Hsp70h and the N-terminal domain of p64. In the former case, it appears obvious that an ancestral closterovirus captured a cellular mRNA for Hsp70 (1) whereas the origin of the upstream portion of the p64 gene remains obscure. In addition to being an MP and an essential virion component (5, 36), Hsp70h provides a docking site for long-distance transport factor p20, which is required for systemic virus spread through the plant vasculature (38). Although p20 is associated with virions, unlike other virion proteins, it is not essential for assembly or cell-to-cell movement. The evolutionary scenario for tailed closterovirus virions can be interpreted as a hierarchical buildup of virion functions from merely protecting the genome to driving cell-to-cell movement to mediating long-distance transport of the virus. It seems likely that, during closterovirus evolution, the selective advantage conferred on the virus by these increasingly complex devices for virus-host interaction was a driving force behind the evolution of the mechanisms of subgenomic RNA synthesis and its regulation and, accordingly, the overall increase in genome size.
Identification of the third protein that is required for the tail assembly and cell-to-cell movement of BYV further advances the concept of the closterovirus tail as a specialized movement device. What could be the architectural and functional roles of p64 in BYV virions? The virion-embedded, CP-like domain of p64 may fit into the helical assembly of the CP and/or CPm subunits. Because p64 is required for tail assembly, it is likely to be physically associated with the tail. One of the possible functions of p64 is formation of the connector between the body and the tail of closterovirus virions. Such connector proteins form collar regions between the head and the tail in some bacteriophages (31, 48). It seems likely that, in addition to an architectural role, the unique N-terminal domain of p64 provides additional activities required for the cell-to-cell movement of BYV. The next challenge is to determine the exact molecular architecture of closterovirus virions and to characterize the mechanistic contribution of each of the five virion proteins.
Although closterovirus virions are the most complex of nonenveloped plant virus virions, recruitment of additional virion-associated proteins for cell-to-cell movement, systemic transport, or plant-to-plant transmission is rather common. Among the helical plant viruses, one example is provided by an RNA helicase-related MP of a Potexvirus that is capable of binding and modifying virions (6). Another example involves the VPg protein of a Potyvirus that is present at one copy per virion and is required for systemic virus transport (44). In a Benyvirus and a Pomovirus, a minor CP generated via readthrough of the gene for CP is incorporated at one end of the virion and is required for virion assembly and transmission (11, 45). Thus, evolutionarily diverse helical viruses have independently evolved the ability to utilize virions as a structural platform for the buildup of additional functional units.
Acknowledgments
We thank Bryce Falk and Mike Taliansky for helpful discussions and critical reading of the manuscript.
This research was supported by grants from the National Institutes of Health (R1GM53190B) and the U.S. Department of Agriculture (CSREES 2001-35319-10875) to V.V.D.
REFERENCES
- 1.Agranovsky, A. A., V. P. Boyko, A. V. Karasev, E. V. Koonin, and V. V. Dolja. 1991. Putative 65 kDa protein of beet yellows closterovirus is a homologue of HSP70 heat shock proteins. J. Mol. Biol. 217:603-610. [DOI] [PubMed] [Google Scholar]
- 2.Agranovsky, A. A., D. E. Lesemann, E. Maiss, R. Hull, and J. G. Atabekov. 1995. “Rattlesnake” structure of a filamentous plant RNA virus built of two capsid proteins. Proc. Natl. Acad. Sci. USA 92:2470-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alzhanova, D. V., Y. Hagiwara, V. V. Peremyslov, and V. V. Dolja. 2000. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 268:192-200. [DOI] [PubMed] [Google Scholar]
- 5.Alzhanova, D. V., A. Napuli, R. Creamer, and V. V. Dolja. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J. 20:6997-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atabekov, J. G., N. P. Rodionova, O. V. Karpova, S. V. Kozlovsky, and V. Y. Poljakov. 2000. The movement protein-triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology 271:259-263. [DOI] [PubMed] [Google Scholar]
- 7.Boyko, V. P., A. V. Karasev, A. A. Agranovsky, E. V. Koonin, and V. V. Dolja. 1992. Coat protein gene duplication in a filamentous RNA virus of plants. Proc. Natl. Acad. Sci. USA 89:9156-9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callaway, A., D. Giesman-Cookmeyer, E. T. Gillock, T. L. Sit, and S. A. Lommel. 2001. The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39:419-460. [DOI] [PubMed] [Google Scholar]
- 9.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman, S., G. Hills, J. Watts, and D. Baulcombe. 1992. Mutational analysis of the coat protein gene of potato virus X: effects on virion morphology and viral pathogenicity. Virology 191:223-230. [DOI] [PubMed] [Google Scholar]
- 11.Cowan, G. H., L. Torrance, and B. Reavy. 1997. Detection of potato mop-top virus capsid readthrough protein in virus particles. J. Gen. Virol. 78:1779-1783. [DOI] [PubMed] [Google Scholar]
- 12.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 13.Dolja, V. V., V. P. Boyko, A. A. Agranovsky, and E. V. Koonin. 1991. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology 184:79-86. [DOI] [PubMed] [Google Scholar]
- 14.Dolja, V. V., R. Haldeman, N. L. Robertson, W. G. Dougherty, and J. C. Carrington. 1994. Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 13:1482-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doneanu, C. E., D. A. Griffin, E. L. Barofsky, and D. F. Barofsky. 2001. An exponential dilution gradient system for nanoscale liquid chromatography in combination with MALDI or nano-ESI mass spectrometry for proteolytic digests. J. Am. Soc. Mass Spectrom. 12:1205-1213. [DOI] [PubMed] [Google Scholar]
- 16.Gowda, S., T. Satyanarayana, M. A. Ayllon, M. R. Albiach-Marti, M. Mawassi, S. Rabindran, S. M. Garnsey, and W. O. Dawson. 2001. Characterization of the cis-acting elements controlling subgenomic mRNAs of citrus tristeza virus: production of positive- and negative-stranded 3′-terminal and positive-stranded 5′-terminal RNAs. Virology 286:134-151. [DOI] [PubMed] [Google Scholar]
- 17.Harrison, S. C. 2001. Principles of virus structure, p. 53-85. In D. M. Knipe and P.M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 18.Jagadish, M. N., D. Huang, and C. W. Ward. 1993. Site-directed mutagenesis of a potyvirus coat protein and its assembly in Escherichia coli. J. Gen. Virol. 74:893-896. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanamaru, S., P. G. Leiman, V. A. Kostyuchenko, P. R. Chipman, V. V. Mesyanzhinov, F. Arisaka, and M. G. Rossmann. 2002. Structure of the cell-puncturing device of bacteriophage T4. Nature 415:553-557. [DOI] [PubMed] [Google Scholar]
- 21.Karasev, A. V. 2000. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38:293-324. [DOI] [PubMed] [Google Scholar]
- 22.Kasteel, D. T., J. Wellink, R. W. Goldbach, and J. W. van Lent. 1997. Isolation and characterization of tubular structures of cowpea mosaic virus. J. Gen. Virol. 78:3167-3170. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy, R. T., and J. W. Jorgenson. 1989. Quantitative analysis of individual neurons by open tubular liquid chromatography with voltammetric detection. Anal. Chem. 61:436-441. [DOI] [PubMed] [Google Scholar]
- 24.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 25.Lazarowitz, S. G., and R. N. Beachy. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11:535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lough, T. J., N. E. Netzler, S. J. Emerson, P. Sutherland, F. Carr, D. L. Beck, W. Lucas, and R. L. S. Forster. 2000. Cell-to-cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant-Microbe Interact. 13:962-974. [DOI] [PubMed] [Google Scholar]
- 27.Lucas, W. J., and S. Wolf. 1999. Connections between virus movement, macromolecular signalling and assimilate allocation. Curr. Opin. Plant Biol. 2:192-197. [DOI] [PubMed] [Google Scholar]
- 28.Marchler-Bauer, A., A. R. Panchenko, B. A. Shoemaker, P. A. Thiessen, L. Y. Geer, and S. H. Bryant. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medina, V., V. V. Peremyslov, Y. Hagiwara, and V. V. Dolja. 1999. Subcellular localization of the HSP70-homolog encoded by beet yellows closterovirus. Virology 260:173-181. [DOI] [PubMed] [Google Scholar]
- 30.Melzer, M. J., A. V. Karasev, D. M. Sether, and J. S. Hu. 2001. Nucleotide sequence, genome organization and phylogenetic analysis of pineapple mealybug wilt-associated virus-2. J. Gen. Virol. 82:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Molineux, I. J. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 40:1-8. [DOI] [PubMed] [Google Scholar]
- 32.Napuli, A. J., B. W. Falk, and V. V. Dolja. 2000. Interaction between HSP70 homolog and filamentous virions of the beet yellows virus. Virology 274:232-239. [DOI] [PubMed] [Google Scholar]
- 33.Oparka, K. J., and A. G. Roberts. 2001. Plasmodesmata: a not so open-and-shut case. Plant Physiol. 125:123-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng, C. W., V. V. Peremyslov, A. R. Mushegian, W. O. Dawson, and V. V. Dolja. 2001. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 75:12153-12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1998. Genes required for replication of the 15.5-kilobase RNA genome of a plant closterovirus. J. Virol. 72:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1999. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc. Natl. Acad. Sci. USA 96:14771-14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ploubidou, A., and M. Way. 2001. Viral transport and the cytoskeleton. Curr. Opin. Cell Biol. 13:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prokhnevsky, A. I., V. V. Peremyslov, A. J. Napuli, and V. V. Dolja. 2002. Interaction between long-distance transport factor and Hsp70-related movement protein of beet yellows virus. J. Virol. 76:11003-11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed, J. C., K. D. Kasschau, A. I. Prokhnevsky, K. Gopinath, G. P. Pogue, J. C. Carrington, and V. V. Dolja. Suppressor of RNA silencing encoded by beet yellows virus. Virology, in press. [DOI] [PubMed]
- 40.Roberts, I. M., D. Wang, K. Findlay, and A. J. Maule. 1998. Ultrastructural and temporal observations of the potyvirus cylindrical inclusions (Cls) show that the Cl protein acts transiently in aiding virus movement. Virology 245:173-181. [DOI] [PubMed] [Google Scholar]
- 41.Santa Cruz, S., A. G. Roberts, D. A. M. Prior, S. Chapman, and K. J. Oparka. 1998. Cell-to-cell and phloem-mediated transport of potato virus X: the role of virions. Plant Cell 10:495-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satyanarayana, T., S. Gowda, M. A. Ayllon, M. R. Albiach-Marti, S. Rabindran, and W. O. Dawson. 2002. The p23 protein of citrus tristeza virus controls asymmetrical RNA accumulation. J. Virol. 76:473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satyanarayana, T., S. Gowda, M. Mawassi, M. R. Albiach-Marti, M. A. Ayllon, C. Robertson, S. M. Garnsey, and W. O. Dawson. 2000. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 278:253-265. [DOI] [PubMed] [Google Scholar]
- 44.Schaad, M. C., A. D. Lellis, and J. C. Carrington. 1997. VPg of tobacco etch potyvirus is a host genotype-specific determinant for long-distance movement. J. Virol. 71:8624-8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt, C., E. Balmori, G. Jonard, K. E. Richards, and H. Guilley. 1992. In vitro mutagenesis of biologically active transcripts of beet necrotic yellow vein virus RNA 2: evidence that a domain of a 75-kDa readthrough protein is important for efficient virus assembly. Proc. Natl. Acad. Sci. USA 89:5715-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuler, G. D., S. F. Altschul, and D. J. Lipman. 1991. A workbench for multiple alignment construction and analysis. Proteins 9:180-190. [DOI] [PubMed] [Google Scholar]
- 47.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 48.Simpson, A. A., Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage φ29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas, C. L., and A. J. Maule. 1995. Identification of structural domains within the cauliflower mosaic virus movement protein by scanning deletion mutagenesis and epitope tagging. Plant Cell 7:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian, T., L. Rubio, H.-H. Yeh, B. Crawford, and B. W. Falk. 1999. Lettuce infectious yellows virus: in vitro acquisition analysis with partially purified virions and the whitefly, Bemisia tabaci. J. Gen. Virol. 80:1111-1117. [DOI] [PubMed] [Google Scholar]