Abstract
The degree of genetic heterogeneity of DNA virus populations in nature and its consequences for disease control are virtually unknown. The parvovirus minute virus of mice (MVMi) was used here to investigate (i) the frequency of antibody-escape mutants in populations of a DNA virus and (ii) the ability of a DNA virus to evade in the long-term a passive monoclonal antibody (MAb) therapy in an immunodeficient natural host. Independent clonal populations of MVMi harbored a high proportion of mutants resistant to neutralizing MAb (mutant frequency = [2.8 ± 0.5] × 10−5) that rapidly evolved under antibody pressure in culture to become mixtures dominated by genotypically diverse escape mutants. Immunodeficient mice naturally infected with clonal populations of MVMi and subsequently treated by intravenous injections of MAb were initially protected from the characteristic viral induced lethal leukopenia. However, some treated animals developed a delayed severe leukopenic syndrome associated with the emergence of genetically heterogeneous populations of MAb-resistant mutants in the MVMi main target organs. The 11 plaque-purified viruses analyzed from an antibody-resistant population obtained from one animal corresponded to four different mutant genotypes, although their consensus sequence remained wild type. All cloned escape mutants harbored single radical amino acid changes within a stretch of seven residues in a surface-exposed loop at the threefold axes of the capsid. This antigenic site, which can tolerate radical changes preserving MVMi pathogenic potential, may thereby allow the virus to evade the immune control. These findings indicate a high genetic heterogeneity and rapid adaptation of populations of a mammal DNA virus in vivo and provide a genetic basis for the failure of passive immunotherapy in the natural host.
Many RNA virus populations are extremely heterogeneous and dynamic distributions of mutant genomes termed viral quasispecies (for reviews, see references 18, 19, 20, 25, and 33). Viral quasispecies constitute large reservoirs of variants that are the basis for the extreme adaptability and rapid evolution of many RNA viruses. The quasispecies concept and its biological implications are increasingly being taken into account in the development of preventive measures and therapies against diseases caused by RNA viruses (20, 21). The high heterogeneity and adaptability of RNA virus populations are partly due to the high mutation rates during RNA replication in the absence of proofreading mechanisms and mismatch repair systems. In contrast, DNA virus genomes are replicated by the action of either cellular or their own polymerases, for which proofreading activities have been either proved (35) or assumed. This has led, despite the absence of any conclusive experimental evidence, to the generally held view that the DNA virus genomes may be no more variable than cellular genes (27, 61) and that genetic heterogeneity may be not an important concern regarding diseases caused by DNA viruses.
Passive antibody administration is being used or tested as a preventive or therapeutic measure against a number of RNA virus infections, including those caused by important human pathogens such as human immunodeficiency virus, hepatitis A virus, Ebola virus, measles virus, or respiratory syncytial virus (12, 37, 42, 47). Animal models of viral infections, mainly mice with severe immunodeficiency (SCID [9]), have been used to evaluate the efficacy of passive immunity (29, 46). The use in humans of monoclonal antibodies (MAbs) is now being recommended for biosafety reasons, as exemplified by the licensing of a neutralizing MAb against respiratory syncytial virus for passive immunotherapy in children (52). Unfortunately, the quasispecies nature of RNA virus populations may severely hamper the effectiveness of both vaccination and passive immunotherapy. RNA viruses can easily evade the action of neutralizing antibodies in vitro through the rapid selection from their heterogeneous populations of MAb-resistant (MAR) mutants with amino acid substitutions at the corresponding epitopes (21). In vivo, the emergence of escape mutants in individuals either vaccinated or treated with passive immunotherapy has also been documented (10, 14, 26, 48, 50, 57). Thus, some current protocols of passive immunotherapy against human virus pathogens contemplate the use of a cocktail of MAbs targeting a wide repertoire of antigenic sites (12, 15, 32, 41, 65), an approach that may reduce the emergence of antibody-resistant variants.
In spite of the wide use of passive immunotherapy for the treatment of diseases caused by DNA viruses, the major uncertainties about the genetic heterogeneity of DNA virus populations leave the long-term emergence of escape mutants as an open question. Several members of the Parvoviridae, a family of viruses with a single-stranded DNA (ssDNA; 5-kb) genome packaged into a 25-nm-diameter icosahedral capsid (45), cause persistent and chronic diseases in animals and immunocompromised humans that are often treated by passive immunotherapy (2, 5, 38). Persistent parvovirus B19 infection of anemic and human immunodeficiency virus type 1-immunodepressed patients is commonly treated with antisera (31, 38), and the generalized use of MAbs is also being recommended in this case (30). The present study addresses the role of genetic variation in the effectiveness of a passive immunotherapy against a model DNA virus, the strain i of the autonomous parvovirus minute virus of mice (MVMi) (16), an important mouse pathogen that causes a lethal hematological disease in the immunodeficient host (55). The results reveal a high genetic heterogeneity in populations of a DNA virus and the ability of MVMi to escape passive immunotherapy through the in vivo selection of antibody-resistant mutants in the virus capsid surface. Virus population heterogeneity may be an important factor to be considered for the control of DNA virus infections.
MATERIALS AND METHODS
Virus and cells.
Clonal viral stocks of the immunosuppressive strain of MVMi were prepared from an infectious cDNA clone through a minimal number of passages in cell culture. A total of 5 × 106 cells of the EL-4 lymphoma in which this virus was originally isolated (8) were transfected by electroporation with 20 μg of an infectious molecular DNA clone of MVMi (28) grown from a single transformed colony of Escherichia coli JC8111, a bacterial strain that permits deletion-resistant propagation of MVM plasmid clones bearing terminal palindromes (7). Newly formed intracellular virus was harvested 48 h afterward and used to infect growing EL-4 cultures at a multiplicity of infection of 0.01 PFU/cell. Cultures were harvested 4 days postinfection (dpi) with incipient cytopathic effect and virus purified through sucrose cushion and cesium chloride gradient centrifugations as previously described (54) and sterile filtered. Virus titers were determined by plaque assay on monolayers of the fully permissive NB324K cell line (59). Cells were cultured in Dulbecco modified Eagle medium (Gibco Laboratories) supplemented with 5% inactivated fetal bovine serum (FCS).
Neutralization assays.
A collection of hybridoma cell lines (kindly provided by C. R. Parrish) elicited against MVM capsid was tested for the production of neutralizing MAb against MVMi. The hybridoma designated B7 producing a relative high neutralization titer was cultured to confluence in Dulbecco modified Eagle medium supplemented with 10% FCS, and the clarified supernatants used as a source of antibody. When a higher antibody titer was required, the hybridoma cells were grown in the peritoneal cavity of mice and ascites fluid recovered 20 days after inoculation. MAb B7 specifically recognizes the MVM intact capsid as previously demonstrated by different immunological tests (40), although its epitope had not been located. Neutralization titers were determined by incubation of serial dilutions of the antibody with 150 PFU of MVMi in 400 μl for 30 min at 37°C. The proportion of nonneutralized virus was obtained by a plaque assay in duplicate, and one neutralization unit (Nu) was defined as the amount of MAb required to neutralize 50% of the virus in this assay (see Fig. 3A). To determine the frequency of MAR mutants in the viral populations, suspensions of infected cultures or mouse organ homogenates were incubated with 25 Nu of MAb/ml, and the mixtures were serially diluted and inoculated onto cell monolayers for plaque assay. After virus adsorption, the inoculum was removed, and 2 Nu of MAb/ml was added to the overlaid semisolid medium. Plaques were counted from duplicates 6 days afterward, and some candidate MAR mutants were shown by DNA sequencing to harbor amino acid changes (see Results).
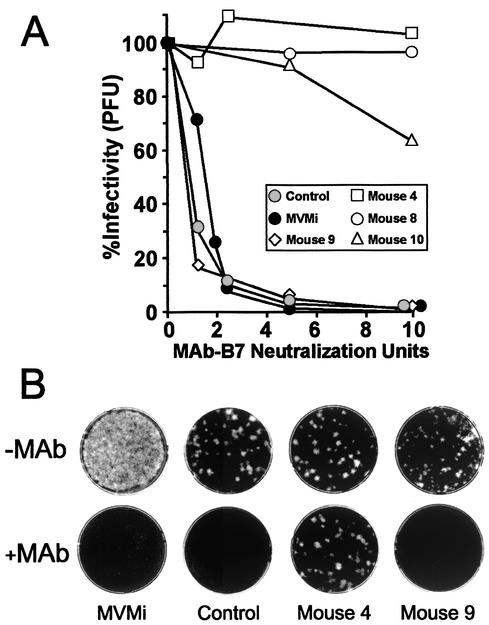
FIG. 3.
Phenotype of viral populations recovered from MVMi-infected SCID mice. (A) Neutralization kinetics comparing a MVMi stock with virus populations obtained from the spleen of untreated mice (control) and of some MAb-treated mice (animals 4, 8, 9, and 10). Shown are the percent residual infectivity values after incubation with or without the indicated units of MAb B7. The experiment was repeated three times with similar outcomes. (B) Representative plaque assay showing MAR mutants recovered from spleen homogenates of the indicated mice. Note the different proportion of MAR mutants between samples.
Passive immunotherapy.
Eight- to ten-week-old females of the severe combined immunodeficient (SCID) C.B-17 inbred mouse strain (9) were used. Breeding pairs, originally obtained from Jackson Laboratories (Bar Harbor, Maine), were allowed autoclaved food and water ad libitum, routinely handled under sterile conditions, and housed in sterile microisolators under a light-dark cycle from 9 a.m. to 9 p.m. Mice were inoculated intranasally with 106 PFU/mouse of a purified stock of parvovirus MVMi in 10 μl of phosphate-buffered saline (PBS) as previously described (55). At the indicated number of days postinfection, mice were treated on a weekly or fortnight basis by intravenous injection at the tail vein of 12 Nu from supernatant of the B7 hybridoma culture diluted in 100 μl of PBS and sterile filtered. Control mice receiving injection of culture medium with 10% FCS diluted in PBS were not protected (not shown). Mice were weekly monitored for pathological signs (ruffled fur and hunched posture) and white blood cell counts as described previously (55) and then euthanized as moribund according to European Union guidelines (86/609/CEE).
Viral genetic analysis.
MVMi replicative and genomic DNA forms were isolated from mouse organs by a modified Hirt procedure (44). Briefly, organ homogenates were adjusted to 20 mM NaCl-50 mM Tris-HCl (pH 8.0)-10 mM EDTA and incubated with 100 μg of proteinase K (Boehringer)/ml for 2 h at 37°C. Low-molecular-weight DNA was precipitated and resuspended in 10 mM Tris-HCl (pH 7.5)-1 mM EDTA. When indicated, DNA was similarly recovered from viral plaques that were pick isolated and extracted in 100 μl of PBS by vigorous shaking and left overnight at 4°C. Virus DNA was amplified by PCR according to published protocols, using a Perkin-Elmer DNA Cetus (GeneAmp PCR system 9600) or Bio-Rad gene cycler with the two sets of oligonucleotides (either VVP1 [nucleotides {nt} 2221 to 2238]) and pser 388 [nt 3950 to 3967] or VVP6 [nt 3444 to 3460] and VPSeq0 [nt 4688 to 4706]) in order to encompass the VP coding region. The amplified fragments were purified from agarose gels by concert rapid gel extraction system (Gibco-BRL), and the sequence was determined in a Perkin-Elmer 377 automated sequencer. The VP gene was sequenced by using the following oligonucleotides at the specified positions on the MVM genome (3): VVP1 (nt 2221 to 2238), VVP2 (nt 2503 to 2518), VVP4 (nt 2945 to 2961), VVP6 (nt 3444 to 3460), VVP8 (nt 3901 to 3917), and VPSeq0 (nt 4706 to 4688). All mutations were confirmed by performing at least two independent PCR amplifications.
RESULTS
High mutant frequency in clonal populations of MVMi.
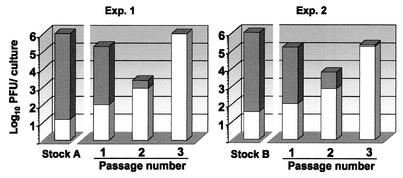
Populations derived from a single molecular clone of MVMi were obtained from infected cultured cells, and the proportion of infectious virions able to escape MAb B7 neutralization was determined. Despite minimal viral amplification in the absence of immune pressure or other detectable selective forces, a high mutant frequency was obtained for the two viral populations tested (Fig. 1). Four independent determinations resulted in an average mutant frequency value of (2.8 ± 0.5) × 10−5. Cultures of permissive cells infected with either viral stock and passaged in the presence of neutralizing doses of the same MAb resulted in a rapid enrichment of MAb-resistant (MAR) mutants (Fig. 1), which dominated the populations upon only two serial passages.
FIG. 1.
Proportion of MAR mutants in populations of MVMi. Independent clonal populations of MVMi (stocks A and B) obtained in the absence of antibody as described in Materials and Methods were used to infect 106 NB324K cells at a multiplicity of infection of 1. At 4 h postadsorption, MAb B7 was added to the medium (2 Nu/ml), and the intracellular virus was harvested at 72 hpi (passage 1). Passages 2 and 3 were similarly obtained in the presence of antibody by inoculating 106 cells with one-tenth of the virus obtained in the previous passage. For each population, the total infectious virus titer (bars) and the MAR mutants titer (white portion of each bar) were determined by plaque assay in the absence or presence of antibody, respectively. The phenomenon was also observed with MVM-neutralizing MAb harvested from other hybridoma cell lines (not shown).
MVMi can evade passive immunotherapy in the natural host by selection of antibody-escape mutants.
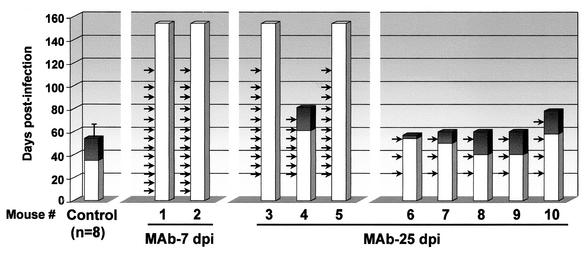
The high MAR mutant frequency and rapid dominance of MVMi escape mutants observed under immune pressure in vitro could imply important biological consequences if also occurring in the natural host. To investigate this point, the efficacy of a passive immunotherapy against a lethal MVMi-induced disease was evaluated in the long-term. Adult SCID mice were oronasally inoculated with a lethal dose (106 PFU) of MVMi and then subjected to diverse passive immunotherapy protocols based in the administration of a neutralizing anti-capsid MAb (Fig. 2). In the SCID mice, the capacity of this virus to target committed and primitive hemopoietic precursors (54) results in a severe leukopenia and dysregulated erythropoiesis that leads to death (55). The mice were monitored weekly for protection against the characteristic leukopenic syndrome associated with this infection that is normally apparent by 40 dpi and precedes the death of the animal, which occurs by 60 dpi (55). Two mice (animals 1 and 2) received weekly MAb B7 injections starting early (7 dpi) after the infection and were both fully protected even in the long-term. Infectious virus and viral DNA (analyzed by PCR) were not detectable in any organ at sacrifice (157 dpi), 45 days after the last MAb dosis. Three mice (animals 3 to 5) also received weekly injections of MAb B7, but the treatment started at 25 dpi, when the virus titer has substantially increased in the bone marrow of the SCID mice (55). Full protection and virus clearance was observed in two cases, but mouse 4 developed a delayed severe leukopenia with concomitant weight loss and evident clinical signs, leading to death about 1 month after that of the untreated infected controls. Five additional mice (animals 6 to 10) were treated with MAb B7 on a fortnightly basis, starting at 25 dpi. All five developed a lethal disease following either a normal or a delayed course (Fig. 2).
FIG. 2.
Protection analysis of a passive immunotherapy in SCID mice lethally infected by MVMi. Animals 1 to 10 and negative controls (n = 8) were oronasally infected with 106 PFU of MVMi and treated at the indicated dates (arrows) with an intravenous injection of 12 Nu of MAb B7. Bars represent the life span of the animals, and the shaded portions indicate the appearance of the leukopenic syndrome (white blood cell count, <5 × 105/ml of peripheral blood).
The delayed leukopenia developing in some mice under a continuous supply of neutralizing levels of MAb in their blood prompted us to evaluate whether MAR mutants of MVMi had been selected in vivo during the antibody therapy. Infectious MVMi populations recovered at high titers from different organs of an untreated mouse were as sensitive to MAb neutralization in vitro as the original MVMi stock (Fig. 3A). In contrast, viral populations obtained from some leukopenic mice undergoing passive immunotherapy clearly showed different levels of resistance to antibody neutralization (Fig. 3). Table 1 shows that MAR mutants reached high titers and dominated the viral populations recovered from the organs of the two mice in which the leukopenia followed a delayed course (animals 4 and 10) and from one mouse (animal 8) of the two analyzed among those becoming moribund before 60 dpi. In summary, these analyses showed that passive humoral immunotherapy protocols may fail to cure SCID mice from MVMi infection and that this failure is associated with the capacity of MVMi to generate pathogenic antibody-escape mutants that become predominant in the main organs, causing a generally delayed lethal disease.
TABLE 1.
Percentage of MAR mutants in viral populations of MAb-treated mouse organs
| Mouse no. | Organa | Titerb | % MAR mutants |
|---|---|---|---|
| 4 | S | 3.7 × 106 | 100 |
| L | 1.5 × 105 | 85 | |
| K | 5 × 104 | 100 | |
| 8 | S | 8.2 × 106 | 85 |
| L | 1.1 × 106 | 90 | |
| K | 5.5 × 104 | 80 | |
| 9 | S | 4.6 × 107 | <1 |
| L | 2.6 × 106 | <1 | |
| K | 2 × 105 | <1 | |
| 10 | S | 6 × 104 | 95 |
| L | 4.7 × 104 | 85 | |
| K | 1.2 × 104 | 65 | |
| Control | S | 3 × 107 | <1 |
| L | 7.5 × 106 | <1 | |
| K | 8 × 105 | <1 |
S, spleen; L, liver; K, kidney.
Titers are given as the mean PFU per gram of wet tissue from duplicate assays.
Genetic heterogeneity of MVMi populations derived from animals subjected to passive immunotherapy.
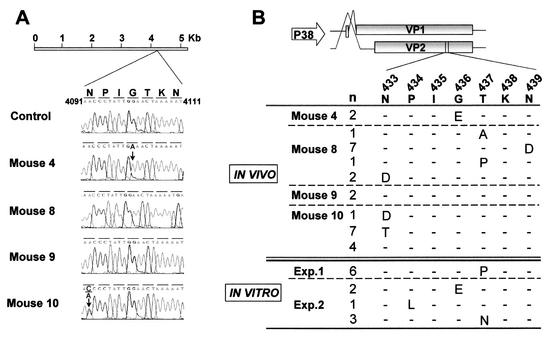
To determine the genetic basis of the MAR phenotype found in treated mice, DNA isolated from the MVMi populations recovered from different organs was subjected to sequence analysis across the entire VP gene, which codes for the proteins VP1 and VP2 forming the MVMi capsid (Fig. 4A). Consensus VP sequences of the antibody-sensitive populations derived from the spleen of untreated mice or of mouse 9 were identical to that of the original MVMi inocula. In contrast, consensus sequences of antibody-resistant populations derived from unprotected mice 10 and 4 showed single point mutations at nt 4091 or 4110 of the MVMi genome (3), which respectively determined a N433T or G436E amino acid substitution (VP2 numbering) in the capsid. Interestingly, no nucleotide changes with respect to control mice were detected in the consensus sequence of the fully antibody-resistant population derived from mouse 8 (Fig. 4A).
FIG. 4.
Sequence analysis of mutations conferring MAb B7 resistance in vivo or in vitro. (A) Nucleotide sequence of MVMi populations recovered from the spleen of the indicated mice. Shown is the VP region of MVM genome where VP coding changes map. The predominant point mutation in the population of mouse 4 and mouse 10 is indicated (arrows). A punctual change V551A found in some mouse organs but not correlating with MAb resistance is not indicated in the figure. (B) Scheme of the VP1 and VP2 structural proteins translated from alternative spliced messengers expressed from the P38 viral promoter. The sequence of the segment of both polypeptides where MAR mutations were located is shown below by using the VP2 numbering. Amino acid differences relative to the parental sequence in viral clones isolated from the indicated mice or from populations independently obtained in vitro (experiments 1 and 2 [see Fig. 1, Exp. 1 and Exp. 2]) are indicated by using the single-letter code. The letter n refers to the number of clones from each population showing the corresponding amino acid substitution.
This later, paradoxical observation was suggestive of the population derived from mouse 8 being genetically heterogeneous. Thus, several individual viral clones of each of the MVMi populations, obtained by plaque purification in the absence of antibody, were sequenced across a region of the VP gene (nt 3700 to 4300 of the MVMi genome) encompassing the positions mutated in the viral populations (Fig. 4A). Consistent with the consensus sequence analyses, a single substitution G436E was found in the two viral clones derived from mouse 4 (Fig. 4B). The substitution N433T was found in 7 of 12 clones derived from mouse 10, 1 clone showed an N433D change, and the remaining 4 clones showed no mutations. Most remarkable, each of the 11 sequenced clones from mouse 8 showed one of four different amino acid substitutions (N433D, T437A, T437P, or N439D) within the VP sequence. Although each viral clone was a MAR mutant, their consensus sequences remained wild type. This result solves the paradox of a viral population with a nonmutated average genotype associated with an antibody-resistant phenotype and provides strong evidence for a highly heterogeneous MVMi population being present in the mouse, the natural host.
These results prompted a search for genetic heterogeneity also in clonal populations of MVMi obtained in vitro. Several viral clones were plaque purified in the absence of antibodies from the two independent MVMi populations that had been obtained under immune pressure (Fig. 1, passage 3). All clones harbored single-amino-acid changes mapping at or close to the VP amino acid residues found substituted in the MAR mutants obtained from mice (Fig. 4B, bottom). Single substitutions at any of three different positions were found in each of six plaque-purified clones of the same population (Fig. 4B, experiment 2), and again the consensus VP sequence of the whole population was the same as for the wild type (not shown), a finding further supporting a remarkable MVMi population heterogeneity also arising in cell culture.
Mapping the antibody-escape mutations in the MVMi capsid.
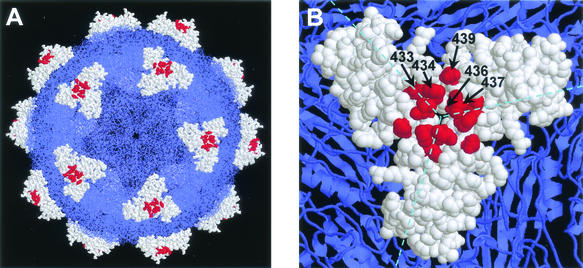
Collectively, the sequencing results also allowed the precise mapping of the MVMi epitope recognized by MAb B7 within the amino acid stretch encompassing residues 433 to 439 (VP2 numbering) in the capsid of MVMi. When displayed on the crystallographic model of the MVMi capsid structure (1), this segment was found to be very close to the threefold axis of symmetry, as a part of loop 4 in the uppermost region of a protrusion called the spike (Fig. 5). The side chains of N433, P434, T437, and N439 are highly exposed to solvent and may have direct contact with the antibody. G436 is immediately underneath T437, but the introduction of a Glu side chain at position 436 may severely disrupt the local conformation and the interaction with the antibody. Major epitopes have been found also in the equivalent capsid region of other related parvoviruses (5, 6, 39, 51, 56, 62). In this regard, our observations with MVMi indicate that this antigenic loop can tolerate drastic changes without major alterations of parvovirus pathogenicity.
FIG. 5.
Localization in the MVMi capsid structure of the amino acid residues whose replacement conferred the MAR phenotype. (A) Model of the complete capsid. (B) Close-up of one of the protruding spikes located at the threefold axes. The amino acid residues that form the spike are represented as space-filling models and colored white, except for those involved in antibody recognition (Fig. 4), which are colored red. All other capsid residues are colored blue and are shown either as wireframe (A) or ribbon (B) models. The figure was prepared by using the program RasMol (53) and the MVMi coordinates (1MVM) deposited in the PDB (1).
DISCUSSION
The results presented here reveal an unprecedented high frequency of antibody-resistant mutants and a remarkable heterogeneity in populations derived from a single clone of a ssDNA virus, both in vitro and in vivo. Although a high frequency of MAR mutants was previously found in vitro in clonal populations of herpesvirus, a double-stranded DNA virus (34, 63), the heterogeneity of the herpesvirus populations in vivo was not investigated. A complex distribution of mutant DNA genomes has been also found in laboratory populations of maize streak virus, a geminivirus (36). It is remarkable that such high mutant frequencies are in the range found for most RNA viruses (20). Extensive work with a number of RNA viruses has shown that their genetically highly heterogeneous populations (quasispecies) are a dynamic but balanced phenomenon resulting from a number of factors, which include very high mutation rates and large population sizes, on the one hand, and not particularly restrictive negative selection pressures, on the other hand (18-20, 24, 25, 33). For DNA viruses however, these factors are generally ill-defined or essentially unknown.
Average mutation rates (substitutions per nucleotide per round of replication) for most RNA genomes range from 10−5 to 10−3 (23). In comparison, mutation rates for cellular genes usually range from 10−10 to 10−9 (22). Interestingly, mutation rates for double-stranded DNA viruses (T2, T4, and lambda) range from ca. 10−8 to 10−7, and for the ssDNA virus M13 the rate is close to 10−6 (22). These rates are clearly higher than those determined for cellular genes. However, it must also be considered that determination of mutation rates is subject to some experimental uncertainty, and some values may be off by 1 order of magnitude or more. Furthermore, mutation rates depend substantially on the biochemical context and the particular nucleotide positions analyzed (22). The replication of the ssDNA genomes through rolling-hairpin mechanisms (17) may proceed with a fidelity different from that determined for replication of cellular genes. The MVM genome uses a cellular machinery that involves at least the DNA polymerase δ and the factors PCNA, RPA, RFC, and cyclin A, besides the viral protein NS1 (4, 13). The fidelity and proofreading activity of this enzymatic complex, compared to that of DNA polymerase α and the other main polymerases involved in the replication of the cellular genome, remains to be established. In addition, the efficient intermolecular recombination of parvovirus genomes (64) may also operate as a common genetic mechanism introducing high genetic heterogeneity in these viruses.
It must also be noted that both in vitro and in vivo populations of MVM are large (e.g., ca. 106 PFU in a small plaque derived from infection of a cell monolayer with a single virion, or more than 108 infectious particles per infected mouse), and this may be an additional important factor in explaining the observed population heterogeneity (20). Moreover, the restrictive qualitative and quantitative effects of negative selection on the heterogeneity of MVM populations are essentially unknown. Epitopes on a virus capsid, even of highly variable RNA viruses, may be subjected to severe structural and/or functional restrictions to variation (43). It is thus possible that the high mutant frequency observed for the B7-resistant phenotype may arise partly because of a particularly high structural and functional tolerance to amino acid substitutions of this particular epitope. Thus, it is important to further analyze whether high mutant frequencies can be obtained for MVM when other epitopes and phenotypes are probed. Irrespective of the molecular mechanisms involved, the results obtained here clearly reveal a surprisingly high genetic heterogeneity in populations of a DNA virus. Such populations harbor a high proportion of escape mutants (and probably many other mutants). This heterogeneity provides this DNA virus with a remarkable adaptability through the rapid selection of antibody-escape mutants, which is clearly associated with the failure of the delayed immunotherapy protocols attempted here. The capacity of the parvovirus capsid to evade immune pressure exerted at major antigenic sites may contribute to the evolution and host range drift of canine parvovirus during epidemics (11), leading to an estimated rate of retained sequence substitution of 1.69 × 10−4/nt/year in the VP gene (49, 60), to the persistence of ADV (2), and to human B19 chronic diseases (38, 58).
One of the three mice treated weekly with MAb was not protected and developed a delayed leukopenia. This mouse (animal 4) yielded a relatively homogeneous antibody-resistant population in which the consensus sequence had the same mutation observed in individual plaque-purified clones. This population thus probably emerged in a bottleneck event from a limited pool of replicating viral genomes not cleared by the MAb. The immunotherapy was less effective in curing mice when administered on a fortnightly basis, and the individual responses were diverse. One of the mice analyzed (animal 9) died from a leukopenia that followed a normal course and yielded high titers of nonmutated virus. In contrast, mouse 8 yielded a highly heterogeneous antibody-resistant population composed of a wide range of MAR genotypes (Fig. 4). In the absence of any treatment, the level of MVMi multiplication in the bone marrow of mice increases sharply by 20 dpi (55). This event may have greatly facilitated the independent selection of a range of escape mutations in mouse 8. An intermediate result was found for mouse 10. Several aspects, including the timing in the emergence of the MAR mutants, may eventually shape the adaptation of the virus and the course of the disease. The diversity of outcomes found in mice infected with MVMi and subjected to immunotherapy suggests that protection against DNA virus infections by systemic administration of antibodies may require (i) early and frequent treatment to limit as much as possible the pool of replicating genomes and (ii) the use of a cocktail of MAbs directed against independent epitopes to minimize the selection of mutants able to evade such therapy. Similar conclusions have been outlined for rapidly evolving RNA viruses (12, 15, 32, 41, 65). To our knowledge, the present study is the first description for a DNA virus of the emergence of a highly heterogeneous population of antibody-resistant mutants in a natural host.
Acknowledgments
We are indebted to E. Domingo (CBMSO, Madrid) and L. Villarreal (Irvine, Calif.) for critical advice and helpful comments on the manuscript, to J. Rubal and J. Dean (MIRT Program, UCI, Irvine, Calif.) for experimental support, to P. Tattersall (Yale University, New Haven, Conn.) for the MVMi infectious molecular clone, and to C. Parrish (Cornell University, Ithaca, N.Y.) for the generous gift of the MAb-producing hybridoma cell lines.
This work was supported by grant SAF 2001-1325 CICYT from the Spanish MCyT and EU contract QLK3-CT-2001-01010 to J.M.A., grant 08.2/0008/2000 from the Comunidad de Madrid to J.M.A. and M.G.M., grant BI02000-0408 from the MCyT to M.G.M., and an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.”
REFERENCES
- 1.Agbandje-McKenna, M., A. LLamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 2.Alexandersen, S., S. Larsen, A. Cohn, A. Uttenthal, R. E. Race, B. Aasted, M. Hansen, and M. E. Bloom. 1989. Passive transfer of antiviral antibodies restricts replication of Aleutian mink disease parvovirus in vivo. J. Virol. 63:9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astell, C. R., M. E. Gardinerd, and P. Tattersall. 1986. DNA sequence of the lymphotropic variant of minute virus of mice, MVM(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J. Virol. 57:656-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashir, T., R. Horlein, R. Rommelaere, and R. K. Willwand. 2000. Cyclin A activates the DNA polymerase delta-dependent elongation machinery in vitro: a parvovirus DNA replication model. Proc. Natl. Acad. Sci. USA 97:5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloom, M. E., S. M. Best, S. F. Hayes, R. D. Wells, J. B. Wolfinbarger, R. McKenna, and M. Agbandje-McKenna. 2001. Identification of Aleutian mink disease parvovirus capsid sequences mediating antibody-dependent enhancement of infection, virus neutralization, and inmune complex formation. J. Virol. 75:11116-11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom, M. E., D. A. Martin, L. L. Oie, M. E. Huhtanen, F. Costello, J. B. Wolfinbarger, S. F. Hayes, and M. Agbandje-Mckenna. 1997. Expression of Aleutian mink disease parvovirus capsid proteins in defined segments: localization of immunoreactive sites and neutralizing epitopes to specific regions. J. Virol. 71:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boissy, R., and C. R. Astell. 1985. An Escherichia coli recBC sbc BrecF host permits the deletion-resistant propagation of plasmid clones containing the 5′-terminal palindrome of minute virus of mice. Gene 35:179-185. [DOI] [PubMed] [Google Scholar]
- 8.Bonnard, G. D., E. K. Manders, D. A. Campbell, R. B. Herberman, and M. J. Collins. 1976. Immunosuppressive activity of a subline of the mouse EL-4 lymphoma. J. Exp. Med. 143:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosma, G. C., R. P. Custer, and M. J. Bosma. 1983. A severe combined immunodeficiency mutation in the mouse. Nature 301:527-530. [DOI] [PubMed] [Google Scholar]
- 10.Carman, W. F., A. R. Zanetti, P. Karayiannis, J. Waters, G. Manzillo, E. Tanzi, A. J. Zuckerman, and H. C. Thomas. 1990. Vaccine-indiced escape mutant of hepatitis B virus. Lancet 336:325-329. [DOI] [PubMed] [Google Scholar]
- 11.Chang, S.-F., J.-Y. Sgro, and C. R. Parrish. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 66:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chanock, R. M., J. E. Crowe, Jr., B. R. Murphy, and D. R. Burton. 1993. Human monoclonal antibody Fab fragments clones from combinatorial libraries: potential usefulness in prevention and/or treatment of major human viral diseases. Infect. Agents Dis. 2:118-131. [PubMed] [Google Scholar]
- 13.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciurea, A., L. Hunziker, P. Klenerman, H. Hengartner, and R. M. Zinkernagel. 2001. Impairment of CD4+ T cell responses during chronic virus infection prevents neutralizing antibody responses against virus escape mutants. J. Exp. Med. 193:297-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conley, A. J., J. A. Kessler II, L. J. Boots, P. M. McKenna, W. Schleif, E. A. Emini, G. E. Mark III, H. Katinger, E. K. Cobb, S. M. Lunceford, S. R. Rouse, and K. K. Murthy. 1996. The consequence of passive administration of an anti-human immunodeficiency virus type 1 neutralizing monoclonal antibody before challenge of chimpanzees with a primary virus isolate. J. Virol. 70:6751-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91-174. [DOI] [PubMed] [Google Scholar]
- 17.Cotmore, S. F., and P. Tattersall. 1996. Parvovirus DNA replication, p. 799-813. In M. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Domingo, E., and J. J. Holland. 1988. High error rates, population equilibrium, and evolution of RNA replication systems, p. 3-36. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics, vol III. CRC Press, Inc., Boca Raton, Fla.
- 19.Domingo, and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 20.Domingo, E., C. K. Biebricher, M. Eigen, and J. J. Holland. 2001. Quasispecies and RNA virus evolution: principles and consequences. Landes Bioscience, Georgetown, Tex.
- 21.Domingo, E., L. Menéndez-Arias, M. E. Quiñones-Mateu, A. Holguín, M. Gutiérrez-Rivas, M. A. Martínez, J. Quer, I. S. Novella, and J. J. Holland. 1997. Viral quasispecies and the problem of vaccine escape and drug-resistant mutants. Prog. Drug Res. 48:99-128. [DOI] [PubMed] [Google Scholar]
- 22.Drake, J. W. 1991. A constant rate of spontaneous mutations in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drake, J. W. 1993. Rates of spontaneous mutations among RNA viruses. Proc. Natl. Acad. Sci. USA 90:4171-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eigen, M., and C. K. Biebricher. 1988. Sequence space and quasispecies distribution, p. 211-245. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics, vol. III. CRC Press, Inc., Boca Raton, Fla.
- 26.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 27.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka (ed.). 2000. Principles of virology: molecular biology, pathogenesis, and control, p. 323-324. ASM Press, Washington, D.C.
- 28.Gardiner, E. M., and P. Tattersall. 1988. Evidence that developmentally regulated control of gene expression by a parvoviral allotropic determinant is particle mediated. J. Virol. 62:1713-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauduin, M. C., P. W. H. I. Parren, R. Weir, C. F. Barbas III, D. R. Burton, and R. A. Koup. 1997. Passive immunization with a human monoclonal antibody protects hu-PBL-SCID mice against challenge by primary isolates of HIV-1. Nat. Med. 3:1389-1393. [DOI] [PubMed] [Google Scholar]
- 30.Gigler, A., S. Dorsch, A. Hemauer, C. Williams, S. Kim, N. S. Young, S. Zoll-Pazner, H. Wolf, M. K. Gorny, and S. Modrow. 1999. Generation of neutralizing human monoclonal antibodies against parvovirus B19 proteins. J. Virol. 73:1974-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heegaard, E. D., and K. E. Brown. 2002. Human parvovirus B19. Clin. Microbiol. Rev. 15:485-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hezareh, M., A. J. Hessell, R. C. Jensen, J. G. J. van de Winkel, and P. W. H. I. Parren. 2001. Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J. Virol. 75:12161-12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland, J. J., J. C. de la Torre, and D. Steinhauer. 1992. RNA virus populations as quasispecies. Curr. Top. Microbiol. Immunol. 176:1-20. [DOI] [PubMed] [Google Scholar]
- 34.Holland, T. C., S. D. Marlin, M. Levine, and J. Glorioso. 1983. Antigenic variants of herpes simplex virus selected with glycoprotein-specific monoclonal antibodies. J. Virol. 45:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang, Y. T., B.-Y. Liu, D. M. Coen, and C. B. C. Hwang. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 71:7791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isnard, M., M. Granier, R. Frutos, B. Reynaud, and M. Peterschmitt. 1998. Quasispecies nature of three maize streak virus isolates obtained through different modes of selection from a population used to assess response to infection of maize cultivars. J. Gen. Virol. 79:3091-3099. [DOI] [PubMed] [Google Scholar]
- 37.Keller, M. A., and E. R. Stiehm. 2000. Passive immunity in prevention and treatment of infectious diseases. Clin. Microbiol. Rev. 13:602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurtzman, G. J., K. Ozawa, B. Cohen, G. Hanson, R. Oseas, and N. S. Young. 1987. Chronic bone marrow failure due to persistent B19 parvovirus infection. N. Engl. J. Med. 317:287-294. [DOI] [PubMed] [Google Scholar]
- 39.Langeveld, J. P. M., J. I. Casal, C. Vela, K. Dalsgaard, S. H. Smale, W. C. Puijk, and R. H. Meloen. 1993. B-cell epitopes of canine parvovirus: distribution on the primary structure and exposure on the viral surface. J. Virol. 67:765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lombardo, E., J. C. Ramírez, M. Agbandje-McKenna, and J. M. Almendral. 2000. A β-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74:3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mascola, J. R., M. Lewis, G. Stiegler, D. Harris, T. C. Van Cott, D. Hayes, M. K. Louder, C. R. Brown, C. V. Sapan, S. S. Frankel, Y. Lu, M. L. Robb, H. Katinger, and D. L. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 73:4009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mascola, J. R., G. Stiegler, T. C. Van Cott, H. Katinger, C. B. Carpenter, C. E. Hansos, H. Beary, D. Hayes, S. S. Frankel, D. L. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207-210. [DOI] [PubMed] [Google Scholar]
- 43.Mateu, M. G. 1995. Antibody recognition of picornaviruses and escape from neutralization: a structural view. Virus Res. 38:1-24. [DOI] [PubMed] [Google Scholar]
- 44.McMaster, G. K., P. Beard, H. D. Engers, and B. Hirt. 1981. Characterization of an immunosuppressive parvovirus related to the minute virus of mice. J. Virol. 38:317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2359. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 46.Palladino, G., K. Mozdzanowska, G. Washko, and W. Gerhard. 1995. Virus-neutralizing antibodies of immunoglobulin G (IgG) but not of IgM or IgA isotypes can cure influenza virus pneumonia in SCID mice. J. Virol. 69:2075-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parren, P. W. H. I., and D. R. Burton. 2001. The antiviral activity of antibodies in vitro and in vivo. Adv. Immunol. 77:195-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parren, P. W. H. I., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13:137-162. [PubMed] [Google Scholar]
- 49.Parrish, C. R., C. F. Aquadro, M. L. Strassheim, J. F. Evermann, J.-Y. Sgro, and H. O. Mohammed. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 65:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poignard, P., R. Sabbe, G. R. Picchio, M. Wang, R. J. Gulizia, H. Katinger, P. W. H. I. Parren, D. E. Mosier, and D. R. Burton. 1999. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 10:431-438. [DOI] [PubMed] [Google Scholar]
- 51.Sato, H., J. Hirata, M. Furukawa, N. Kuroda, H. Shiraki, I. Maeda, and K. Okochi. 1991. Identification of the region including the epitope for a monoclonal antibody which can neutralize human parvovirus B19. J. Virol. 65:1667-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawyer, L. A. 2000. Antibodies for the prevention and treatment of viral diseases. Antiviral Res. 47:57-77. [DOI] [PubMed] [Google Scholar]
- 53.Sayle, R. A., and E. J. Milner-White. 1995. RasMol: biomolecular graphics for all. Trends Biochem. Sci. 20:374-376. [DOI] [PubMed] [Google Scholar]
- 54.Segovia, J. C., A. Real, J. A. Bueren, and J. M. Almendral. 1991. In vitro myelosuppressive effects of the parvovirus minute virus of mice (MVMi) on hematopoietic stem and committed progenitor cells. Blood 77:980-988. [PubMed] [Google Scholar]
- 55.Segovia, J. C., J. M. Gallego, J. A. Bueren, and J. M. Almendral. 1999. Severe leukopenia and dysregulated erythropoiesis in SCID mice persistently infected with the parvovirus minute virus of mice. J. Virol. 73:1774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strassheim, M. L., A. Gruenberg, P. Veijalainen, J.-Y. Sgro, and C. R. Parrish. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198:175-184. [DOI] [PubMed] [Google Scholar]
- 57.Taboga, O., C. Tami, E. Carrillo, J. I. N(acute)uñez, A. Rodríguez, J. C. Saiz, E. Blanco, M. L. Valero, X. Roig, J. Camarero, D. Andreu, M. G. Mateu, E. Giralt, E. Domingo, F. Sobrino, and E. L. Palma. 1997. A large-scale evaluation of peptide vaccines against foot-and-mouth disease: lack of solid protection in cattle and isolation of escape mutants. J. Virol. 71:2606-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi, Y., C. Murai, S. Shibata, Y. Munakata, T. Ishii, K. Ishii, T. Saitoh, T. Sawai, K. Sugamura, and T. Sasaki. 1998. Human parvovirus B19 as a causative agent for rheumatoid arthritis. Proc. Natl. Acad. Sci. USA 95:8227-8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interaction of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46:944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Truyen, U., A. Gruemberg, S.-F. Chang, B. Obermaier, P. Veijalainen, and C. R. Parrish. 1995. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 69:4702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villarreal, L. P., and V. R. De Filippis. 2001. Virus evolution, p. 353-370. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 62.Wobus, C. E., B. Hugle-Dorr, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, C.-T. B., M. Levine, F. Homa, S. L. Highlander, and J. C. Glorioso. 1990. Characterizarion of the antigenic structure of herpes simplex virus type 1 glycoprotein C through DNA sequence analysis of monoclonal antibody-resistant mutants. J. Virol. 64:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan, Z., Y. Zhang, D. Duan, and J. F. Engelhardt. 2000. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. USA 97:6716-6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zwick, M. B., M. Wang, P. Poignard, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Neutralization synergy of human immunodeficiency virus type 1 primary isolates by cocktails of broadly neutralizing antibodies. J. Virol. 75:12198-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]