Abstract
Alkaline nuclease (AN) of the Autographa californica multiple-capsid nucleopolyhedrovirus (AcMNPV) (open reading frame 133) was expressed in recombinant baculovirus as a His6-tagged fusion and purified by sequential chromatography on Ni-NTA-agarose, DEAE-Toyopearl, and heparin-Sepharose. At all stages of purification, AcMNPV AN was found to copurify with a 44-kDa polypeptide which was identified as the baculovirus single-stranded DNA (ssDNA)-binding (SSB) protein, LEF-3. Sedimentation analysis in glycerol gradients of highly purified samples suggested that AN and LEF-3 are associated in a complex (designated *AN/L3), predominantly as heterodimers, although oligomeric forms containing both proteins were evident. In reactions with single- or double-stranded 62-mer oligonucleotides that were labeled with 32P at the 5′ or 3′ ends, *AN/L3 carried out exonucleolytic hydrolysis of both substrates exclusively in a 5′→3′ direction. Saturation of ssDNA with an excess of LEF-3 prior to the addition of *AN/L3 resulted in a marked decrease in the rate of ssDNA hydrolysis. This suggests that excess LEF-3 may protect ssDNA from digestion by a AN-LEF-3 complex, thus regulating its activity in infected cells. The association of baculovirus AN with the viral SSB LEF-3 and the 5′→3′ exonuclease activity of this complex suggests that AN and LEF-3 may participate in homologous recombination of the baculovirus genome in a manner similar to that of exonuclease (Redα) and DNA-binding protein (Redβ) of the Red-mediated homologous recombination system of bacteriophage λ.
Members of the Baculoviridae contain double-stranded (ds) circular DNA genomes of 100 to 180 kb, depending on the strain of virus (13). The family includes two genera, the granuloviruses (GVs) and the nucleopolyhedroviruses (NPVs). One NPV, Autographa californica multinucleocapsid NPV (AcMNPV), is widely used for the generation of recombinant viruses for the expression of foreign genes. Production of such expression vectors was based on efficient homologous recombination for the incorporation of the gene of interest into the virus genome (30). Although homologous recombination of the AcMNPV genome has been widely employed, its mechanism is unknown and the viral gene products involved in this process have not been identified. In a previous report, it was noted that AcMNPV and other GVs and NPVs encode proteins related to the alkaline nucleases (ANs) of the Herpesviridae (22). The AN encoded by AcMNPV (open reading frame 133) was found to possess divalent cation-dependent DNase activity, and it preferentially digested single-stranded DNA (ssDNA) and had an optimum at pH 9 to 10 (22).
Homologs of baculovirus ANs are widely distributed in eubacteria and archaea and are also found in eukaryotes (1). These enzymes participate in the repair of double-strand breaks and in homologous recombination and therefore play a vital role in the maintenance of genome integrity. The best-studied recombination system is from bacteriophage λ and is called the Red system (for a review, see references 19, 31, and 33). It includes λ exonuclease (Redα), which degrades double-stranded DNA (dsDNA) from the 5′ ends, producing 3′ overhangs which serve as intermediates in recombination (24). The λ exonuclease forms a toroidal-shaped trimer in solution, with a channel in the center which can accommodate dsDNA at one end but only ssDNA at the other (18). During recombination, λ exonuclease interacts with a ssDNA-binding (SSB) protein (Redβ), which promotes renaturation of complementary strands, thereby mediating strand annealing and strand exchange reactions (5, 23). Recently, comprehensive sequence analyses indicated that ANs of large dsDNA viruses of the Herpesviridae and Baculoviridae belong to a family of enzymes which includes λ exonuclease (1, 3). The herpes simplex virus type 1 AN is not essential for viral DNA synthesis (36). However, when the gene encoding AN is deleted, complex branched viral DNAs accumulate, suggesting that AN either cleaves or prevents the generation of these structures (10, 26). Because large concatemeric intermediates are likely produced in DNA replication of baculoviruses (21, 29), it was suggested that AN may be involved in the resolution of replication intermediates and maturation of the baculovirus genomes (22). The herpes simplex virus AN interacts with the major viral DNA-binding protein (mDBP) (34, 35), and recent evidence suggests that both these proteins may be directly involved in homologous recombination of the herpesvirus genome (R. Reuven, A. Staire, R. Myers, and S. Weller, submitted for publication). This observation suggested that the baculovirus homolog of herpesvirus AN and an unidentified DNA-binding protein may also participate in the baculovirus genome recombination.
In this report we describe further characterization of AcMNPV AN. We have focused mainly on two questions concerning the possible involvement of baculovirus AN in homologous recombination. The first question was to determine whether baculovirus AN interacts with a viral DNA-binding protein to produce a complex that is typically found in diverse recombination systems. In our original report (22), we identified a protein present in our preparations that was smaller than AN and that our evidence indicated might be a breakdown product of AN. In this report, we demonstrate that AcMNPV AN tightly associates with the SSB LEF-3 (in a complex that we call *AN/L3) and that LEF-3 was likely a component of the smaller band that we originally observed. In addition, we expected that if AN is involved in recombination, its enzymatic activity should conform to the generation of the 3′ overhangs as prerequisites for the strand exchange reaction. Therefore, the second question involved the determination of the polarity of hydrolysis catalyzed by the baculovirus AN/LEF-3 complex; we demonstrate that it digests DNA strands in a 5′→3′ direction. These properties are in agreement with presumed involvement of AN and LEF-3 in homologous recombination of the baculovirus genome.
MATERIALS AND METHODS
Chemicals and enzymes.
Radiolabeled nucleotides, [γ-32P]ATP, and [α-32P]dATP (cordycepin) were from Perkin-Elmer, terminal deoxynucleotidyl transferase and the 10-bp DNA ladder were from Gibco-BRL, and T4 polynucleotide kinase was from New England Biolabs.
Cells and recombinant baculovirus.
Spodoptera frugiperda 9 (Sf9) cells were cultured in Sf900II serum-free medium (Invitrogen) with penicillin G (50 units/ml), streptomycin (50 μg/ml; Whittaker Bioproducts), and fungizone (amphotericin B; 375 ng/ml) (Flow Laboratories) as previously described (12). Recombinant baculovirus vAcHISAN for overexpression of an AcMNPV His6-tagged AN (open reading frame 133) under the control of the polyhedrin promoter was previously described (22). It has a predicted molecular mass of 52.6 kDa (22).
DNA substrates.
The 62-mer oligonucleotide (TGGGTGAACCTGCAGGTGGGCAAAGATGTCCTAGCAATGTAATCGTCAAGCTTTATGCCGTT) was used as an ss substrate for analysis of exonuclease activity. It was labeled with 32P at the 5′ end by using T4 polynucleotide kinase and [γ-32P]ATP or at the 3′ end by using terminal deoxynucleotidyl transferase and [α-32P]dATP (cordycepin). To obtain a ds substrate, the 32P-labeled 62-mer was annealed to a complementary oligonucleotide (AACGGCATAAAGCTTGACGATTACATTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCCA). Escherichia coli DNA was labeled with [3H]thymidine (14).
Purification of *AN/L3.
Sf9 cells at a density of 1.5 × 106/ml in shaker flasks were infected with the recombinant baculovirus vAcHISAN at a multiplicity of infection of about 4 and incubated with shaking for 48 h at 28°C. *AN/L3 was purified routinely from 100-ml cultures of infected cells by sequential liquid chromatography on Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen), DEAE-Toyopearl 650 (TosoHaas), and heparin-Sepharose CL-6B (Amersham Pharmacia Biotech) columns. The infected cells were pelleted by centrifugation for 5 min at 500 × g and resuspended in 8 ml of lysis buffer containing 50 mM Tris-HCl (pH 8.5), 200 mM KCl, 1% Nonidet P-40, 5 mM 2-mercaptoethanol, and a set of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, 5 μg of leupeptin per ml, 5 μg of aprotinin per ml, 2 μg of E64 per ml). After extraction for 15 min at 4°C on a rotating shaker, the preparation was clarified by centrifugation at 30,000 × g for 30 min. The supernatant was loaded onto a Ni-NTA-agarose column (0.8 ml), equilibrated with buffer A (20 mM Tris-HCl [pH 8.5], 0.5 M KCl, 10% [vol/vol] glycerol, 5 mM 2-mercaptoethanol, 20 mM imidazole). The column was washed successively with 10 ml of buffer A, 2 ml of buffer B (20 mM Tris-HCl [pH 8.5], 1 M KCl, 10% [vol/vol] glycerol, 5 mM 2-mercaptoethanol), 1 ml of buffer A, and finally with 2 ml of buffer C (20 mM Tris-HCl [pH 8.5], 75 mM KCl, 10% [vol/vol] glycerol, 5 mM 2-mercaptoethanol) containing 20 mM imidazole.
Protein was eluted from the column with 4 ml of buffer C containing 150 mM imidazole. The sample was loaded at a rate of 4 ml per h onto a DEAE-Toyopearl column (0.5 by 2.5 cm) equilibrated with buffer D (10 mM Tris-HCl [pH 7.5], 20% [vol/vol] glycerol, 1 mM dithiothreitol [DTT], 1 mM EDTA) containing 75 mM KCl. The column was washed successively with 1 ml of buffer D containing 75 mM KCl and 2 ml of buffer D containing 110 mM KCl, and the protein was then eluted with 3 ml of the same buffer containing 200 mM KCl. The sample was loaded on a 0.5-ml column of heparin-Sepharose equilibrated with buffer D containing 200 mM KCl. The column was washed with several volumes of this buffer, and *AN/L3 was then eluted with sequential 1-ml portions of buffer D containing KCl in final concentrations of 0.25, 0.30, 0.35, 0.40, and 0.5 M. Proteins from each fraction were analyzed by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) followed by staining with Coomassie brilliant blue and Western blot analysis. The fractions collected at 0.30, 0.35, and 0.4 M KCl were combined or dialyzed separately against buffer E (0.1 M KCl, 10 mM Tris-HCl [pH 7.5], 50% [vol/vol] glycerol, 1 mM DTT, 0.2 mM EDTA) and stored at −20°C for periods of 1 to 2 months or at −80°C for long-term storage. Protein concentrations were determined by SDS-PAGE followed by optical densitometry of the gel stained with Coomassie brilliant blue. Bovine serum albumin (BSA) loaded in different amounts on separate lanes of the same gel was used for generation of the calibration curve.
AcMNPV LEF-3 was purified from Sf21 cells infected with wild-type (wt) AcMNPV by column chromatography by using ssDNA cellulose and DEAE-Toyopearl 650 resins as described previously for BmNPV LEF-3 (27).
PAGE and Western blotting.
SDS-10% PAGE (20) gels were either fixed and stained or electrophoretically transferred to PVDF-Plus transfer membranes (Micron Separations Inc) by using a Trans-blot SD semidry transfer cell (Bio-Rad) in accordance with the manufacturer's guidelines. Western blots were probed with a 1:2,000 dilution of rabbit polyclonal antiserum to AcMNPV AN (22) or with a 1:3,000 dilution of rabbit polyclonal antiserum to AcMNPV LEF-3 (7). The resulting membranes were washed, incubated with a 1:2,500 dilution of goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Promega), and developed by using ECL detection reagents (Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions.
Assay for *AN/L3 exonuclease activity with [3H]DNA.
The specific exonuclease activity of *AN/L3 was determined in reaction mixtures containing 1.6 μg of 3H-labeled E. coli ssDNA (120 × 103 cpm per μg), 25 mM Tris-HCl (pH 9.1), 5 mM MgCl2, 50 mM KCl, 100 μg of BSA per ml, and 1 mM DTT. Purified *AN/L3 (10 to 50 ng) was added in 5 μl of buffer E to a final volume of 20 μl. After incubation for 30 min at 37°C, reactions were terminated by the addition of 10 μl of tRNA (10 mg/ml) and 120 μl of 10% trichloroacetic acid. The samples were incubated for 10 min on ice and microcentrifuged for 5 min. The supernatant was mixed with 3.5 ml of liquid scintillation counting cocktail formula 989 (Packard, Inc.) and counted in a liquid scintillation counter.
Analysis of *AN/L3 digestion products in polyacrylamide gels.
Reaction mixtures (10 μl) contained 0.005 pmol of 32P-labeled (at the 5′ or 3′ end) ss or ds 62-mer oligonucleotide, 25 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 50 mM KCl, 100 μg of BSA per ml, 2 mM DTT, and various amounts of *AN/L3 added with 2.5 μl of buffer E. Reactions were carried out at 30°C for 2 min and were terminated by chilling on ice and adding 7 μl of stop solution (95% formamide, 20 mM EDTA, 0.05% each bromophenol blue and xylene cyanol). After heating for 3 min at 100°C, a 5-μl portion of each reaction mixture was loaded onto a 15% polyacrylamide-8 M urea slab gel (17 by 14.7 by 0.08 cm). Electrophoresis was performed in TBE (Tris-borate-EDTA) buffer (32) at 600 V for 1.5 to 2 h until the bromophenol blue migrated 4 cm above the bottom of the gel. Size standards, generated by 5′-end labeling a 10-bp DNA ladder (Gibco-BRL) with 32P, were electrophoresed in a parallel lane on the same gel. The gel was transferred onto a polymer support and exposed to X-ray film at −80°C with an intensifying screen.
Sedimentation in glycerol gradients.
Linear 15 to 30% glycerol gradients were prepared in buffer F (0.4 M KCl, 10 mM Tris-HCl [pH 7.5], 1 mM DTT, 1 mM EDTA). After dialysis against buffer F containing 5% glycerol, protein sample (100 μg in 90 μl) was layered over 4.8 ml of a gradient prepared in nitrocellulose tubes. Ovalbumin (45 kDa, 3.5S), BSA (66 kDa, 4.3S), aldolase (158 kDa, 7.7S), and catalase (220 kDa, 11.3S) (0.2 mg of each) were centrifuged in individual tubes as sedimentation standards. After centrifugation in the SW 50.1(Beckman) rotor at 46,000 rpm and 4°C for 19 h, the gradients were fractionated from the bottom with a peristaltic pump. The presence of *AN/L3 was monitored by SDS-10% PAGE, followed by staining with Coomassie brilliant blue.
RESULTS
Purification of AcMNPV AN.
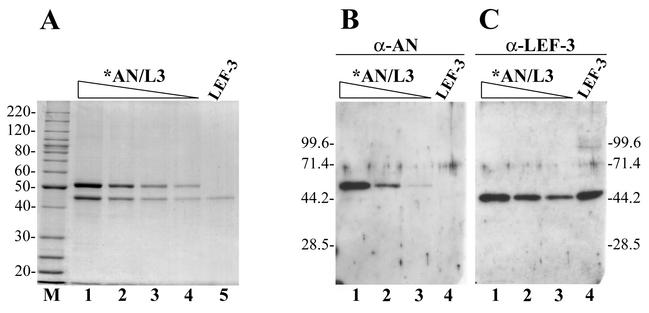
We used the recombinant baculovirus vAcHISAN previously described (22) for the overexpression of AcMNPV AN (open reading frame 133) as a His6-tagged fusion. Chromatography on a Ni-NTA column was the first step in the purification procedure and involved washing the bound enzyme with buffers containing KCl concentrations up to 1 M, followed by elution with 150 mM imidazole. Analysis of the protein fractions collected from this resin by SDS-10% PAGE followed by the Coomassie blue staining revealed the presence of two proteins with apparent molecular masses of 52 kDa and 44 kDa. The larger protein had a molecular mass expected for the His-tagged AN (22) and was immunoreactive with polyclonal antibodies against AcMNPV AN (see below). These data permitted us to confirm the identity of the 52-kDa protein as His-tagged AN. The 44-kDa protein did not react with anti-AN serum under Western blotting, and therefore its identity was unclear. The 44-kDa protein copurified with AN under subsequent chromatography on both anion exchange (DEAE-Toyopearl) and cation exchange (heparin-Sepharose) columns, indicating that they likely form a complex. The highly purified samples collected from the heparin-Sepharose contained only these two proteins (Fig. 1A, lanes 1 to 4). The electrophoretic mobility of the 44-kDa protein corresponded exactly to the mobility of baculovirus SSB protein LEF-3 (lane 5). LEF-3 was considered to be a good candidate to form a complex with AN, because a homolog of the baculovirus nuclease, AN of herpes simplex virus, interacts with an mDBP (34, 35). To determine if LEF-3 was the 44-kDa polypeptide associated with AN, we carried out Western blot analysis by using polyclonal antibodies to AN and LEF-3 (Fig. 1B and C). The antibodies to AN reacted specifically with the 52-kDa protein (Fig. 1B), whereas the antibodies to LEF-3 reacted specifically with the 44-kDa protein (Fig. 1C). These data permitted us to identify the 44-kDa protein in the purified samples of AN as the baculovirus SSB protein LEF-3. We have given the preparations of His6-tagged AN containing LEF-3 the designation *AN/L3. The total yield of purified *AN/L3 from 100-ml cultures of Sf9 cells infected with vAcHISAN was about 700 μg. Optical densitometry of several Coomassie blue-stained polyacrylamide gels with different preparations of *AN/L3 showed that the 52-kDa band was usually about 1.5-fold more intense than the 44-kDa band. The more intense staining of the 52-kDa protein may be partially due to its higher molecular mass. The composition of the protein may also affect the efficiency of its staining. Therefore, the staining pattern did not allow us to determine the exact molar ratio of AN to LEF-3 in *AN/L3, but it appears to be close to 1:1.
FIG. 1.
Analysis of *AN/L3 purified from infected Sf9 cells by SDS-10% PAGE, followed by staining with Coomassie brilliant blue (A) or by Western blotting with polyclonal antibodies against AN (B) or LEF-3 (C). (A) Lanes 1 to 4, purified *AN/L3: lane 1, 1.25 μg; lane 2, 0.5 μg; lane 3, 0.25 μg; lane 4, 0.13 μg. Lane 5, LEF-3 (0.1 μg); lane M, 10-kDa protein ladder. (B and C) Lanes 1 to 3, purified *AN/L3: lane 1, 130 ng; lane 2, 50 ng; lane 3, 20 ng. Lane 4, LEF-3 (25 ng). Panel C represents the same blot as the panel B after stripping and reprobing with antibodies against LEF-3. The molecular masses of protein markers (in kilodaltons) are shown on the sides of the panels.
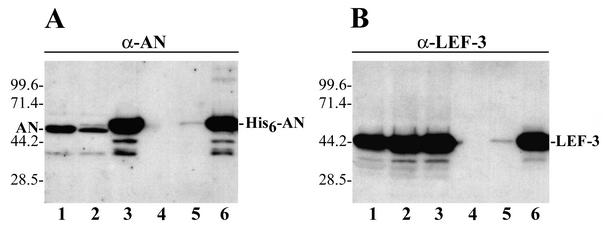
The appearance of LEF-3 in preparations of His6-tagged AN collected from the Ni-NTA resin was presumed to result from a specific interaction between the two proteins, because LEF-3 does not contain a His tag and therefore would not specifically interact with Ni-NTA. However, we could not exclude that LEF-3 might bind nonspecifically to the Ni-NTA resin. To determine factors crucial for LEF-3 binding to the Ni-NTA resin, we compared the LEF-3 yield after passing three different extracts from infected cells through identical Ni-NTA columns (Fig. 2). The first extract was prepared from cells infected with wt AcMNPV that lacks His6-tagged AN and was collected at 48 h postinfection (p.i.) (Fig. 2A, lane 1). The second extract was from cells collected early (18 h) after infection with recombinant virus vAcHISAN, and it contained a low amount of His6-tagged AN which is present above the main AN band (Fig. 2A, lane 2). The third extract was also from cells infected with recombinant virus vAcHISAN but was collected late (48 h p.i.) and contained a large amount of His6-tagged AN (Fig. 2A, lane 3). All three extracts contained a large amount of LEF-3 due to its normal high level of expression (Fig. 2B, lanes 1 to 3). Each extract was passed through a separate Ni-NTA column, the bound proteins were eluted with 150 mM imidazole, and the amount of AN and LEF-3 in the collected samples was determined by Western blot analysis by using antiserum to AN or LEF-3 (lanes 4 to 6). Both AN and LEF-3 were absent in the sample from cells infected with wt AcMNPV (lanes 4), and they were both present in very low amounts in the sample from the cells infected with recombinant virus vAcHISAN and collected at 18 h p.i. (lanes 5). In contrast, the amount of both proteins in the sample from the cells infected with the recombinant virus and collected at 48 h p.i. was high (lanes 6). These data indicated that LEF-3 does not bind the Ni-NTA resin in the absence of His6-tagged AN (Fig. 2B, lane 4). In addition, the amount of LEF-3 bound to the Ni-NTA resin appeared to be dependent on the amount of His6-tagged AN that was produced in infected cells and then bound to the Ni-NTA resin (compare lanes 4 to 6 in Fig. 2A and 2B). These data confirmed that LEF-3 was retained on the Ni-NTA column due to its association with His6-tagged AN and therefore suggest that His6-AN normally forms a complex with LEF-3 in infected cells.
FIG. 2.
Western blot analysis of the binding of AN and LEF-3 to a Ni-NTA resin. Equal portions (3 μl) from the crude extracts and the samples collected from Ni-NTA columns were analyzed by SDS-10% PAGE followed by Western blot analysis with polyclonal antibodies to AN (A) or LEF-3 (B). The extracts (5 ml) from 0.9 × 108 Sf9 cells infected (multiplicity of ∼4) with wt AcMNPV (48 h p.i.), vAcHISAN (18 h p.i.), or vAcHISAN (48 h p.i.) were analyzed in lanes 1, 2, and 3, respectively. The extracts were clarified by centrifugation and processed on identical Ni-NTA columns (0.6 ml), as described in Materials and Methods. The bound proteins were eluted with 3 ml of buffer C containing 150 mM imidazole, and the samples obtained from the cells infected with wt AcMNPV (48 h p.i.), vAcHISAN (18 h p.i.), or vAcHISAN (48 h p.i.) were analyzed in lanes 4, 5, and 6, respectively. The molecular masses of protein markers (in kilodaltons) are shown on the left sides of the panels.
Sedimentation analysis of *AN/L3 in glycerol gradients.
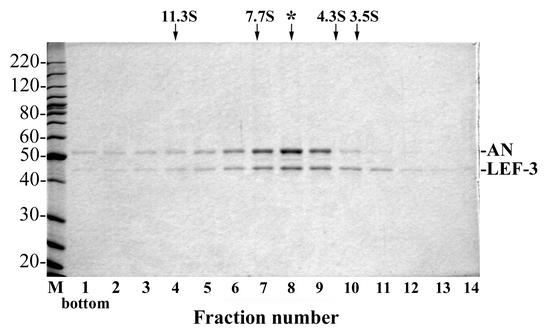
To obtain more information about the molecular structure of *AN/L3, we analyzed its sedimentation in a 15 to 30% glycerol gradient. To prevent nonspecific aggregation of protein during centrifugation, potassium chloride at a final concentration of 0.4 M was added to the solutions used for the gradients. Both components of *AN/L3, AN and LEF-3, showed overlapping sedimentation distributions with maxima close to the same position in the gradient (Fig. 3). In contrast to the sharp and unimodal distributions of the marker proteins catalase (11.3S), aldolase (7.7S), BSA (4.3S), and ovalbumin (3.5S) (data not shown), the *AN/L3 distribution was relatively broad, indicating an apparent heterogeneity in the molecular structure. The main fraction of *AN/L3 had a sedimentation coefficient of about 6.3S (the position marked by the asterisk in Fig. 3), which corresponds to a globular protein of approximately 110 kDa. Although this value is higher than would be predicted for an AN-LEF-3 heterodimer (96 kDa), the most straightforward interpretation is that the major fraction of *AN/L3 contains both proteins associated predominantly in a complex with a molar ratio of AN to LEF-3 equal to 1:1. The sedimentation pattern of *AN/L3 also revealed oligomeric forms which sedimented even faster than the catalase marker (11.3S, 220 kDa). When the high-molecular-weight material near the bottom of the gradient (fractions 1 to 3) (Fig. 3) was combined, concentrated, and recentrifuged in a 15 to 30% glycerol gradient, a major portion of AN again sedimented faster than the catalase (11.3S) marker. In contrast, the material from the peak fractions 7 to 9 (Fig. 3) did not reveal the presence of such oligomeric forms under recentrifugation (data not shown). Thus, the sedimentation pattern of *AN/L3 in the glycerol gradients suggests the capacity of *AN/L3 to form the oligomeric structures.
FIG. 3.
Sedimentation analysis of *AN/L3. Purified *AN/L3 (100 μg in 90 μl) was layered over 4.8 ml of a 15 to 30% glycerol gradient in buffer containing 0.4 M KCl, 10 mM Tris-HCl (pH 7.5), 1 mM DTT, and 1 mM EDTA and was centrifuged in an SW 50.1 rotor at 46,000 rpm at 4°C for 19 h. The gradient was fractionated from the bottom into 15 fractions, and 5-μl portions from fractions 1 to 14 were analyzed by SDS-10% PAGE followed by Coomassie blue staining. The positions of the standards centrifuged in separate tubes are shown by arrows: catalase (220 kDa, 11.3S), aldolase (158 kDa, 7.7S), BSA (66 kDa, 4.3S), and ovalbumin (45 kDa, 3.5S). The peak fraction of *AN/L3 is indicated by the asterisk.
Exonucleolytic digestion of oligonucleotides with *AN/L3.
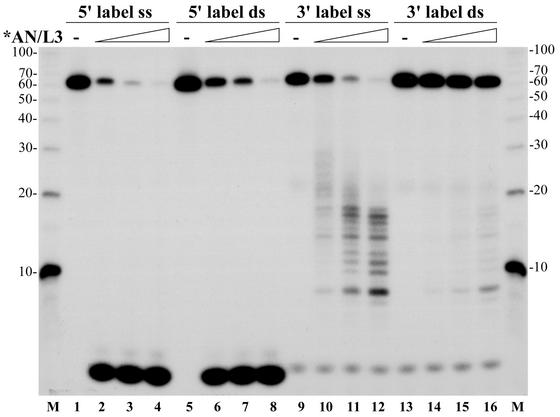
Purified *AN/L3 possesses a potent DNase activity. In reactions with 3H-labeled ssDNA from E. coli, 1 ng of *AN/L3 hydrolyzed approximately 1.2 ng of DNA during a 1-min incubation at 37°C. It was shown earlier that AcMNPV AN is capable of cleaving closed circular DNA, but its preferred substrate is ssDNA, and it displays a maximum activity at pH 9 to 10 in the presence of Mg2+ ions and in the absence of monovalent salts (22). In our study, we analyzed the polarity of the exonucleolytic hydrolysis catalyzed by *AN/L3 by using oligonucleotide substrates. A 62-mer oligonucleotide was labeled with 32P at the 5′ or 3′ end and used as the ssDNA substrate. To obtain the ds probes, the labeled 62-mers were annealed to the complementary oligonucleotides (see Materials and Methods). The digestion products obtained after incubation of four DNA substrates with increasing amounts of *AN/L3 were separated by electrophoresis in a 15% polyacrylamide-8 M urea gel (Fig. 4). After incubation for 2 min at 30°C, *AN/L3 was found to efficiently remove the 5′-terminal nucleotides from both ss and ds probes labeled at the 5′ end (lanes 1 to 8), while the 3′-terminal label was retained in oligonucleotides which gradually decreased in size down to 7 nucleotides (nt) (lanes 9 to 16). The absence of 5′-labeled digestion products with an intermediate mobility between deoxynucleoside monophosphates and the full-size 62-mers suggests that the DNA substrates were hydrolyzed exonucleolytically in a 5′→3′ direction.
FIG. 4.
Dose dependence of exonucleolytic digestion with *AN/L3 of ss and ds oligonucleotide substrates labeled at the 5′ or 3′ end. The assay was carried out in 10-μl reaction mixtures containing 0.005 pmol of the 62-mer oligonucleotide labeled with 32P at the 5′ or 3′ end and added in an ss or ds form obtained by annealing to the complementary oligonucleotide. Lanes 1 to 4, ss substrate labeled at the 5′ end; lanes 5 to 8, ds substrate labeled at the 5′ end; lanes 9 to 12, ss substrate labeled at the 3′ end; lanes 13 to 16, ds substrate labeled at the 3′ end. The mixtures also contained 25 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 50 mM KCl, 100 μg of BSA per ml, and 2 mM DTT. The purified *AN/L3 was added in the following amounts: 1 ng (lanes 2, 6, 10, and 14), 2.5 ng (lanes 3, 7, 11, and 15), and 6.3 ng (lanes 4, 8, 12, and 16). Lanes 1, 5, 9, and 13 represent control reactions lacking *AN/L3. After incubation for 2 min at 30°C, the reactions were terminated and the samples were processed for electrophoresis in a 15% polyacrylamide-8 M urea gel as described in Materials and Methods. Lanes M represent the 32P-labeled 10-nt ladder. The sizes of oligonucleotide markers (in nucleotides) are shown on the left and right sides.
Comparison of the full-size 62-mers retained in the reactions shown in Fig. 4 indicates that *AN/L3 utilized 5′-labeled ss 62-mers with a similar or slightly higher rate than 3′-labeled ss 62-mers (compare lanes 1 to 4 with lanes 9 to 13). In contrast, *AN/L3 utilized the 5′-labeled ds 62-mers at a much higher rate than the 3′-labeled ds 62-mers (compare lanes 5 to 8 with lanes 13 to 16). The more efficient hydrolysis of the 5′-labeled 62-mers by *AN/L3 may be caused by the presence of the 5′-terminal phosphates which can facilitate the interaction of 5′→3′ exonucleases with DNA (24). We have not analyzed a possible role of the 5′-terminal phosphates on *AN/L3 interaction with DNA in this report.
The effect of LEF-3 on DNA digestion with *AN/L3.
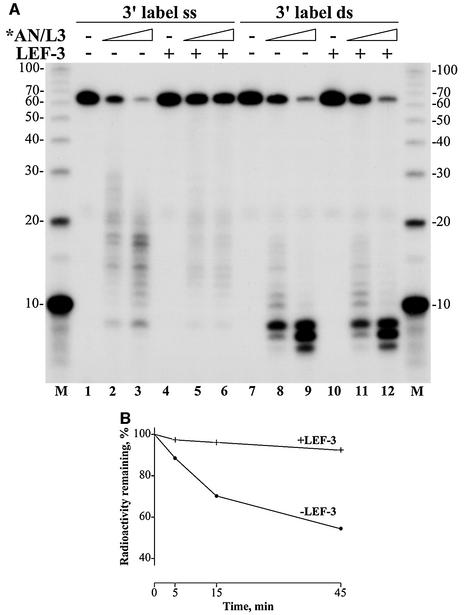
As shown above, *AN/L3 efficiently digested ss 62-mers labeled with 32P at either the 5′ or 3′ end (Fig. 4). These data suggest that ssDNA might be a preferred substrate for viral AN in infected cells. However, it is unlikely that a large portion of viral or host DNA in the infected cells is present in a ss form and free of bound SSB proteins, either the host cell RP-A or viral LEF-3. The baculovirus SSB protein LEF-3 appears to be one of the most abundant viral proteins in infected cells (11, 28). One of its putative roles is to bind and saturate the transient regions of ssDNA in the replicative fork, thus facilitating the action of replicative enzymes such as helicase and DNA polymerase. The abundance of LEF-3 in infected cells suggests that in vivo viral AN interacts with ssDNA that is presumably covered by this protein. To elucidate the effect of LEF-3 associated with ssDNA on hydrolysis catalyzed by *AN/L3, we performed the digestion reactions with 32P-labeled ss 62-mers bound to LEF-3 as a substrate for *AN/L3 (Fig. 5A). The amount of LEF-3 added to the reaction mixtures was calculated to be enough to saturate all ss 62-mers (11, 27). The saturation of ss 62-mers with LEF-3 resulted in a marked inhibition of hydrolysis by *AN/L3 (compare lanes 4 to 6 with lanes 1 to 3). In contrast, LEF-3 did not affect the hydrolysis of ds 62-mers (lanes 7 to 12). The relatively short size of the 62-mer oligonucleotide might restrict LEF-3 binding in a cooperative mode that is typical for interaction of SSB proteins with long polynucleotides. Therefore, we assayed the LEF-3 effect on digestion of highly polymeric ssDNA obtained by heat denaturation of 3H-labeled E. coli DNA (Fig. 5B). The LEF-3 added into the reaction mixture was enough to achieve approximately 50% saturation of ssDNA. The calculation is based on the assumption that at saturation, the LEF-3 monomer interacts approximately with 10 nt of ssDNA (11, 27). Even at this subsaturation level, LEF-3 caused a severalfold decrease in the rate of the ssDNA hydrolysis by *AN/L3 (Fig. 5B). These data suggest that LEF-3 may efficiently protect ssDNA from exonucleolytic digestion by viral AN, thus regulating its action on ssDNA in the infected cells.
FIG. 5.
Inhibition effect of LEF-3 on digestion of ssDNA with *AN/L3. (A) Electrophoresis in a 15% polyacrylamide-8 M urea gel of oligonucleotides digested with *AN/L3 in the reaction with added LEF-3. The assay was carried out in 10-μl reaction mixtures containing 0.005 pmol of 3′- 32P-labeled 62-mer oligonucleotide in an ss form (lanes 1 to 6) or in a ds form obtained by annealing to the complementary 62-mer (lanes 7 to 12). The mixtures also contained 25 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 50 mM KCl, 100 μg of BSA per ml, and 2 mM DTT. LEF-3 (15 ng) was added to each reaction shown in lanes 4 to 6 and 10 to 12. The reaction mixtures were assembled and preincubated for 20 min on ice. The purified *AN/L3 was added in the following amounts: 1 ng (lanes 2 and 5), 2.5 ng (lanes 3 and 6), 10 ng (lanes 8 and 11), and 25 ng (lanes 9 and 12). Lanes 1, 4, 7, and 10 represent control reactions lacking *AN/L3. After incubation for 2 min at 30°C, the reactions were terminated and the samples were processed for electrophoresis as described in Materials and Methods. Lanes M represent 32P-labeled 10-nt ladder. The sizes of oligonucleotide markers (in nucleotides) are shown on the left and right sides. (B) Digestion of 3H-labeled ssDNA of E. coli with *AN/L3 in the presence of LEF-3 and in its absence. Two 70-μl reaction mixtures, each containing 70 ng of 3H-labeled (5,600 cpm) ssDNA of E. coli, 25 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 50 mM KCl, 100 μg of BSA per ml, and 2 mM DTT, were preincubated for 15 min at room temperature in the presence of 0.45 μg of LEF-3 or in its absence. The mixtures were transferred at 30°C, and reaction was initiated by the addition of 2.5 ng of *AN/L3 to each sample. After incubation for 5, 15, and 45 min, 20-μl portions were taken from the samples and processed for determination of acid-soluble radioactivity as described in Materials and Methods. The acid-insoluble radioactivity remaining in the probes at each time point is shown as a percentage of the initial radioactivity.
DISCUSSION
ANs of baculoviruses belong to a family of enzymes typified by phage λ exonuclease (1, 3). λ exonuclease, also called Redα, was originally identified from a phage λ mutant defective in recombination as a component of the recombination defective (or red) system (reviewed in reference 31). This system also includes the SSB protein Red β, which promotes renaturation of complementary strands. It forms complexes with λ exonuclease and modulates its nucleolytic and recombination activities. The primary function of λ exonuclease is to generate ss overhangs which serve as intermediates in the repair and recombination of phage chromosomes (for a review, see references 19, 31, and 33). λ exonuclease forms a toroidal-shaped structure consisting of three identical subunits that form a ring surrounding the DNA substrate (18), and it is capable of hydrolyzing thousands of nucleotides in a 5′→3′ direction in a processive manner (4). Baculovirus ANs contain all three motifs conserved in exonucleases of the λ family (1, 3). The data presented in this paper confirm that AcMNPV AN hydrolyzes DNA in a 5′→3′ direction as was shown previously for λ exonuclease. Based on the structure of λ exonuclease (18), it is possible to predict that the conserved aspartate D153 in motif II and two glutamates Q221 and Q223 in motif III of AcMNPV AN coordinate a divalent cation (Mg2+) required for cleavage of a phosphodiester bond. The conserved lysine in motif III (K169) likely contacts a phosphate of the DNA backbone, which has been shown for the analogous lysine in EcoRV endonuclease (37). Despite possessing the conserved motifs of λ exonuclease, the baculovirus AN may differ from it in its enzymatic activities and spatial structure. Lambda exonuclease does not possess an endonuclease activity, and therefore it does not digest closed circular DNA (4). The rigid ring-shaped λ exonuclease trimer has its active sites located in a channel in the center (18). This structure permits λ exonuclease to react with the free ends of DNA but prevents endonucleolytic attack of the internal phosphodiester bonds in DNA. AcMNPV AN was shown to hydrolyze closed circular DNA (22), and we confirmed the presence of an endonuclease activity in highly purified preparations of AcMNPV *AN/L3 (data not shown). The ANs of the Herpesviridae also possess both 5′→3′ exonuclease and endonuclease activities (2, 6, 9, 15, 17). The endonuclease activity does not conform to a rigid toroidal structure of the enzyme, suggesting that an alternative structure might be possible. Trimerization of λ exonuclease is mediated mainly by two α-helical regions, one of which is conserved in members of the λ family, whereas the other is poorly conserved (1). Although these regions may support oligomerization of the proteins, the toroidal structure might not necessarily extend to the entire family. The sedimentation analysis of *AN/L3 (Fig. 3) suggests the capacity of AN to form oligomers but does not indicate the presence of stable trimers as a major species. The structures adopted by *AN/L3 in the course of exonucleolytic hydrolysis and under endonucleolytic action remain to be determined. It also remains to be shown how these structures incorporate LEF-3 subunits and how this association interferes with both enzymatic activities. The presence of a small bridging molecule linking LEF-3 to AN can also not be ruled out. The herpes simplex virus AN interacts with the mDBP (34, 35). mDBP mutants have been isolated that have altered AN activity at nonpermissive temperatures, thus indicating functional interaction between the two proteins (25). In the case of the nucleases stably or transiently associated with SSB proteins, it is possible to imagine other mechanisms that would ensure the processive exonucleolytic hydrolysis besides the physical encirclement of the DNA substrate. The SSB protein associated with the AN may stabilize its association with the DNA strand due to a high binding affinity of SSB for ssDNA. It may also facilitate melting dsDNA ahead of the nuclease that moves in a 5′→3′ direction. Interestingly, the 5′→3′ direction of the *AN/L3 movement along the DNA strand during hydrolysis coincides with the preferred direction for LEF-3 entry into duplex DNA during unwinding (27). Therefore, LEF-3 might play an accessory role in the reactions catalyzed by *AN/L3. In addition to its accessory role in the AN-LEF-3 complex, LEF-3 may regulate activity of this complex in infected cells by protecting ssDNA from digestion. Saturation or even subsaturation of ssDNA with LEF-3 markedly inhibited the hydrolysis of ssDNA by *AN/L3 (Fig. 5). The protection of ssDNA with the abundant LEF-3 might cause dsDNA to be the preferred substrate for AN in the infected cells, despite more efficient hydrolysis of naked ssDNA by *AN/L3 in vitro.
Although we did not detect the presence of an AN fraction that was free of LEF-3 in infected cells, we cannot rule out that it may be present. Therefore, the interaction of LEF-3 with AN may alter the nuclease activity of AN or modify its specificity. In addition, since we did not test the specificity of the inhibition of AN activity on ssDNA in the presence of LEF-3, it is possible that cellular SSBs could also influence the activity of the baculovirus nuclease under these conditions, and, conversely, LEF-3 might influence the activity of other nucleases.
LEF-3 appears to play a central role in metabolism of viral DNA in infected cells. It interacts with viral helicase (8) and mediates transfer of helicase to the nucleus (38), where it serves presumably as an accessory factor for both helicase and DNA polymerase. The data presented in this report suggest that LEF-3 also interacts with AN and plays accessory and regulatory roles in its activity. The presence of LEF-3 was not detected in the preparations of His-tagged AcMNPV AN that were characterized earlier (22). This was likely due to the use of the less-specific INDIA HisProbe-HRP reagent for Western blot analysis of isolated proteins. In the present report, we used another method for AN purification and specific polyclonal antibodies generated against AN and LEF-3 for Western blotting. That allowed us to identify unambiguously a 44-kDa polypeptide in the AN samples as LEF-3. Reconsidering the published data (22) and the data presented in this report, we have concluded that the general composition of the AN samples and their enzymatic properties are the same in both reports.
Although baculovirus AN has been shown to be related to a number of enzymes that belong to a diverse family which includes λ exonuclease (1, 3), LEF-3 does not demonstrate sequence relationships with other known SSBs. Three evolutionarily distinct superfamilies of single-strand annealing proteins have been described (16). One of these includes the Redβ protein of bacteriophage λ. Our computer analyses suggest that LEF-3 may be a member of a fourth superfamily of this type of protein (data not shown).
AcMNPV AN was originally predicted to be involved in the processing of replicative intermediates during maturation of viral genomes (22). The data obtained in this paper extend the possible roles of AN in the baculovirus replication cycle. The 5′→3′ exonuclease activity of viral AN and its association with viral SSB protein LEF-3 are in agreement with a putative central role of the AN-LEF-3 complex in both the repair and recombination of viral genomes. Experiments to determine a recombination potential of AcMNPV alkaline nuclease and LEF-3 are in progress.
Acknowledgments
We are indebted to Rik Myers and Sandy Weller for suggestions and advice relating to this research. We thank Scott Given for assistance with computer analysis.
This research was supported by grants from the NIH (GM9982536) to G.F.R. and from the Russian Foundation for Basic Research (00-04-49237) to V.S.M.
Footnotes
This is Technical Report no. 11932 from the Oregon State University Agricultural Experiment Station.
REFERENCES
- 1.Aravind, L., K. Makarova, and E. Koonin. 2000. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronstein, J. C., and P. C. Weber. 1996. Purification and characterization of herpes simplex virus type 1 alkaline exonuclease expressed in Escherichia coli. J. Virol. 70:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bujnicki, J., and L. Rychlewski. 2001. The herpesvirus alkaline exonuclease belongs to the restriction endonuclease PD-(D/E)XK superfamily: insight from molecular modeling and phylogenetic analysis. Virus Genes 22:219-230. [DOI] [PubMed] [Google Scholar]
- 4.Carter, D., and C. Radding. 1971. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J. Biol. Chem. 246:2502-2512. [PubMed] [Google Scholar]
- 5.Cassuto, E., T. Lash, K. Sriprakash, and C. Radding. 1971. Role of exonuclease and beta protein of phage lambda in genetic recombination. V. Recombination of lambda DNA in vitro. Proc. Natl. Acad. Sci. USA 68:1639-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Draper, K., G. Devi-Rao, R. Costa, E. Blair, R. Thompson, and E. Wagner. 1986. Characterization of the genes encoding herpes simplex virus type 1 and type 2 alkaline exonucleases and overlapping proteins. J. Virol. 57:1023-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, J. T., and G. F. Rohrmann. 1997. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J. Virol. 71:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, J. T., G. S. Rosenblatt, D. J. Leisy, and G. F. Rohrmann. 1999. Characterization of the interaction between the baculovirus ssDNA-binding protein (LEF-3) and putative helicase (P143). J. Gen. Virol. 80:493-500. [DOI] [PubMed] [Google Scholar]
- 9.Francke, B., H. Moss, M. Timbury, and J. Hay. 1978. Alkaline DNase activity in cells infected with a temperature-sensitive mutant of herpes simplex virus type 2. J. Virol. 26:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein, J. N., and S. K. Weller. 1998. In vitro processing of herpes simplex virus type 1 DNA replication intermediates by the viral alkaline nuclease, UL12. J. Virol. 72:8772-8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hang, X., W. Dong, and L. A. Guarino. 1995. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J. Virol. 69:3924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harwood, S. H., L. Li, P. S. Ho, A. K. Preston, and G. F. Rohrmann. 1998. AcMNPV late expression factor-5 interacts with itself and contains a zinc ribbon domain that is required for maximal late transcription activity and is homologous to elongation factor TFIIS. Virology 250:118-134. [DOI] [PubMed] [Google Scholar]
- 13.Hayakawa, T., G. F. Rohrmann, and Y. Hashimoto. 2000. Patterns of genome organization and content in lepidopteran baculoviruses. Virology 278:1-12. [DOI] [PubMed] [Google Scholar]
- 14.Hays, J. B., S. J. Martin, and K. Bhatia. 1985. Repair of nonreplicating UV-irradiated DNA: cooperative dark repair by Escherichia coli Uvr and Phr functions. J. Bacteriol. 161:602-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann, P., and Y. Cheng. 1979. DNase induced after infection of KB cells by herpes simplex virus type 1 or type 2. II. Characterization of an associated endonuclease activity. J. Virol. 32:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer, L., E. Koonin, and L. Aravind. 2002. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redβ, ERF and RAD52. BMC Genomics 3:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kehm, E., M. Goksu, S. Bayer, and C. W. Knopf. 1998. Herpes simplex virus type 1 DNase: functional analysis of the enzyme expressed by recombinant baculovirus. Intervirology 41:110-119. [DOI] [PubMed] [Google Scholar]
- 18.Kovall, R., and B. Matthews. 1997. Toroidal structure of lambda-exonuclease. Science 277:1824-1827. [DOI] [PubMed] [Google Scholar]
- 19.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Leisy, D. J., and G. F. Rohrmann. 1993. Characterization of the replication of plasmids containing hr sequences in baculovirus-infected Spodoptera frugiperda cells. Virology 196:722-730. [DOI] [PubMed] [Google Scholar]
- 22.Li, L., and G. F. Rohrmann. 2000. Characterization of a baculovirus alkaline nuclease. J. Virol. 74:6401-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Z., G. Karakousis, S. Chiu, G. Reddy, and C. Radding. 1998. The beta protein of phage lambda promotes strand exchange. J. Mol. Biol. 276:733-744. [DOI] [PubMed] [Google Scholar]
- 24.Little, J. 1967. An exonuclease induced by bacteriophage lambda. II. Nature of the enzymatic reaction. J. Biol. Chem. 242:679-686. [PubMed] [Google Scholar]
- 25.Littler, E., D. Purifoy, A. Minson, and K. Powell. 1983. Herpes simplex virus non-structural proteins. III. Function of the major DNA-binding protein. J. Gen. Virol. 64:683-695. [DOI] [PubMed] [Google Scholar]
- 26.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikhailov, V. 2000. Helix-destabilizing properties of the baculovirus single-stranded DNA-binding protein (LEF-3). Virology 270:180-189. [DOI] [PubMed] [Google Scholar]
- 28.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oppenheimer, D. I., and L. E. Volkman. 1997. Evidence for rolling circle replication of Autographa californica M nucleopolyhedrovirus genomic DNA. Arch. Virol. 142:2107-2113. [DOI] [PubMed] [Google Scholar]
- 30.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. W. H. Freeman & Co., New York, N.Y.
- 31.Poteete, A. 2001. What makes the bacteriophage lambda Red system useful for genetic engineering: molecular mechanism and biological function. FEMS Microbiol. Lett. 201:9-14. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Stahl, F. 1998. Recombination in phage lambda: one geneticist's historical perspective. Gene 223:95-102. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, M., M. Gao, D. Knipe, and K. Powell. 1992. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J. Virol. 66:1152-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaughan, P., L. Banks, D. Purifoy, and K. Powell. 1984. Interactions between herpes simplex virus DNA-binding proteins. J. Gen. Virol. 65:2033-2041. [DOI] [PubMed] [Google Scholar]
- 36.Weller, S., M. Seghatoleslami, L. Shao, D. Rowse, and E. P. Carmichael. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 71:2941-2952. [DOI] [PubMed] [Google Scholar]
- 37.Winkler, F., D. Banner, C. Oefner, D. Tsernoglou, R. Brown, S. Heathman, R. Bryan, P. Martin, K. Petratos, and K. Wilson. 1993. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 12:1781-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, Y., and E. B. Carstens. 1998. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology 247:32-40. [DOI] [PubMed] [Google Scholar]