Abstract
Polycomb and trithorax group genes maintain the appropriate repressed or activated state of homeotic gene expression throughout Drosophila melanogaster development. We have previously identified the batman gene as a Polycomb group candidate since its function is necessary for the repression of Sex combs reduced. However, our present genetic analysis indicates functions of batman in both activation and repression of homeotic genes. The 127-amino-acid Batman protein is almost reduced to a BTB/POZ domain, an evolutionary conserved protein-protein interaction domain found in a large protein family. We show that this domain is involved in the interaction between Batman and the DNA binding GAGA factor encoded by the Trithorax-like gene. The GAGA factor and Batman codistribute on polytene chromosomes, coimmunoprecipitate from nuclear embryonic and larval extracts, and interact in the yeast two-hybrid assay. Batman, together with the GAGA factor, binds to MHS-70, a 70-bp fragment of the bithoraxoid Polycomb response element. This binding, like that of the GAGA factor, requires the presence of d(GA)n sequences. Together, our results suggest that batman belongs to a subset of the Polycomb/trithorax group of genes that includes Trithorax-like, whose products are involved in both activation and repression of homeotic genes.
In Drosophila melanogaster, segmental identity along the anterior-posterior axis is specified by homeotic genes, whose expression is established early in development by a combination of maternal, pair-rule, and gap gene products (50, 59, 67, 70). Since these regulators are only transiently expressed during early embryogenesis, a second system of regulation takes over to maintain heritable activation or repression of homeotic gene expression later in development. The maintenance genes have been separated in two groups, the Polycomb group (PcG) of repressors (37) and the trithorax group (trxG) of activators (66). Loss-of-function mutations of PcG genes lead to posterior segmental transformations in embryos, as a result of the ectopic expression of homeotic genes anterior to their normal expression domain (20, 46, 68, 74, 82). In contrast, loss-of-function mutations of trxG genes lead to anterior transformations of abdominal segments (34), as a result of the loss of sustained expression of homeotic genes (9, 10). Synergistic interactions are observed between mutant alleles of PcG genes, as well as between mutant alleles of trxG genes. In contrast, suppressive interactions between mutations of PcG and trxG genes are usually encountered, consistent with their opposite regulatory functions on homeotic gene expression (38).
PcG proteins regulate homeotic genes at the transcriptional level (74). Characterization of homeotic gene regulatory sequences led to the identification of PcG response elements (PREs) that are required to maintain a spatially restricted pattern of homeotic gene expression in a PcG-dependent manner (49, 69). Consistent with this, the products of PcG genes were characterized as chromosomal proteins associated to specific regions of larval salivary polytene chromosomes (10, 44, 85), including already-defined PREs. Although many of these specific regions are common target sites for all the PcG proteins (85), some are specific to particular PcG proteins (43, 60, 71). These results led to the proposition that PcG proteins are involved in complexes of overlapping but distinct composition, depending on their targets. Evidence for the participation of PcG proteins to macromolecular complexes came from in vitro and Saccharomyces cerevisiae interaction assays, as well as from coimmunoprecipitation studies (35, 39, 40, 55, 77). Polycomb (PC), Extra sex comb (ESC), Enhancer of zeste (EZ), Pleiohomeotic (PHO), and Polyhomeotic (PH) were recently shown to interact transiently during early embryonic development (58). In late embryos, these factors are found in two separate complexes, PRC1 (including PH, PC, PSC, and Sex comb on midleg [SCM] [65]) and another complex involving ESC, EZ (35), and PHO (58, 78).
PcG proteins may exert their repressive effect at different levels of transcriptional regulation, including interaction with the transcription machinery, histone modifications, and nucleosomal organization, as well as higher order of chromatin structure. The presence in PC of a conserved domain called the chromodomain, also found in HP1, a heterochromatin-associated protein, led to the proposal that a higher-order chromatin structure is induced by PcG multimeric complexes (53). This view is supported by the phenotypic similarities between the silencing of a miniwhite transgene by heterochromatin and that of PRE-miniwhite constructs by PcG proteins (25), the local spreading of PcG proteins over the PRE (75), and the reduced accessibility of a locus when repressed by PcG genes (7, 26, 45). Recent data also point towards a localized effect on nucleosomal organization at the level of the PRE: PC interacts in vitro with nucleosomes (11), PcG repression correlates with a modification of histone acetylation (16), and both the ESC/EZ/PHO and PRC1 complexes contain the histone deacetylase RPD3 (58, 63, 78). In addition, PcG proteins interact physically with general transcription factors and may thereby directly inhibit transcription at bound promoters (12, 63).
Many trxG genes were recovered as suppressors of Pc loss-of-function mutations or of gain-of-function mutations of the homeotic Antennapedia gene (38). The trxG proteins, as is the case for PcG proteins, are involved in several separable complexes. One of them, the Brahma complex, contains homologues of the SWI/SNF chromatin remodeling complex (19, 76, 81) necessary for maintaining an active state of transcription in mammals and yeast. At least two other complexes are found in Drosophila, one including Trithorax (TRX) and ASH1 (62) and another containing ASH2 (52).
The functional antagonism between genes of the PcG and trxG suggests that the remodeling properties of trxG proteins must somehow compete with the properties of PcG proteins at homeotic loci. Molecular characterization of TRX showed that it contains a SET protein-protein interaction domain that is shared by the PcG protein EZ (36), which suggests that antagonism of PcG and trxG factors may result from opposite effects on common partners. Furthermore, the binding of PRC1 on a DNA template prevents SWI/SNF ATP-dependent chromatin remodeling (65). Symmetrically, TRX is necessary to counteract the silencing effect of PcG proteins on a PRE (57). In addition, PHO was shown to bind PcG proteins from the PRC1 complex (PC and PH) as well as trxG proteins from the Brahma complex through independent domains (48). The maintenance of the expression status of their targets by PcG and trxG proteins may thus in part act at the same level, that is, the control of access to the transcription machinery by remodeling of the nucleosomal organization of their target sequences. Along the same line, the GAGA factor, encoded by the trxG gene Trithorax-like (Trl) (23), binds to both active and inactive PREs (75) and interacts with the NURF remodeling complex (80); with TRX (57); with Sap18 (21), a component of the Sin3 corepressor complex; and with PC, a component of PRC1 (58). In addition, a close association exists between PRE and trxG response elements in homeotic genes (79). Presumably, these composite elements would switch between activation and repression by PcG or trxG proteins, depending on the preestablished transcriptional state. The vision of PcG and trxG proteins as completely antagonistic factors may not reflect all their regulatory properties as inferred from genetic data (41; reviewed in reference 13). Therefore, they may collectively be addressed as PcG/trxG proteins in order to include all the potential functions of each member.
Elucidation of the precise mechanisms by which PcG/trxG proteins regulate their diverse targets will require the identification of all the PcG/trxG members and the functional characterization of the interaction network between these proteins. With this aim, we characterized batman (ban), previously identified as an enhancer of ph (24). We showed that ban mutant flies display phenotypes that indicate a function of ban in both activation and repression of homeotic genes, identifying ban as a PcG/trxG member. We found that the Batman protein contains a BTB/POZ domain, and associates to polytene chromosomes at several hundreds sites, including many PcG/trxG sites. We identified a Batman partner as the BTB/POZ-containing GAGA factor, a trxG protein (TRL/GAF), that also has a dual, activating and repressing function in the regulation of homeotic genes.
MATERIALS AND METHODS
Fly strains and original cDNAs.
Flies were grown on standard corn-agar medium at 25°C except when otherwise mentioned. All the markers and stocks used in this study are referred to in Flybase (http://flybase.bio.indiana.edu/). Most of the stocks were provided by the Bloomington or Umea stock centers, and other providers are referred to below: ph410w, Pc1, and Pc3 were provided by J.-M. Dura, and Pc16 was provided by R. Paro. The ubiquitous driver Da:Gal4 refers to GAL4daG32 (83). The PH-FLAG line is a kind gift of W. Bender. hsp83:GAGA519 and the hsp83:GAGA581 lines were kindly provided by A. Greenberg.
LD14505, LD08847, GM10385, LD08692, LD15257, LD06695, LD08823, and LD16436 cDNAs were provided to us by the Berkeley Drosophila Genome Project (BDGP). Sequencing of their 3′ ends and alignment to the genomic sequence showed that seven cDNAs correspond to a false initiation of reverse transcription due to an internal stretch of poly(A) and that the LD16436 cDNA is complete for its 3′ end. At the 5′ end, LD16436 is not complete, but LD14505 starts where the predicted ban gene starts. LD14505served as a template for the synthesis of riboprobes to be used for in situ hybridization.
batman alleles.
In the BDGP P[LacW] collection, two insertions are described as not complementing l(2)k02512: l(2)k11212 and l(2)k07907 (73). By sequencing the flanking regions of the P[LacW] insertion sites, we showed that the transposon is inserted in the same orientation in the three cases, with l(2)k11212 being located at the same position as l(2)k02512 and l(2)k07907 located 133 bp upstream. Viable revertants of banl(2)k02512 and banl(2)k11212 were recovered after excision of P[LacW], indicating that the lethality of these alleles is due to the P-element insertion. Df(2R)311a, a lethal P[MTW] insertion associated with a 14-kb deletion (24) also affects the ban gene (Fig. 1A). banl(2)k02512, banl(2)k11212, banl(2)k07907, and banDf(2R)311a were out-crossed for 10 generations over a w1118 stock before complementation analysis. The original l(2)k07907 stock contained an additional lethal mutation that could be separated from the P-element insertion. Combinations of Df(2R)311a with any of the three P[LacW] insertions are viable, showing rough eyes in rare cases, thereby indicating that Df(2R)311a is a weak ban hypomorphic allele. New alleles of ban were generated by excision of the P-element from the banl(2)k02512, banl(2)k11212, and banDf(2R)311a insertions using Delta2-3 as a source of transposase (61) and were recovered based on their white eye phenotype and lethality over the parental insertion. The Polycomblike (Pcl) gene is located in 55B5-7, 18 kb away from ban. Therefore, all new ban alleles were assayed for complementation with PclX21 or PclE90. Complementation of lethality was observed in all cases. Furthermore, Southern blot analysis did not reveal any alteration of the Pcl genomic region, showing that Pcl is not affected in any of the chromosomes carrying the ban alleles used in this study.
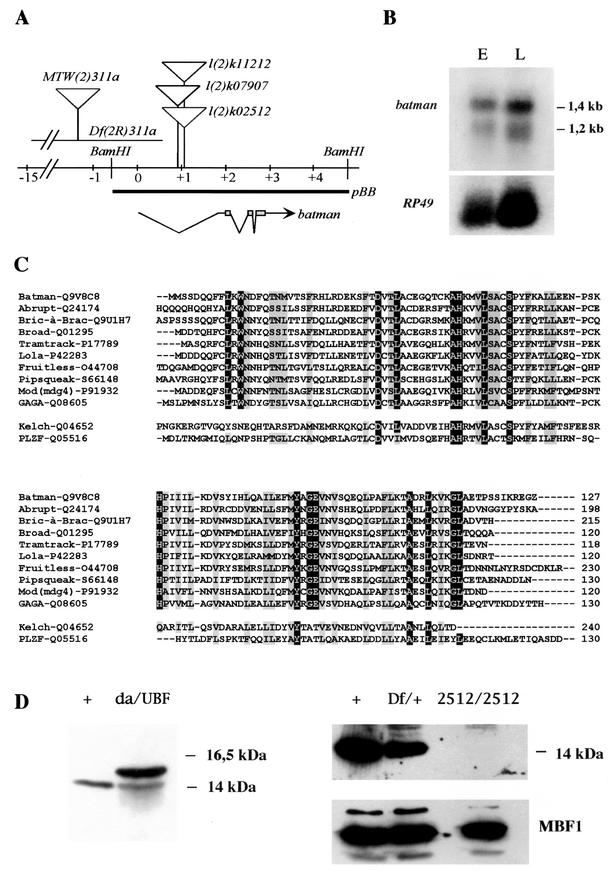
FIG. 1.
The batman transcription unit. (A) Molecular map of the ban locus. The ban gene is transcribed from distal to proximal relative to the centromere. The two BamHI sites delimit the ban transcription unit as inferred from rescue assays using the pBB fragment (5,405 bp). Three lethal noncomplementing P[LacW] insertions are positioned in the first intron of ban. Df(2R)311a (24) uncovers the 5′ region of ban up to the first intron. This deficiency is the only one available that affects ban without affecting the neighboring Pcl gene, located 18 kb downstream of ban. The sequence corresponding to ORF127 is boxed on the ban transcript. (B) Northern blot analysis of embryonic (E) and third-instar larva (L) RNA probed with ban cDNA (top) and RP49 cDNA (bottom) as a control. (C) Multiple sequence alignment of D. melanogaster BTB/POZ proteins ordered according to decreasing similarity to BAN as calculated with the BLASTP algorithm. The alignment was generated using CLUSTALW. The cytoplasmic D. melanogaster Kelch protein and the human PLZF protein, for which a crystal structure has been determined (1), are set apart. The N-terminal nuclear BTB signature, characteristic of the Tramtrack subfamily and not conserved in Kelch (4, 87), is boxed. Conserved amino acids are boxed in black, and similarities are boxed in gray. The coordinates on the right refer to the position of the last amino acid included in the alignment from the sequences listed in GenBank. (D) Western blot analysis of larval proteins using the batC11 antibody. Extracts from third-instar larvae of the appropriate genotype were separated on sodium dodecyl sulfate-18% polyacrylamide gels. +/, w1118; da/UBF:da:Gal4/UBF larvae were obtained by crossing da:Gal4 homozygous flies to UBF homozygotes; Df/+, Df(2R)PC4/+ larvae; 2512/2512, banl(2)k02512/banl(2)k02512 escaper third-instar larvae were selected among the progeny of banl(2)k02512/CyO-GFP heterozygous flies based on the absence of GFP expression. Anti-MBF1 antibody (bottom panel) was used as a control for gel loading.
Genomic and tagged constructs.
The ban gene is contained in the DS08860 genomic P1 clone (BDGP). The DS08860 BamHI fragment overlapping the ban gene (Fig. 1A) was inserted into PCaSpeR in opposite orientation to the miniwhite gene (56). The resulting construct was named pBB. Standard transformation protocols were used to transform a w1118 strain. Eight lines were obtained and are referred to in the text as pBB with a superscript number indicating the line.
In order to generate the UAS:ban-enhanced green fluorescent protein (EGFP) (UBG) transgene, a linker was inserted into the LD14505 cDNA, thereby replacing the stop codon of the ban open reading frame (ORF) with an EcoRI site. This modified cDNA was then inserted into a pUAS:EGFP construct, previously generated by insertion of the EGFP cDNA (Clontech, Palo Alto, Calif.) into pUAST (8). The UAS:BAN-FLAG construct (UBF) was generated by PCR insertion of the FLAG epitope DYKDDDDK immediately upstream of the stop codon in the ban ORF. The modified cDNA was then inserted in pUAST. Twenty lines were obtained for the UBG construct, and seven were obtained for the UBF construct. Several lines of each construct were assayed in each experiment.
Genetic interactions between ban and ph, Pc, Trl, or Ubx. (i) esc phenotype.
For quantifying the number of sex comb teeth, males of the appropriate genotype were mated to homozygous ph410w or Pc16Ki/TM6 females. Crosses were performed under uncrowded conditions at 25°C. Males were sorted according to their genotype. An average of 100 metathoracic legs were analyzed. A categorical χ2 test was performed on each set of data (degrees of liberty = 2). The null hypothesis was that the probability of having 0 to 2, 3 to 5, or more than 6 sex comb teeth is the same for the two compared genotypes. Each genotype was given a score that corresponds to the average number of sex comb teeth by metathoracic leg.
(ii) Ubx phenotype.
Flies carrying the Ubx130 allele from the TM2 balancer chromosome show a weak transformation of halter to wing. When assaying for interaction between ban, Trl, and Ubx, the expressivity was quantified as basal when halters were similar to those observed in Ubx130/+ flies, weak when halters were swollen, and strong when wing and/or notal structures were present on the metathoracic segment. At 25°C, Trl13c/TM2 Ubx130 flies often (one of four) had swollen halters. The expressivity of the Ubx phenotype in banl(2)k02512/+; Trl13c/TM2 Ubx130 flies when compared to that of Trl13c/TM2 Ubx130 flies was measured in siblings from the progeny of banl(2)k02512/+; Trl13c/TM3Sb females crossed to Dr/Ubx130 males.
Rescue assays. (i) Rescue of lethality.
Recombinant banl(2)k02512 pBB10/CyO flies carrying an insert of the ban genomic construct were crossed to their siblings. Homozygous banl(2)k02512 pBB10/banl(2)k02512 pBB10 flies were recovered from this cross in a proportion relative to heterozygous banl(2)k02512 pBB10/CyO flies, which is indicative of a rescue of banl(2)k02512 lethality. A recombinant UBG da:Gal4 chromosome was selected and allowed us to obtain banl(2)k02512/banl(2)k02512; UBG da:Gal4/+ rescued flies. Both banl(2)k02512 pBB10/banl(2)k02512 pBB10 and banl(2)k02512/banl(2)k02512; UBG da:Gal4/TM3 constitute fertile stocks.
(ii) Rescue of the E(ph) phenotype.
ph410 w/ph410 w; banl(2)k02512/CyO females were crossed to pBB10/+ males. ph410 w/Y; banl(2)k02512/pBB10 males had an average score of 2.4 sex comb teeth per metathoracic leg, whereas ph410w/Y; banl(2)k02512/+ males had a score of 5.8 sex comb teeth per metathoracic leg and ph410/Y; +/CyO males had a score of 2.6 sex comb teeth per metathoracic leg in the same cross (Table 1, lines 3, 2, and 1, respectively). Similar results were obtained with the pBB22 insert for rescue of lethality and E(ph) phenotype of banl(2)k02512 (Table 1, lines 7, 6, and 5 and data not shown). ph410w/ph410w; banl(2)k02512/CyO; da:Gal4/da:Gal4 females were crossed to UBG/+ males. Presence of the da:Gal4 UBG combination was tested by UV fluorescence examination of the progeny of this cross. ph410w/Y; banl(2)k02512/UBG; da:Gal4/+ males had the same score as that of ph410w/Y; CyO/+; da:Gal4/+ males for the five UBG lines tested (Table 1, lines 11 and 9, respectively, and data not shown). Dependency on the da:Gal4 driver for rescue was shown for the UBG47 line (Table 1, lines 18 and 19). Similar results were obtained with the UBF construct (Table 1 and data not shown).
TABLE 1.
Effect of ban transgenes on ph410 ESC phenotypea
| Line no. | Cross | Genotype | Mean no. of sex comb teeth | Legs analyzed (n)b | Distribution of legsc
|
||
|---|---|---|---|---|---|---|---|
| 0-2 | 3-5 | ≥6 | |||||
| 1 | ph410w/ph410w; banl(2)k02512/CyO × pBB10/+ | ph410w/Y; CyO/+ | 2.6 | 112 | 62 | 42 | 8 |
| 2 | ph410w/Y; pBB10/CyO | 1.3 | 111 | 92 | 16 | 3 | |
| 3 | ph410w/Y;banl(2)k02512/+ | 5.8 | 110 | 5 | 43 | 62 | |
| 4 | ph410w/Y;banl(2)k02512/pBB10 | 2.4 | 106 | 54 | 49 | 3 | |
| 5 | ph410w/ph410w; banl(2)k02512/+ × pBB22/CyO | ph410w/Y; CyO/+ | 3.1 | 110 | 47 | 53 | 10 |
| 6 | ph410w/Y; pBB22/+ | 2.0 | 109 | 74 | 32 | 3 | |
| 7 | ph410w/Y;banl(2)k02512/CyO | 5.8 | 115 | 9 | 39 | 67 | |
| 8 | ph410w/Y;banl(2)k02512/pBB22 | 3.8 | 109 | 34 | 51 | 24 | |
| 9 | ph410w/ph410w; banl(2)k02512/CyO; da:Gal4/da:Gal4 × UBG47/+ | ph410w/Y; CyO/+; da:Gal4/+ | 3.6 | 55 | 13 | 33 | 9 |
| 10 | ph410w/Y; UBG47/CyO; da:Gal4/+ | 1.7 | 80 | 56 | 23 | 1 | |
| 11 | ph410w/Y; banl(2)k02512/+; da:Gal4/+ | 7.7 | 56 | 0 | 4 | 52 | |
| 12 | ph410w/Y; banl(2)k02512/UBG47; da:Gal4/+ | 2.5 | 76 | 40 | 33 | 3 | |
| 13 | ph410w/ph410w; banl(2)k02512/+; da:Gal4/da:Gal4 × UBFc/CyO | ph410w/Y; CyO/+; da:Gal4/+ | 3.4 | 119 | 39 | 57 | 23 |
| 14 | ph410w/Y; UBFc/+; da:Gal4/+ | 1.2 | 118 | 104 | 14 | 0 | |
| 15 | ph410w/Y; banl(2)k02512/CyO; da:Gal4/+ | 6.0 | 80 | 1 | 33 | 46 | |
| 16 | ph410w/Y; banl(2)k02512/UBFc; da:Gal4/+ | 2.1 | 117 | 81 | 32 | 4 | |
| 17 | ph410w/ph410w; banl(2)k02512/CyO; da:Gal4/+ × UBG47/UBG47 | ph410w/Y; UBG47/CyO | 3.7 | 50 | 13 | 27 | 10 |
| 18 | ph410w/Y; UBG47/CyO: da:Gal4/+ | 1.4 | 34 | 27 | 6 | 1 | |
| 19 | ph410w/Y; banl(2)k02512/UBG47 | 6.9 | 52 | 1 | 13 | 38 | |
| 20 | ph410w/Y; banl(2)k02512/UBG47; da:Gal4/+ | 2.1 | 39 | 29 | 6 | 4 | |
| 21 | ph410w/ph410w; da:Gal4/+ × UBFc/UBFc | ph410w/Y; UBFc/+ | 3.0 | 118 | 53 | 47 | 18 |
| 22 | ph410w/Y; UBFc/+: da:Gal4/+ | 0.8 | 119 | 112 | 6 | 1 | |
The esc phenotype was scored as the average number of sex comb teeth on the metathoracic legs of the males. Sibling males of the appropriate genotype were compared. Males were sorted according to their CyO phenotype, their eye color, and, when necessary, the expression of GFP.
n, number of legs analyzed for each genotype.
The distributions of legs in the three classes (0 to 2, 3 to 5, or ≥6 sex comb teeth per leg) were compared for each pair of genotypes analyzed using the chi-square test (P < 0.001).
(iii) Rescue of the lethal phenotype of banl(2)k02512/+; Trl13c/Trl13c flies.
Both the hsp83:GAGA519 and the hsp83:GAGA581 transgenes were previously shown to rescue the lethality of Trl13c/TrlR67 mutant combinations at 22°C (29). In control experiments at 22°C, the maternal and zygotic presence of the hsp83:GAGA519 transgene allowed the recovery of up to 97% of the Trl13c/Trl13c flies, whereas 16% of their siblings that only received the maternal contribution of thee transgenes survived. When the hsp83:GAGA581 was present both maternally and zygotically, 52% of the Trl13c/Trl13c flies survived, whereas this transgene did not seem to affect the number of Trl13c escapers when it was only maternally provided (12%). In addition these flies (hsp83:GAGA581/+; Trl13c/Trl13c) displayed rough eyes that are not observed when the similar experiment is done using hsp83:GAGA519. Thus, in this Trl13c context, the hsp83:GAGA519 transgenes appear more efficient than the hsp83:GAGA581 to rescue the maternal haploinsufficiency and the zygotic requirement of Trl. Females of the hsp83:GAGA519/CyO; Trl13c/TM6Tb genotype were crossed to males of the (banl(2)k02512; Trl13c)/T(2,3) apXa genotype at 22°C. The frequency of the hsp83:GAGA519/banl(2)k02512; Trl13c/Trl13c escapers was calculated based on expected Mendelian ratios. Control crosses were run in the same experimental series in order to compare the lethality of Trl13c homozygotes and that of banl(2)k02512/+; Trl13c/TM6Tb to that obtained in the presence of the transgenes.
Antibodies, Western blotting, and immunostaining.
A BLAST analysis of the BAN sequence indicates that the last 10 C-terminal residues, which are not included in the BTB/POZ domain, are specific for the BAN protein. A peptide corresponding to the last 11 amino acids (aa) was synthesized (Eurogentec, Liege, Belgium) and injected in rabbits using standard immunization procedures. Preimmune serum was used for the control of the specificity of the BAN antiserum (11-residue, C-terminal-most peptide [batC11]). Western blot analysis was performed using the crude serum at a 10−4 dilution in phosphate-buffered saline-Tween 20 (0.1%)-nonfat dry milk (5%) and developed with enhanced chemiluminescence using the superSignal West Pico chemiluminescent substrate (Pierce).
FLAG-tagged BAN protein was revealed in Western blots using monoclonal anti-FLAG M2 antibody (1/1,000) (Stratagene, La Jolla, Calif.). The Drosophila multiprotein bridging factor 1 (MBF1) protein was revealed using rabbit polyclonal anti-MBF1 antibody (5 × 10−4) kindly provided by Marek Jindra. MBF1 is ubiquitously expressed at a constant level throughout development (M. Jindra, personal communication). Affinity-purified rabbit anti-TRL/GAF polyclonal antibody was kindly provided by Peter Becker. Rabbit anti-PH polyclonal antibody was a kind gift of Jean Maurice Dura.
Immunostaining of polytene chromosomes was performed according to the method of Giacomo Cavalli (http://www.igh.cnrs.fr/equip/cavalli/link.labgoodies.html). batC11 was used at a 1/100 dilution, anti-FLAG M2 was used at a 1/40 dilution, anti-TRL/GAF was used at a 1/40 dilution, and anti-PH was used at a 1/40 dilution. Secondary antibodies—anti-rabbit (Alexafluor 594; catalog no. A-11037; Molecular Probes) and anti-mouse (Alexafluor 488; catalog no. A-11001; Molecular Probes)—were used at a 10−2 dilution. Detection of fluorescence was performed using a Nikon Eclipse E800 microscope, captured using a CoolSnap video camera, and processed using Adobe Photoshop 5.5 software.
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts from 0- to 18-h-old embryo collections were prepared as previously described (78) with a modification of the nuclear extraction buffer composition (20 mM HEPES [pH 7.5], 220 mM KCl, 300 mM NaCl, 0.1% Tween 20, 10% glycerol, 1 mM dithiothreitol, 0.1 mM EDTA), supplemented with protease inhibitors (1 mM 4-[2-aminoethyl]-benzenesulfonyl fluoride hydrochloride and complete protease inhibitor tablets [3 mg/ml; Roche]). DNA binding reactions and subsequent gel electrophoresis were performed using 10 μg of nuclear protein extract as described previously (3). Sequences of MHS-70 and LS-1/9 are described in reference 31. Double-stranded GAGA oligonucleotide competitor was obtained by self-annealing of the palindromic single-strand oligonucleotide 5′TCTCTCTGCAGAGAGATGCATCTCTCTGCAGAGAGATGCATCTCTCTGCAGAGAGATGCATCTCTCTGCAGAGAGA3′.
One to two microliters of the appropriate antibody was added to the binding reaction mixture for the supershift experiments.
Protein immunoprecipitation.
Embryonic extracts were prepared according to reference (6). Twenty-five late-third-instar larvae were dissected to remove gut and homogenized in 600 μl of lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% NP-40, 50 mM sodium fluoride, 1 mM dithiothreitol, 1 mM AEBSF [4-{2-aminoethyl}-benzenesulfonyl fluoride hydrochloride]) and 1.9 mg of a complete protease inhibitors tablet (Roche) using a pestle. The homogenate was incubated at 4°C with gentle agitation for 1 h. Debris were pelleted by centrifugation (10 min at 10,000 × g), and supernatant was recovered. An aliquot of 20 μl of extract was kept for the Western blot and the rest was incubated with 20 μl of anti-FLAG M2 beads (Sigma) by gentle inversion overnight at 4°C. The supernatant was removed, and the beads were washed four times with 1 ml of lysis buffer. The beads were resuspended in 20 μl of 2× sodium dodecyl sulfate gel loading buffer, boiled 5 min, and centrifuged 1 min at 10,000 × g. Supernatant was analyzed by Western blotting with the appropriate antibody.
Two-hybrid assays.
The yeast strain EGY48 containing the lacZ reporter plasmid pSH18-34 was transformed with a pEG202 derivative (22) containing the LexA DNA binding domain sequence fused in frame to either the BAN or the Ultraspiracle (USP) coding sequences. The yeast strain RFY206 was transformed with a pJG4-5 derivative (22) containing the B42 activation domain sequence fused in frame with either full-length BAN, EcR-B1, TRL/GAF519, or TRL/GAF581 or truncated coding sequences from TRL/GAF519 or TRL/GAF581. The pJG4-5-derived constructs containing coding sequences for the TRL/GAF BTB/POZ domain (aa 1 to 121) or the full-length TRL/GAF519 isoform were kindly provided by Frédérique Peronnet. All junctions in constructs were sequenced. EGY48 and RFY206 transformants were mated, and diploids were assayed for β-galactosidase activity using the overlay method as described in reference 27.
RESULTS
Molecular characterization of batman.
The ban gene was previously identified based on the enhancement of the ESC phenotype of ph410 [E(ph)] associated with several noncomplementing P-element mutations (24). Four ban alleles were available at the beginning of this study. Three are lethal P[LacW] insertions [l(2)k02512, l(2)k11212, and l(2)k07907] located in the first intron of the ban gene, whereas the fourth, Df(2R)311a, is a lethal P[MTW] insertion associated with a 14-kb flanking deletion that eliminates several genes as well as the ban 5′-most sequences (Fig. 1A) (24). Viable revertants of banl(2)k02512 and banl(2)k11212 were recovered after excision of P[LacW], indicating that the lethality of these alleles is due to the P-element insertion.
The ban transcription unit was delimited by the ability of the 5.4-kb pBB genomic fragment (Fig. 1A) to rescue both the lethality and the E(ph) effect of the banl(2)k02512 mutation in pBB transgenic flies (Table 1, lines 2, 3, 6, and 7). A major transcript of ca. 1.4 kb was detected by Northern blot analysis of embryonic and larval total RNA using the full LD14505 cDNA from the BDGP cDNA library that is included in the pBB genomic fragment as a probe (Fig. 1B). In situ hybridization on whole embryos using the same cDNA as a probe reveals that ban is ubiquitously expressed at all stages of embryogenesis (data not shown). Sequencing of the 1,308-bp LD14505 cDNA (GenBank AF308476) indicated that it includes an ORF of 127 codons (ORF127) (Fig. 1A).
Comparison of the sequence of ORF127 with databank sequences revealed the presence of a BTB/POZ domain (Broad Complex, Tramtrack and Bric-à-brac proteins, Poxvirus and Zinc finger [5, 87]) located from aa 6 to 117. This BTB/POZ domain shows conservation over its entire length with other fly BTB/POZ domains from the Tramtrack subfamily (Fig. 1C) (87). In order to test whether ORF127 includes all ban functions, we established transgenic lines with constructs designed to express ORF127 fused to a FLAG epitope (UBF construct) or to the EGFP (UBG construct) at the C terminus. These constructs were placed under the control of the UAS/GAL4 expression system (8). Ubiquitous expression of either construct using the da:Gal4 driver rescued both the lethality and the E(ph) phenotype of banl(2)k02512(Table 1), thereby indicating that the BTB/POZ domain followed by 10 aa contained in ORF127 is sufficient to fulfill known ban functions. This makes the BAN protein unique among BTB/POZ proteins, since previously known BTB/POZ domains are usually found as part of larger multidomain zinc finger-containing or actin-binding proteins (2).
An antiserum was raised against batC11 which covered the only region of BAN that is not conserved among known proteins. Upon using this antiserum in Western blot experiments, a ca. 14-kDa band was detected in wild-type larvae (Fig. 1D). Both the 14-kDa band and a strong band with slightly lower mobility are detected in da:Gal4/UBF larvae. The latter band is detected by the anti-FLAG antibody, indicating that it corresponds to the BAN polypeptide flanked by the 8-aa FLAG epitope (data not shown). The intensity of the 14-kDa band was reduced in larvae heterozygous for the Df(2R)PC4 deficiency uncovering ban, and the 14-kDa band was undetectable in homozygous banl(2)k02512 escaper third-instar larvae, indicating that the banl(2)k02512 allele is a strong hypomorphic allele (Fig. 1D). Immunostaining experiments using the batC11 antiserum indicated that the BAN protein is ubiquitously expressed in embryos as well as in larvae, a result that is in good agreement with in situ hybridization experiments using a ban cDNA probe (data not shown).
Requirement of batman function during embryonic development.
All three P[LacW] insertions in ban are homozygous lethal predominantly at the first-instar larval stage, with very few escapers (7.2%) surviving until adulthood. Since ban transcripts are detected in presyncytial embryos (data not shown), a maternal contribution of ban may be sufficient to allow embryonic development to proceed until hatching of first-instar larvae. To generate embryos lacking a maternal contribution, we took advantage of the absence of expression of the pUAST-derived constructs UBF and UBG in the female germ line (8). We generated banl(2)k02512/banl(2)k02512; da:Gal4, UBG/+ rescued adults, and observed in females that no EGFP-tagged BAN protein was indeed detected in oocytes, whereas follicular cell nuclei were labeled (data not shown). These females were crossed to sibling males of identical genotype and embryos showing no fluorescent signal were identified as homozygous banl(2)k02512/banl(2)k02512 embryos, thus lacking both the maternal and the zygotic contribution of ban. These embryos did not hatch, and all displayed cuticular defects compared to the wild type (Fig. 2). All embryos showed incomplete head involution, and most of them showed defects in dorsal closure as well (Fig. 2B and C). In contrast, embryos derived from banl(2)k02512/banl(2)k02512; da:Gal4, UBG/+ rescued females crossed to wild-type males hatched and did not display cuticular defects (data not shown), thereby indicating that the zygotic expression of a wild-type allele inherited from the father is sufficient to rescue the maternal loss of ban function.
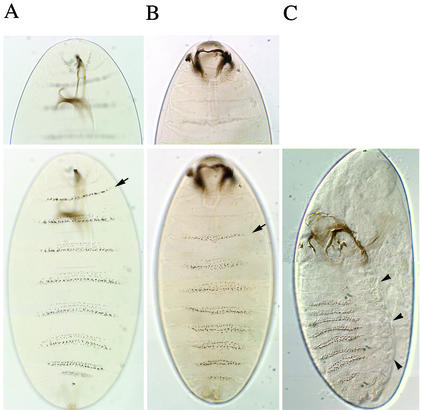
FIG. 2.
Cuticular defects in banl(2)k02512/banl(2)k02512 embryos deprived of the maternal contribution of ban. (A and B) Focus is on the anterior part in the top panels and on the denticular belts in the bottom panels. The two mutant embryos (B and C) illustrate the various ranges of cuticular defects. When compared to wild-type embryos (A), the position of the first abdominal denticle belt (arrow) is moved posteriorly in ban mutant embryos (B), indicating defects in head involution associated with altered mouth hooks. Dorsal closure defects were always observed, however, with various degrees of severity, from a hole in the dorsal cuticle (not shown) to the absence of closure (C). (C) Arrowheads point to the edge of the dorsal cuticle.
batman interacts genetically with PcG genes.
The four ban alleles tested [banDf(2R)311a, banl(2)k02512, banl(2)k11212 and banl(2)k07907] enhance the esc phenotype of both ph410 and Pc16 (24) (Table 1 and data not shown). This phenotype represents a homeotic transformation of mesothoracic or metathoracic legs toward prothoracic legs due to the deregulation of Sex combs reduced (54). In contrast, one pBB extra copy of ban in an otherwise wild-type context for ban suppressed the ESC phenotype of ph410 (Table 1, lines 1, 4, and 5, 8). The UBF and the UBG constructs had a similar effect that in both cases depended on the presence of the da:Gal4 driver (Table 1).
A Contrabithorax (Cbx) phenotype, which is a wing-to-halter transformation due to ectopic expression of Ubx in the posterior part of the wing disk (15), was also detected in ban mutants heterozygous for alleles banl(2)k02512, banl(2)k11212, and banl(2)k07907when also heterozygous for ph410, Pc16, Pc1, Pc3, or Pc15. The Cbx phenotype was further enhanced in ban heteroallelic viable combinations [banl(2)k02512/banDf(2R)311a, banl(2k11212/banDf(2R)311a, and banl(2)k07907/banDf(2R)311a] when also heterozygous for any of these Pc or ph alleles (Fig. 3A and data not shown).
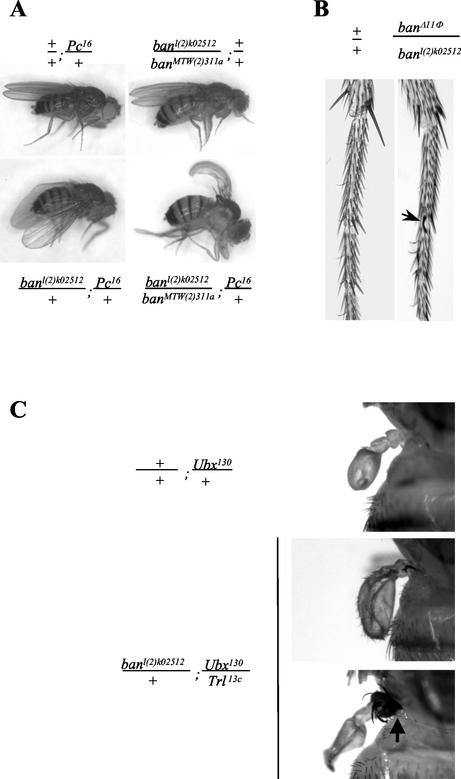
FIG. 3.
ban homeotic phenotypes. (A) Cbx phenotype of heteroallelic ban combinations in a Pc16 heterozygous context. Heteroallelic banDf(2R)311a/banl(2)k02512 flies as well as Pc16/+ flies have wild-type wings. Double-heterozygous banl(2)k02512/+; Pc16/+ flies display a curving of the wings that is indicative of the Cbx phenotype. This phenotype is enhanced in heteroallelic banDf(2R)311a/banl(2)k02512; Pc16/+ flies. (B) Sex comb teeth normally absent in wild-type males (left) are observed on the mesothoracic tarsus in banΔ11Φ/banl(2)k02512 males (right). (C) Enhancement of the genetic interaction between Trl and Ubx. Ubx130/+ flies display slightly enlarged halters (top panel) compared to the wild type. The expressivity of this transformation is higher in Trl13c/Ubx130 flies, in which a clear transformation of halter to wing was present in 6.3% (n = 142) of the flies, whereas it was never found in Ubx130/+ flies. In banl(2)k02512/+; Trl13c/Ubx130 flies, this transformation was much more frequent (18% [n = 204]; middle and bottom panels) and was often accompanied by a notal transformation (arrow in bottom panel).
New alleles of ban were generated by excision of the P-element from the l(2)k02512 and l(2)k11212 insertions. The new alleles were selected as lethal or subviable in combination with banl(2)k02512. For two of these (Δ11φ and Δ11ɛ′) ectopic sex comb teeth were observed on meso- or metathoracic legs in combination with banl(2)k02512 in about one-fifth of the male escapers (Fig. 3B). Thus, ban not only enhances PcG mutant phenotypes but also displays the esc phenotype characteristic of PcG mutations.
In summary, the analysis of adult ban mutant phenotypes, together with that of the interactions between ban and ph or Pc, demonstrates a dose-dependent function for ban in the repression of the homeotic genes Scr and Ubx.
batman interacts genetically with Trithorax-like.
The presence of a conserved BTB/POZ domain in both BAN and TRL/GAF (Fig. 1C) prompted us to test genetic interactions between the ban and Trl genes. The hypomorphic allele Trl13c is semilethal (29). Flies of the banl(2)k0251/+; Trl13c/± genotype were viable and did not show any visible phenotype. We assayed the viability of Trl13c mutant flies in a banl(2)k02512 heterozygous background. Whereas 12% of +/+; Trl13c/Trl13c flies hatched, only 4% of banl(2)k0251/+; Trl13c/Trl13c flies were recovered. Since the increase in the lethality could be due to second site mutations in either stock, we tested the possibility of rescuing the lethality specifically with Trl and ban transgenes. Two major isoforms of 519 and 580 aa are encoded by Trl. Both the hsp83:GAGA519 and the hsp83:GAGA581 transgenes have been shown to rescue the lethality of Trl13c/TrlR67 mutant combinations (29). Adding at least one copy of either the hsp83:GAGA519 or the hsp83:GAGA581 transgenes allowed the recovery of a much higher frequency of homozygous Trl13c escapers that are also heterozygous for banl(2)k02512 (up to 80% with four copies of hsp83:GAGA519, see Materials and Methods). Symmetrically, the addition of an extra copy of batman using the pBB transgene also rescued banl(2)k02512/+; Trl13c/Trl13c flies (data not shown). Together these experiments indicate that the increase in the lethality of flies that are mutant for Trl in a heterozygous banl(2)k02512 context results from the synergistic effect of ban and Trl loss of function mutations.
The Trl13c allele was identified as a dominant enhancer of the weak homeotic transformation of halter toward wing found in Ubx130 heterozygotes (23). Flies that are doubly heterozygous for banl(2)k02512 and Ubx130 do not show a significant increase in the expressivity of the Ubx phenotype. In contrast, banl(2)k02512/+; Trl13c/Ubx130 flies display a higher frequency (18%) of strong halter-to-wing transformations (Fig. 3C and Materials and Methods) compared to their +/+; Trl13c/Ubx130 siblings (6.3%). The synergistic interaction between ban and Trl on the Ubx phenotype indicates that ban function is required together with that of Trl for the activation of the homeotic gene Ubx.
Trl dosage was also reported to be important for the pairing-sensitive silencing that is induced by PREs when they are placed next to a white derivative reporter gene. Two independent constructs containing a PRE fragment (260.1 iab-7 PRE [47] and 5F24 25,2 Fab-7 PRE [86]) were tested for a modification of eye pigmentation in Trl and in ban heterozygous mutant backgrounds (Fig. 4). For both PRE constructs, we found that reducing the dose of ban using several ban loss of function alleles leads to derepression of the white reporter gene, an effect similar to that observed when reducing the dose of Trl (Fig. 4) (47). When both Trl and ban functions were reduced in double heterozygotes (Fig. 4), the pairing-sensitive silencing was further suppressed, thereby indicating that ban and Trl genetically interact for silencing the iab-7 PRE.
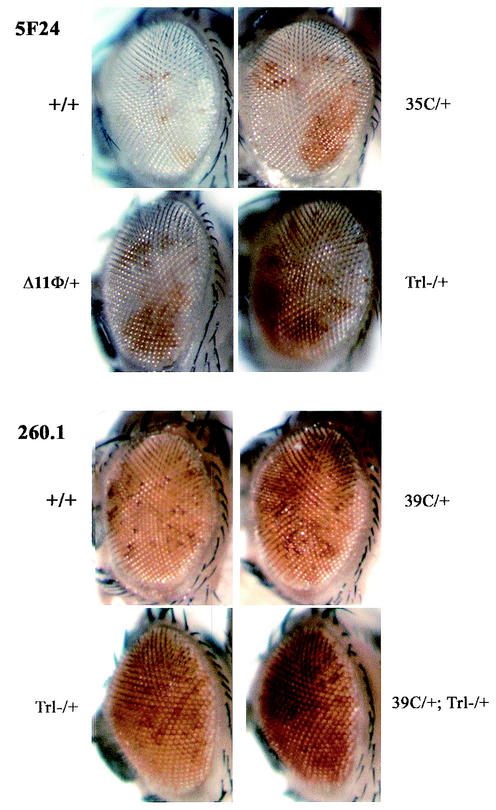
FIG. 4.
Similar effects of ban and Trl loss-of-function mutations on the PRE-dependent pairing-sensitive silencing of white reporters. Three white deletion mutant alleles generated from banDf(2R)311a (39C and 35C) and banl(2)k02512 (Δ11Φ) were assayed in a heterozygous context for the pairing-sensitive silencing of either the X-linked, 3.8-kb Fab-7 PRE line 5F24 25,2 (5F24) (86), or the 260-bp iab-7 PRE line 260.1 (47). In the latter case, ban alleles were first recombined with the 260.1 insertion on the second chromosome. In all cases (35C/+ and Δ11Φ/+ in 5F24 25,2 females and 39C 260.1 males and females), derepression of the white transgene was observed in ban heterozygotes, closely resembling what is observed in Trl13c (Trl-/+) heterozygotes. Derepression was increased in double heterozygotes 39C 260.1/260.1; Trl13c/+ (39C/+; Trl-/+). For each combination, effects were compared between siblings of the same gender and the appropriate genotype.
Taken together, our results suggest that ban cooperates with Trl in order to maintain the activation or the repression of Trl target genes that are necessary for viability and/or normal development of the flies.
Batman colocalizes with PcG/trxG proteins on polytene chromosomes.
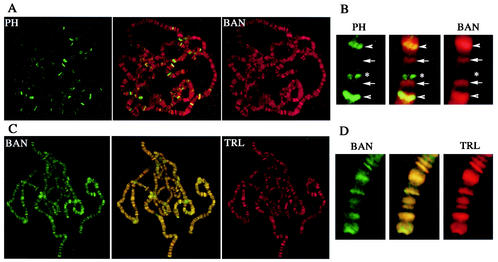
The BAN protein, as detected by indirect fluorescence using the batC11 antibody, accumulates in the nucleus. During early embryonic development, BAN was found associated to condensed chromosomes during mitosis (data not shown). On polytene chromosome spreads of third-instar salivary glands, a discrete banding pattern of more than 300 sites was observed for the BAN protein (Fig. 5A). A similar pattern on polytene chromosomes was observed when using the anti-FLAG antibody in transgenic larvae expressing a BAN-FLAG tagged protein (Fig. 5C). These results indicate that BAN is a chromatin-associated protein.
FIG. 5.
Comparison of the distributions of BAN, PH, and TRL/GAF proteins on larval salivary polytene chromosomes as detected by double immunostaining. Simultaneous immunostaining was performed for the following combinations: PH-FLAG (green) and BAN (red) (A and B) and BAN-FLAG (green) and TRL/GAF (red) (C and D). In each panel, a merge image is shown in the middle. (B) A representative chromosomal region displaying four adjacent BAN binding sites is enlarged. Among these BAN sites, two are shared with PH (arrowheads) and two are specific for BAN (arrows). In the same region, one PH binding site is not shared with BAN (asterisk). (D) Enlargement of a chromosomal region from panel C shows the colocalization of BAN and TRL/GAF binding sites.
The distribution of BAN was compared to that of the PcG protein, PH, and to that of the trxG protein, TRL/GAF, both encoded by genes that show genetic interactions with ban mutations. Since the available antibodies against BAN, TRL/GAF, and PH were obtained in rabbit, double-labeling experiments were performed on transgenic larvae expressing the PH or the BAN proteins fused to the FLAG epitope, which was revealed using a mouse monoclonal antibody. The FLAG-PH protein was previously reported to be functional (65). The BAN-FLAG protein is also functional since it fully rescues both the lethality and the E(ph) phenotype of the loss of function allele banl(2)k02512. In transgenic larvae expressing PH-FLAG, 49 binding sites are shared by BAN and PH-FLAG (Fig. 5A and B). Similar results were obtained in a symmetrical experiment in which BAN-FLAG sites were compared to endogenous PH binding sites (data not shown). Together, these results indicate that BAN codistributes with PH on about half of PH binding sites. The distribution of BAN-FLAG was compared to that of endogenous TRL/GAF in transgenic larvae expressing the BAN-FLAG protein. In contrast to the partial overlap observed between BAN and PH, a perfect codistribution of BAN and TRL/GAF was found on polytene chromosomes (Fig. 5C and D). Consistently, PH-FLAG shares about half of its binding sites with TRL/GAF, as was found for BAN (data not shown). In addition, the position of the strongest binding sites for each of the two factors overlap on polytene chromosomes (Fig. 5C and D), suggesting that the stoichiometry of BAN and TRL/GAF is constant.
Taken together, these experiments indicate that the chromatin-associated protein BAN codistributes with both TRL/GAF and PH on polytene chromosomes. This result is consistent with the genetic interactions observed between ban, Trl, and ph and suggests that BAN may cooperate with both PcG and trxG proteins for the regulation of common target genes.
Batman binds to the MHS-70 element from the bxd PRE.
Since BAN binds to many sites that are also targets of PH and TRL/GAF on polytene chromosomes, we further characterized this binding in vitro on a defined PRE. We chose the 70-bp MHS-70 fragment from the bxd PRE in Ubx. MHS-70 is required in vivo for the maintenance of embryonic silencing of a Ubx enhancer and binds in vitro to both TRL/GAF and PH proteins partially purified from Kc cell nuclear extracts (31).
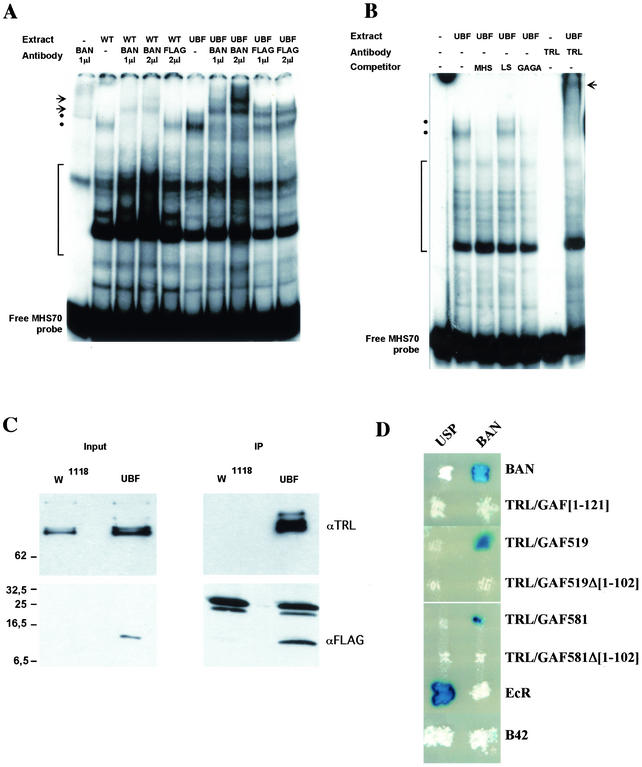
When tested in an EMSA in the presence of a nuclear extract from wild-type embryos, the radiolabeled MHS-70 probe gave rise to the formation of several retarded nucleoprotein complexes (Fig. 6A, lane 2). The two slower-migrating complexes were specifically competed against by an excess of unlabeled MHS-70 probe (Fig. 6B, lane 3). BAN is involved in the formation of these two complexes, since they were specifically supershifted in the presence of the batC11 antibody (Fig. 6A, lanes 3 and 4) but not in the presence of a control anti-FLAG antibody (Fig. 6A, lane 5). These complexes were formed with a much higher efficiency when MHS-70 was used as a probe in the presence of a nuclear extract from embryos expressing BAN-FLAG in an otherwise ban+ context (Fig. 6A, lane 6). They were completely supershifted in the presence of the batC11 antibody (Fig. 6A, lanes 7 and 8), and partially supershifted in the presence of the anti-FLAG antibody (Fig. 6A, lanes 9 and 10), indicating that both endogenous BAN and BAN-FLAG participate in their formation. Together, these results demonstrate that BAN, as well as BAN-FLAG, binds to MHS-70.
FIG. 6.
(A and B) Gel shift analysis of MHS-70 binding activities. (A) Binding of proteins in nuclear extracts from w1118 embryos (WT) or da:Gal4, UBF/+ embryos (UBF) was analyzed in a gel shift assay using the MHS-70 fragment as a probe in the absence (−) or in the presence of BAN or FLAG antibodies as indicated. The positions of BAN-containing low-mobility complexes in the absence (dots) or the presence (arrows) of BAN antibody are indicated. The position of nonspecific complexes that are not displaced by the addition of unlabeled probe is indicated (bracket). (B) Binding of proteins in a nuclear extract from da:Gal4, UBF/+ embryos (UBF) was analyzed in a gel shift assay using the MHS-70 labeled probe in the absence (−) or in the presence of competitors DNAs or TRL/GAF antibody (TRL) as indicated. Positions of the BAN containing complexes in the absence (dots) or the presence (arrow) of TRL/GAF antibody are indicated. The position of unspecific complexes that are not competed against by a 100-fold molar excess of MHS-70 probe is indicated (bracket). (C) Coimmunoprecipitation of BAN-FLAG with theTRL/GAF. Nuclear protein extracts from w1118 or da:Gal4/UBF (UBF) larvae were immunoprecipitated (IP) with anti-FLAG beads and analyzed by Western blotting using anti-TRL/GAF (TRL) or anti-FLAG (FLAG) antibodies. Aliquots (1/60) of the input extracts (input) were analyzed in parallel using the indicated antibody. The positions of protein molecular mass markers (in kilodaltons) are shown on the left. (D) Interaction of BAN and GAF in a yeast two-hybrid assay. Each patch corresponds to diploid yeast colonies coexpressing either the BAN protein (right column) or the USP (left column) as a control; both fused to the DNA binding domain of LexA; and BAN, full-length or truncated TRL/GAF isoforms, or EcR, all fused to the B42 transactivation domain (rows). Interactions resulted in the activation of a lacZ reporter gene placed under the control of LexA binding sites and were revealed in a β-galactosidase assay. Interaction (84) between the EcR-B1 isoform (EcR) and USP provided a positive control, while expression of the B42 domain alone served as a negative control.
Batman participates in a TRL/GAF-containing complex.
Since BAN does not contain much more than a single BTB/POZ protein-protein interaction domain, we reasoned that its binding to DNA most likely requires an interaction with a DNA binding partner. TRL/GAF binds to GAGA sequence motifs found in MHS-70 (31). In addition, TRL/GAF's own BTB/POZ domain provides a putative interface for dimerization with BAN. Thus, TRL/GAF may be the DNA binding partner mediating the binding of BAN to MHS-70. In order to test this hypothesis, we first determined whether TRL/GAF target sequences are required for the binding of BAN to MHS-70. The formation of the BAN-containing complexes in the presence of a nuclear extract from Da:Gal4; UBF transgenic larvae was specifically competed against by a 100-fold molar excess of the double-stranded GAGA oligonucleotide containing eight d(GA)3 motifs (Fig. 6B, lane 5). In contrast, no competition was observed in the presence of a 100-fold molar excess of the MHS-70-derived fragment LS-1/9 (Fig. 6B, lane 4) in which two terminal d(GA)3 sequences have been mutated (31). These results indicate that the BAN-containing complexes bind to GAGA repeats in MHS-70, suggesting that their formation involves a GAGA binding factor such as TRL/GAF. Indeed, the two BAN-containing DNA-binding complexes were specifically supershifted in the presence of anti-TRL/GAF antibody, indicating that they contain TRL/GAF (Fig. 6B, lane 7).
The preceding experiments suggest that the binding of BAN to a bxd PRE fragment involves its interaction with TRL/GAF or a TRL/GAF-containing complex. In order to test whether this interaction occurs independently from binding to a DNA target, we performed protein immunoprecipitation experiments, taking advantage of the tagged BAN-FLAG protein. Anti-FLAG antibody specifically precipitated the BAN-FLAG protein from UBF/+; da:Gal4/+ third instar larvae nuclear extracts (Fig. 6C). Under these conditions, the TRL/GAF protein was efficiently coprecipitated with BAN-FLAG from larval nuclear extracts (Fig. 6C), as well as from embryonic extracts (data not shown). These results provide further evidence that BAN and TRL/GAF are found in the same complexes in vivo, and this independently from binding to DNA.
The BTB/POZ domain has been shown to function as a hetero- or homodimerization interface (1, 18). We therefore tested whether an interaction between BAN and GAF could be mediated by the BTB/POZ domain present in both proteins. In a yeast two hybrid assay (Fig. 6D), the BAN protein interacted with itself as well as with both the 519- and the 581-aa TRL/GAF isoforms. In contrast, BAN did not interact with the TRL/GAF519Δ[1-103] or TRL/GAF581Δ[1-103] variant proteins in which the BTB/POZ domain was deleted. In addition, BAN was not able to interact with the TRL/GAF BTB/POZ domain alone (TRL/GAF[1-121]). Therefore, we conclude that the BTB/POZ domain of TRL/GAF is necessary but not sufficient to mediate the interaction of the two major TRL/GAF isoforms with the BAN BTB/POZ domain.
DISCUSSION
batman encodes a singular type of BTB/POZ protein.
We have characterized ban as a PcG/trxG gene and have shown that most of the BAN protein is composed of a 117-aa BTB/POZ domain (5, 87). The BTB/POZ domain is a homo- and heterophilic protein-protein interaction domain (1, 18) conserved in metazoans in a large family of proteins that includes transcription factors and actin-binding proteins. BTB/POZ proteins exert a wide range of biological functions and generally contain other domains involved in DNA-binding, actin-binding or protein-protein interactions (2, 4). The BAN protein appears unique among members of this family since it is almost reduced to its 117-aa BTB/POZ domain. Therefore, the study of BAN should provide a better understanding of the specific function of a BTB/POZ domain in a biological interaction network.
Our studies show that BAN is a chromatin-associated protein that localizes to more than 300 sites on polytene chromosomes. Therefore, BAN has potentially a wide spectrum of functions outside the regulation of homeotic genes. Since BAN does not contain potential DNA binding domains such as zinc fingers or a Pipsqueak (PSQ) domain (42) found in other nuclear BTB/POZ proteins, specific binding of BAN to its chromosomal targets most likely involves interactions with at least one other protein or a protein complex that can bind DNA. Genetic analyses suggest that PcG and trxG proteins are potential partners of BAN. Among BAN binding sites on polytene chromosomes, only one-sixth correspond to PH binding sites, suggesting that the interaction of BAN with the PcG protein PH is not necessary for its binding to chromatin. In contrast, the perfect codistribution of TRL/GAF and BAN on chromosomes, in addition to their coimmunoprecipitation from nuclear extracts, their interaction in vivo in a yeast 2-hybrid assay, and their cobinding to GAGA target sites in EMSA, strongly suggests that BAN is recruited to DNA by TRL/GAF through heterodimerization between the BTB/POZ domains of the two proteins.
We have shown (J.-Y. Roignant, C. Antoniewski, and J.-A. Lepesant, unpublished results) that BAN also interacts with the BTB/POZ transcription factors Broad and Bric-a-brac. This suggests that, in addition to its participation to a TRL/GAF complex, BAN performs regulatory functions by interacting with a subgroup of nuclear BTB/POZ proteins, at least some of which can bind DNA. Further characterization of the interaction network between BAN and other BTB/POZ partners in Drosophila will be required in order to better understand how the singular BTB/POZ protein BAN exerts its pleiotropic developmental functions.
batman plays a dual function in the control of segmental identity.
The clear-cut distinction between PcG and trxG antagonistic functions has been questioned in functional assays for suppression of trxG mutations by PcG mutations (28, 41). Six genes of the PcG were shown to behave unexpectedly as enhancers of trxG mutations, and therefore constitute the ETP (Enhancers of PcG and trxG mutations) group. Symmetrically, a subgroup of the trxG may play a role in repression. Such is the case for TRL/GAF, first identified as a transcriptional activator (6, 72) and shown to belong to the trxG (23). However, genetic data indicate that loss of function alleles of Trl do not suppress, as expected for trxG mutations, but rather enhance the Pc3 esc phenotype (75), suggesting that Trl function is required for the repression of Scr. In addition, TRL/GAF is required for the repressing activity of the iab-7 PRE (30, 47) and the Mcp silencer (14). Other cases of trxG genes involved in repression have been described: the BRG1 gene, a homologue of the trxG gene brahma in humans (51), and osa and brahma in Drosophila (17). The Drosophila Osa/Brahma chromatin-remodeling complex has been hypothesized to maintain a chromatin conformation that precludes access of the basal transcriptional machinery to target promoters of Wingless signal transduction in the absence of Wingless signal. A role in transcriptional repression or activation may thus in some cases depend on the target, rather than on intrinsic properties of the PcG or trxG proteins.
Our data indicate that ban constitutes a new example of a PcG/trxG candidate that is involved in both the activation and the repression of homeotic genes. Combinations of ban mutant alleles lead to the transformation of mesothoracic legs toward prothoracic legs. This esc phenotype is enhanced by several mutant alleles of Pc and ph. In addition, the overexpression of ban suppresses the esc phenotype of ph410. These data, together with the synergistic interactions between ban and Pc or ph for the Cbx phenotype indicate a function of ban as a repressor of the homeotic genes, Scr and Ubx. However, ban mutant phenotypes also indicate the requirement of ban wild-type function in the activation of homeotic genes. ban enhances the interaction between Trl and Ubx leading to the transformation of halter to wing, which suggests that ban wild-type function is required for the activation of Ubx by Trl. Together, our results provide further support to the idea that at least a subset of PcG/trxG genes, including ban, exerts both positive and negative effects on the regulation of homeotic genes.
Several hypotheses may explain the dual function of ban. It is not possible to exclude the fact that ban mutations may have opposite effects on distinct homeotic genes indirectly through the transcriptional regulation of both activators and repressors of these genes. Alternatively, the dual function of ban may come from its interaction with Trl, whose function in both activation and repression of homeotic genes has already been documented (23, 32, 58, 75). This hypothesis is further supported by our results demonstrating the partnership between BAN and TRL/GAF.
The function of Batman in TRL/GAF-containing complexes.
Several lines of evidence suggest a close association between BAN and TRL/GAF. In 0- to 18-h embryos, increasing the dose of BAN through the use of the Gal4/UAS system increases the formation of the BAN- and TRL/GAF-containing complexes on the MHS-70 Ubx PRE fragment, which are fully displaced by both anti-TRL/GAF and anti-BAN antibodies. This result suggests that BAN may be a TRL/GAF cofactor that modulates its binding to MHS-70. Consistent with this, lowering the dose of ban has the same effect as lowering the dose of TRL/GAF in at least two regulatory pathways, the repression of Scr, and pairing-sensitive silencing of a white reporter gene next to an AbdB PRE. In addition, ban function is necessary for the activity of TRL/GAF in the activation of Ubx. Finally, the increased lethality of Trl13c mutants when the dose of ban is reduced provides additional evidence for the functional significance of the interaction of ban with Trl.
BAN is thus the second BTB/POZ protein that has been shown to participate in a TRL/GAF-containing complex involved in homeotic gene regulation, the other being PSQ. PSQ, like BAN, was previously reported to bind to the bxd MHS-70 PRE fragment (31). More recently, PSQ was found to colocalize and coimmunoprecipitate with TRL/GAF, an interaction that was shown to depend on the BTB/POZ domains of the two proteins (64). In addition, PSQ shares functions with TRL/GAF and BAN, such as its requirement for the activation of Ubx (64), as well as for the repression of Scr (33). However, among the three proteins, BAN appears to display unique functional features since it does not contain a DNA binding domain and since ban mutant phenotypes include the esc phenotype that is characteristic of PcG proteins. It is thus likely that understanding the possible function of d(GA)n binding complexes in tethering PcG or trxG complexes to PREs will require deciphering of the triangular interactions of the three BTB/POZ proteins TRL/GAF, PSQ, and BAN.
Acknowledgments
M.F. and J.-Y.R. contributed equally to this work.
We thank F. Karch, A.-M. Pret, and J. A. Lepesant, for helpful discussions and critical reading of the manuscript and Alexandra Menant for her contribution to the analysis of genetic interactions.
This work was supported by grants to L.T. from the Association pour la Recherche contre le Cancer (grant 6786) and La Ligue contre le Cancer, Comité de l'Essonne (1999 and 2000) and to C.A. from the Association pour la Recherche contre le Cancer (grant 5202). M.F. and J.-Y.R. were supported by the Ministère de l'Education nationale, de la Recherche, et de la Technologie and a fellowship from the Fondation pour la Recherche Médicale. S. Netter was supported by La Ligue contre le Cancer, Comité de l'Essonne.
REFERENCES
- 1.Ahmad, K. F., C. K. Engel, and G. G. Prive. 1998. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 95:12123-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albagli, O., P. Dhordain, C. Deweindt, G. Lecocq, and D. Leprince. 1995. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 6:1193-1198. [PubMed] [Google Scholar]
- 3.Antoniewski, C., M. Laval, A. Dahan, and J. A. Lepesant. 1994. The ecdysone response enhancer of the Fbp1 gene of Drosophila melanogaster is a direct target for the EcR/USP nuclear receptor. Mol. Cell. Biol. 14:4465-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravind, L., and E. V. Koonin. 1999. Fold prediction and evolutionary analysis of the POZ domain: structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285:1353-1361. [DOI] [PubMed] [Google Scholar]
- 5.Bardwell, V. J., and R. Treisman. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8:1664-1677. [DOI] [PubMed] [Google Scholar]
- 6.Biggin, M. D., and R. Tjian. 1988. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53:699-711. [DOI] [PubMed] [Google Scholar]
- 7.Boivin, A., and J. M. Dura. 1998. In vivo chromatin accessibility correlates with gene silencing in Drosophila. Genetics 150:1539-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 9.Breen, T. R., and P. J. Harte. 1991. Molecular characterization of the trithorax gene, a positive regulator of homeotic gene expression in Drosophila. Mech. Dev. 35:113-127. [DOI] [PubMed] [Google Scholar]
- 10.Breen, T. R., and P. J. Harte. 1993. Trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissue-specific, parasegment-specific and promoter-specific effects on each. Development 117:119-134. [DOI] [PubMed] [Google Scholar]
- 11.Breiling, A., E. Bonte, S. Ferrari, P. B. Becker, and R. Paro. 1999. The Drosophila polycomb protein interacts with nucleosomal core particles In vitro via its repression domain. Mol. Cell. Biol. 19:8451-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breiling, A., B. M. Turner, M. E. Bianchi, and V. Orlando. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412:651-655. [DOI] [PubMed] [Google Scholar]
- 13.Brock, H. W., and M. van Lohuizen. 2001. The Polycomb group—no longer an exclusive club? Curr. Opin. Genet. Dev. 11:175-181. [DOI] [PubMed] [Google Scholar]
- 14.Busturia, A., A. Lloyd, F. Bejarano, M. Zavortink, H. Xin, and S. Sakonju. 2001. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development 128:2163-2173. [DOI] [PubMed] [Google Scholar]
- 15.Cabrera, C. V., J. Botas, and A. Garcia-Bellido. 1985. Distribution of Ultrabithorax proteins in mutants of Drosophila bithorax complex and its transregulatory genes. Nature 318:569-571. [Google Scholar]
- 16.Cavalli, G., and R. Paro. 1999. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science 286:955-958. [DOI] [PubMed] [Google Scholar]
- 17.Collins, R. T., and J. E. Treisman. 2000. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 14:3140-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhordain, P., O. Albagli, R. J. Lin, S. Ansieau, S. Quief, A. Leutz, J. P. Kerckaert, R. M. Evans, and D. Leprince. 1997. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc. Natl. Acad. Sci. USA 94:10762-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dingwall, A. K., S. J. Beek, C. M. McCallum, J. W. Tamkun, G. V. Kalpana, S. P. Goff, and M. P. Scott. 1995. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell 6:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dura, J. M., H. W. Brock, and P. Santamaria. 1985. Polyhomeotic: a gene of Drosophila melanogaster required for correct expression of segmental identity. Mol. Gen. Genet. 198:213-220. [DOI] [PubMed] [Google Scholar]
- 21.Espinas, M. L., S. Canudas, L. Fanti, S. Pimpinelli, J. Casanova, and F. Azorin. 2000. The GAGA factor of Drosophila interacts with SAP18, a Sin3-associated polypeptide. EMBO Rep. 1:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Estojak, J., R. Brent, and E. A. Golemis. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farkas, G., J. Gausz, M. Galloni, G. Reuter, H. Gyurkovics, and F. Karch. 1994. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 371:806-808. [DOI] [PubMed] [Google Scholar]
- 24.Faucheux, M., S. Netter, S. Bloyer, M. Moussa, E. Boissonneau, F. Lemeunier, M. Wegnez, and L. Theodore. 2001. Advantages of a P-element construct containing MtnA sequences for the identification of patterning and cell determination genes in Drosophila melanogaster. Mol. Genet. Genomics 265:14-22. [DOI] [PubMed] [Google Scholar]
- 25.Fauvarque, M.-O., and J.-M. Dura. 1993. polyhomeotic regulatory sequences induce developmental regulator-dependent variegation and targeted P-element insertions in Drosophila. Genes Dev. 7:1508-1520. [DOI] [PubMed] [Google Scholar]
- 26.Fitzgerald, D. P., and W. Bender. 2001. Polycomb group repression reduces DNA accessibility. Mol. Cell. Biol. 21:6585-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fromont-Racine, M., J. C. Rain, and P. Legrain. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16:277-282. [DOI] [PubMed] [Google Scholar]
- 28.Gildea, J. J., R. Lopez, and A. Shearn. 2000. A screen for new Trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics 156:645-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenberg, A. J., and P. Schedl. 2001. GAGA factor isoforms have distinct but overlapping functions in vivo. Mol. Cell. Biol. 21:8565-8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hagstrom, K., M. Muller, and P. Schedl. 1997. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics 146:1365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodgson, J. W., B. Argiropoulos, and H. W. Brock. 2001. Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol. Cell. Biol. 21:4528-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horard, B., C. Tatout, S. Poux, and V. Pirrotta. 2000. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20:3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, D. H., Y. L. Chang, C. C. Yang, I. C. Pan, and B. King. 2002. pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Mol. Cell. Biol. 22:6261-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingham, P. W., and R. Whittle. 1980. Trithorax: a new homeotic mutation of Drosophilia Melanogaster causing tranformations of abdominal and thoracic imaginal segments. Mol. Gen. Genet. 179:607-614. [Google Scholar]
- 35.Jones, C. A., J. Ng, A. J. Peterson, K. Morgan, J. Simon, and R. S. Jones. 1998. The Drosophila esc and E(z) proteins are direct partners in polycomb group-mediated repression. Mol. Cell. Biol. 18:2825-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, R. S., and W. M. Gelbart. 1993. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol. Cell. Biol. 13:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jürgens, G. 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316:153-155. [Google Scholar]
- 38.Kennison, J. A., and J. W. Tamkun. 1988. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. USA 85:8136-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kyba, M., and H. Brock. 1998. The Drosophila Polycomb group protein Psc contacts ph and Pc through specific-conserved domains. Mol. Cell. Biol. 18:2712-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyba, M., and H. W. Brock. 1998. The SAM domain of polyhomeotic, RAE28, and scm mediates specific interactions through conserved residues. Dev. Genet. 22:74-84. [DOI] [PubMed] [Google Scholar]
- 41.LaJeunesse, D., and A. Shearn. 1996. E(z): a polycomb group gene or a trithorax group gene? Development 122:2189-2197. [DOI] [PubMed] [Google Scholar]
- 42.Lehmann, M., T. Siegmund, K. G. Lintermann, and G. Korge. 1998. The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem. 273:28504-28509. [DOI] [PubMed] [Google Scholar]
- 43.Martin, E. C., and P. N. Adler. 1993. The Polycomb group gene Posterior Sex Combs encodes a chromosomal protein. Development 117:641-655. [DOI] [PubMed] [Google Scholar]
- 44.Mazo, A. M., D. H. Huang, B. A. Mozer, and I. B. Dawid. 1990. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc. Natl. Acad. Sci. USA 87:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCall, K., and W. Bender. 1996. Probes of chromatin accessibility in the Drosophila bithorax complex respond differently to Polycomb-mediated repression. EMBO J. 15:569-580. [PMC free article] [PubMed] [Google Scholar]
- 46.McKeon, J., and H. W. Brock. 1991. Interactions of the Polycomb group of genes with homeotic loci of Drosophila. Roux's Arch. Dev. Biol. 199:387-396. [DOI] [PubMed] [Google Scholar]
- 47.Mishra, R. K., J. Mihaly, S. Barges, A. Spierer, F. Karch, K. Hagstrom, S. E. Schweinsberg, and P. Schedl. 2001. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 21:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohd-Sarip, A., F. Venturini, G. E. Chalkley, and C. P. Verrijzer. 2002. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 22:7473-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller, J., and M. Bienz. 1991. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 10:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller, J., and M. Bienz. 1992. Sharp anterior boundary of homeotic gene expression conferred by the fushi tarazu protein. EMBO J. 11:3653-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy, D. J., S. Hardy, and D. A. Engel. 1999. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol. Cell. Biol. 19:2724-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papoulas, O., S. J. Beek, S. L. Moseley, C. M. McCallum, M. Sarte, A. Shearn, and J. W. Tamkun. 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125:3955-3966. [DOI] [PubMed] [Google Scholar]
- 53.Paro, R., and D. S. Hogness. 1991. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. USA 88:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pattatucci, A. M., and T. C. Kaufman. 1991. The homeotic gene Sex combs reduced of Drosophila melanogaster is differentially regulated in the embryonic and imaginal stages of development. Genetics 129:443-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peterson, A. J., M. Kyba, D. Bornemann, K. Morgan, H. W. Brock, and J. Simon. 1997. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell. Biol. 17:6683-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pirrotta, V. 1988. Vectors for P-mediated transformation in Drosophila, p. 437-445. In R. L. Rodriguez, and D. T. Denhart (ed.), A survey of molecular cloning vectors and their uses. Butterworths, Boston, Mass.
- 57.Poux, S., B. Horard, C. J. Sigrist, and V. Pirrotta. 2002. The Drosophila trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development 129:2483-2493. [DOI] [PubMed] [Google Scholar]
- 58.Poux, S., R. Melfi, and V. Pirrotta. 2001. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 15:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qian, S., M. Capovilla, and V. Pirrotta. 1991. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 10:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rastelli, L., C. S. Chan, and V. Pirrotta. 1993. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependance on Enhancer of zeste function. EMBO J. 12:1513-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Jonhson-Schlitz, W. K. Benz, and W. R. Engels. 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rozovskaia, T., S. Tillib, S. Smith, Y. Sedkov, O. Rozenblatt-Rosen, S. Petruk, T. Yano, T. Nakamura, L. Ben-Simchon, J. Gildea, C. M. Croce, A. Shearn, E. Canaani, and A. Mazo. 1999. Trithorax and ASH1 interact directly and associate with the trithorax group-responsive bxd region of the Ultrabithorax promoter. Mol. Cell. Biol. 19:6441-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 64.Schwendemann, A., and M. Lehmann. 2002. Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proc. Natl. Acad. Sci. USA 99:12883-12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 66.Shearn, A. 1989. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics 121:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimell, M. J., J. Simon, W. Bender, and M. B. O'Connor. 1994. Enhancer point mutation results in a homeotic transformation in Drosophila. Science 264:968-971. [DOI] [PubMed] [Google Scholar]
- 68.Simon, J., A. Chiang, and W. Bender. 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114:493-505. [DOI] [PubMed] [Google Scholar]
- 69.Simon, J., A. Chiang, W. Bender, M. J. Shimell, and M. O'Connor. 1993. Elements of the Drosophila bithorax complex that mediates repression by Polycomb group products. Dev. Biol. 158:131-144. [DOI] [PubMed] [Google Scholar]
- 70.Simon, J., M. Peifer, W. Bender, and M. O'Connor. 1990. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 9:3945-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sinclair, D. A. R., T. A. Milne, J. W. Hodgson, J. Shellard, C. A. Salinas, M. Kyba, F. Randazzo, and H. W. Brock. 1998. The Additional sex combs gene of Drosophila encodes a chromatin protein that binds to shared and unique Polycomb group sites on polytene chromosomes. Development 125:1207-1216. [DOI] [PubMed] [Google Scholar]
- 72.Soeller, W. C., S. J. Poole, and T. Kornberg. 1988. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 2:68-81. [DOI] [PubMed] [Google Scholar]
- 73.Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty, N. Mozden, S. Misra, and G. M. Rubin. 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153:135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Struhl, G., and M. Akam. 1985. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 4:3259-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strutt, H., G. Cavalli, and R. Paro. 1997. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic genes expression. EMBO J. 16:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamkun, J. W., R. Deuring, M. P. Scott, M. Kissinger, A. M. Pattatucci, T. C. Kaufman, and J. A. Kennison. 1992. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68:561-572. [DOI] [PubMed] [Google Scholar]
- 77.Tie, F., T. Furuyama, and P. J. Harte. 1998. The Drosophila Polycomb Group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development 125:3483-3496. [DOI] [PubMed] [Google Scholar]
- 78.Tie, F., T. Furuyama, J. Prasad-Sinha, E. Jane, and P. J. Harte. 2001. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128:275-286. [DOI] [PubMed] [Google Scholar]
- 79.Tillib, S., S. Petruk, Y. Sedkov, A. Kuzin, M. Fujioka, T. Goto, and A. Mazo. 1999. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol. Cell. Biol. 19:5189-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsukiyama, T., and C. Wu. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83:1011-1020. [DOI] [PubMed] [Google Scholar]
- 81.Vazquez, M., L. Moore, and J. A. Kennison. 1999. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development 126:733-742. [DOI] [PubMed] [Google Scholar]
- 82.Wedeen, C., K. Harding, and M. Levine. 1986. Spatial regulation of Antennapedia and bithorax gene expression by the Polycomb locus in Drosophila. Cell 44:739-748. [DOI] [PubMed] [Google Scholar]
- 83.Wodarz, A., U. Hinz, M. Engelbert, and E. Knust. 1995. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82:67-76. [DOI] [PubMed] [Google Scholar]
- 84.Yao, T. P., W. A. Segraves, A. E. Oro, M. McKeown, and R. M. Evans. 1992. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 71:63-72. [DOI] [PubMed] [Google Scholar]
- 85.Zink, B., and R. Paro. 1989. In vivo binding pattern of a trans-regulator of homeotic genes in Drosophila melanogaster. Nature 337:468-471. [DOI] [PubMed] [Google Scholar]
- 86.Zink, D., and R. Paro. 1995. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 14:5660-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zollman, S., D. Godt, G. G. Prive, J. L. Couderc, and F. A. Laski. 1994. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 91:10717-10721. [DOI] [PMC free article] [PubMed] [Google Scholar]