Abstract
After a two-thirds hepatectomy, normally quiescent liver cells are stimulated to reenter the cell cycle and proliferate to restore the original liver mass. One of the most rapidly and highly induced genes and proteins in regenerating liver is insulin-like growth factor binding protein 1 (IGFBP-1), a secreted protein that may modulate the activities of insulin-like growth factors (IGFs) or signal via IGF-independent mechanisms. To assess the functional role of IGFBP-1 in liver regeneration, mice with a targeted disruption of the IGFBP-1 gene were generated. Although IGFBP-1−/− mice demonstrated normal development, they had abnormal liver regeneration after partial hepatectomy, characterized by liver necrosis and reduced and delayed hepatocyte DNA synthesis. The abnormal regenerative response was associated with blunted activation of mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) and a reduced induction of C/EBPβ protein expression posthepatectomy. Like cell cycle abnormalities observed in hepatectomized C/EBPβ−/− mice, cyclin A and cyclin B1 expression was delayed and reduced in IGFBP-1−/− livers, whereas cyclin D1 expression was normal. Treatment of IGFBP-1−/− mice with a preoperative dose of IGFBP-1 induced MAPK/ERK activation and C/EBPβ expression, suggesting that IGFBP-1 may support liver regeneration at least in part via its effect on MAPK/ERK and C/EBPβ activities. These findings are the first demonstration of the involvement of IGFBP-1 in the regulation of in vivo mitogenic signaling pathways.
The liver, which plays an important role in maintaining metabolic and synthetic homeostasis, constitutes a conditional renewal system in which parenchymal cells normally in G0 may be induced to proliferate following toxic damage, hepatitis, and surgical resection, which culminates in the rapid restoration of hepatic parenchyma (23). Following a 70% partial hepatectomy, preexisting, latent transcription factor complexes such as nuclear factor kappa B (NF-κB) and STAT3 are rapidly activated by means of posttranslational modifications in the remnant liver in response to stimuli such as epidermal growth factor, interleukin-6 (IL-6), hepatocyte growth factor, and tumor necrosis factor alpha (23, 33). Subsequently, during liver regeneration a transcriptional cascade is established that results in the entry of the hepatocytes and nonparenchymal cells into the S phase of the cell cycle.
Among the liver-specific immediate-early genes rapidly induced during regenerating liver and implicated in the maintenance of hepatocyte differentiation and metabolism is that for insulin-like growth factor binding protein 1 (IGFBP-1) (32). As a member of a group of structurally related soluble proteins which specifically bind and modulate the bioavailability and activity of insulin-like growth factors (IGF-I and IGF-II), IGFBP-1 has been shown to either enhance or inhibit the mitogenic effects of IGFs in certain tissues and to play an integral role in glucoregulation by suppressing IGF activity (17). IGFBP-1 may also have IGF-independent actions mediated by its internal RGD sequence, which has been shown to bind to α5β1 integrin (also known as the fibronectin receptor), and affect cell migration and attachment through unknown processes (17).
The role of IGFBP-1 in liver regeneration is unknown. IGFBP-1 is greatly induced at the transcriptional level in the remnant liver, with peak expression at 1 h posthepatectomy (24). Insulin, via inhibition of IGFBP-1 transcription, is a primary determinant of IGFBP-1 expression both in vitro and in vivo (17). However, IL-6 stimulation of hepatic IGFBP-1 expression can supersede the effect of insulin (18). IL-6 transcriptionally upregulates a vast array of genes and is required for normal liver regeneration and repair (4, 19). Evidence for a biologic role of IL-6 in IGFBP-1 upregulation includes increased expression of hepatic IGFBP-1 in IL-6 transgenic animals and following injection of IL-6 into nonfasting animals and its reduced expression in IL-6−/− livers posthepatectomy (18). These findings and other data which showed that hepatic IGFBP-1 mRNA and serum protein levels have a second peak of expression at 36 to 60 h posthepatectomy corresponding to the second round of mitotic activity (16) raised the possibility that IGFBP-1 may have a role in the mitogenic response in regenerating liver (28, 31). Since the level of IGFBP-1 is dynamically regulated by changes in the metabolic state and after hepatic injury, we investigated whether genetic disruption of IGFBP-1 expression could affect the normal hepatic proliferative response following a 70% hepatectomy, in which two larger lobes of the liver are removed intact without injury to remnant liver cells. We found that IGFBP-1 is required for a normal regenerative response, as IGFBP-1−/− livers have defects in the DNA response, increased necrosis, and blunting of expression of cell cycle-regulatory proteins, active mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK), and C/EBPβ transcription factor.
MATERIALS AND METHODS
Construction of targeting vector.
Two mouse IGFBP-1 genomic clones were isolated from a 129SV library (Stratagene, La Jolla, Calif.) as previously described (5). An SpeI IGFBP-1 fragment from position −3384 to +4336 was subcloned into pBluescript to make pISpe. An EcoRI-SalI fragment of pGem7(KJ1)SalI (kindly provided by Theo Danoff) containing PGKNeo (36) was subcloned into pBluescript KS to generate pPGKNeoKS. A NotI-NheI (−3384 to −234) fragment of pISpe was inserted into the NotI and XbaI sites of pPGKNeoKS, producing p5′IKSPGKNeo.pXhoPKSMC1TK. An XbaI-SpeI IGFBP-1 fragment (+2189 to +4336) was inserted into the SpeI and XbaI sites of pXhoPKSMC1TK, which contains MC1TK (kindly provided by Alan Bradley), and XhoI sites flanking the 3′ end of the IGFBP-1 gene and thymidine kinase cassette were used to isolate and insert it into the XhoI site of p5′IKSPGKNeo.
Generation of mutant mice.
The targeting vector was sent to GenomeSystems for electroporation into RW4 embryonic stem (ES) cells and selection with G418. The positive clone was injected into C57BL/6J blastocysts by the Transgenic Mouse Core Facility. Chimera animals were crossed to C57BL/6J females with agouti coat color, indicating germ line transmission by one male. Heterozygous pups were identified by Southern blotting and either backcrossed to C57BL/6 mice or mated brother-sister to generate colonies (B6; 129 hybrid background). Primers IB64 (5′-TGACAATCATTAACCTGTGCCGCAC-3′) and IB546 (5′-ACCTTCATGCTGGGAGCTGAACAAG-3′) were used in PCR analyses to identify the IGFBP-1+/+ animals. PGKNeo (5′-TTCCATTTGTCACGTCCTGCACGAC-3′) and IBKO (5′-GAAACAACTGTGGGCATTGTCACGG-3′) primer pairs were used to genotype the IGFBP-1−/− animals. IB64 begins at position −64 relative to the start site, and IB546 begins at +546. Both of these regions are missing in the knockout mice. IBKO starts at −400 relative to the start site. PGKNeo is within the PGK promoter.
Partial hepatectomy and serological analyses.
For regenerative liver, 12- to 16-week old animals were anesthetized with isoflurane and subjected to midventral laparotomy with ∼70% liver resection (12). For IGFBP-1-treated mice, animals were injected intraperitoneally with 0.3 μg of IGFBP-1/g of body weight for the indicated times or 30 min before surgery. For bromodeoxyuridine (BrdU)-treated mice, animals were injected intraperitoneally with 50 mg of BrdU/kg (0.2% solution in phosphate-buffered saline) at 1 h before fixation (9). Blood was obtained at the time of killing via cannulation of the inferior vena cava; serum was collected and analyzed by Ani Lytics, Inc. (Gaithersburg, Md.).
Immunohistochemistry.
Hepatocyte nuclear staining for BrdU (Roche Diagnostic Corporation/Roche Molecular Biochemicals, Indianapolis, Ind.), a thymidine analogue capable of incorporation into actively replicating DNA, was performed essentially as described previously (9).
Northern and Western analyses.
RNA was isolated from IGFBP-1+/+ and IGFBP-1−/− livers and analyzed by Northern blotting as described previously (9). Whole-cell extracts were prepared as previously described and subjected to Western analyses (18). Primary antibodies used were from Santa Cruz Biotechnology Inc (Santa Cruz, Calif.). Secondary antibodies were from Zymed Laboratories Inc. (South San Francisco, Calif.). The data were scanned densitometrically to quantitate mRNA and protein levels (Image-Quant Software; [Molecular Dynamics] and NIH Image 1.62). StatWorks (Apple Software, Cupertino, Calif.) and Student's t test were used for statistical analyses.
Primary hepatocyte isolation and cell culture.
Primary mouse hepatocytes were isolated as described previously (15). HeLa cells were cultured in Iscove's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin, and 50 U of streptomycin. At 24 h after seeding the cells, the cells were washed and kept in Iscove's medium containing 0.2% fetal bovine serum. After 24 h, the cells were treated with 100 ng of recombinant human IL-6 per ml or 100 ng of IGFBP-1 per ml for the indicated times.
RESULTS
IGFBP-1 gene targeting.
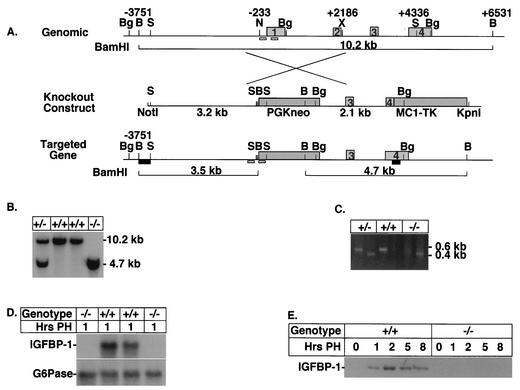
To study the role of IGFBP-1 in regenerating liver, we generated mice harboring a targeted disruption of the IGFBP-1 gene locus by replacing the first 234 bp of the promoter and the first and second exons of the IGFBP-1 gene with PGKNeo (Fig. 1A). After electroporation of mouse ES cells, G418-resistant clones were screened for homologous recombination by Southern blot analyses (data not shown). A single targeted cell line was subsequently isolated and used to generate chimeras, of which only one was germ line. Germ line transmission of the IGFBP-1 knockout allele in the agouti pups was confirmed by Southern blot analyses (Fig. 1B) and by PCR (Fig. 1C). Heterozygous offspring were then bred to homozygosity, and genotypes of the progeny generated from heterozygous crosses were determined by PCR analyses of tail DNA biopsies taken at roughly 3 weeks of age. The absence of IGFBP-1 mRNA in the IGFBP-1−/− mice was confirmed by Northern analyses with 1-h-posthepatectomy total liver RNA isolated from IGFBP-1+/+ and IGFBP-1−/− animals, as there is virtually no expression of IGFBP-1 in the quiescent liver. No IGFBP-1 mRNA induction was detected in IGFBP-1−/− mice following partial hepatectomy (Fig. 1D). In contrast, the expression of glucose-6-phosphatase, another gluconeogenic gene that has a pattern of expression similar to that of IGFBP-1 following partial hepatectomy (10, 24), was normal in the IGFBP-1−/− mice. The absence of IGFBP-1 protein in IGFBP-1−/− mice was verified by Western blotting of whole-cell liver extracts isolated from the quiescent and 1- to 8-h-posthepatectomy animals (Fig. 1E).
FIG. 1.
Targeted disruption of the murine IGFBP-1 gene in ES cells. (A) Schematic representation of the mouse IGFBP-1 gene, targeting vector, and targeted allele. Light gray boxes indicate the locations of the four exons. Darker gray boxes indicate the locations of PGKNeo and thymidine kinase (TK) genes. The homologous IGFBP-1 sequence flanking PGKNeo was 3.2 kb upstream and 2.1 kb downstream of the IGFBP-1 sequences between positions −233 and +2186, relative to the transcription start site. Restriction sites used for making the construct or for Southern screening are as follows: B, BamHI; Bg, BglII; N, NheI; S, SpeI; X, XbaI. Black boxes indicate the locations of the 5′BamHI/SpeI and 3′SpeI/EcoRI external probe fragments. Homologous insertion of PGKNeo results in a change in the size of BamHI fragments from 10.2 kb normally to 3.5 and 4.7 kb. (A change in size of BglII fragments also results.) Small white boxes indicate the locations of PCR primers used for rapid animal screening from tail biopsies. (B) Southern blot analysis of tail DNA by using BamHI and 3′ SpeI/EcoRI probes. Sizes of bands are indicated. (C) PCR screening of tail biopsy DNA. The wild-type primers at the top part are, from left to right, IB64 and IB546 (see Materials and Methods). The KO primers are, from left to right, IBKO and PGKNeo. (D) Northern blot analysis of liver showing lack of IGFBP-1 mRNA in IGFBP-1 knockout mice at 1 h posthepatectomy (PH). G6Pase, glucose-6-phosphatase. (E) Western blot analyses using IGFBP-1+/+ and IGFBP-1−/− whole-cell liver extracts prepared from the indicated times after partial hepatectomy and subjected to immunoblotting with an anti-IGFBP-1 antibody.
Gross phenotype of homozygous mutants.
Mice homozygous for the disrupted IGFBP-1 allele were phenotypically indistinguishable from wild-type or heterozygous littermates. No embryonic lethality or significant developmental defects were observed in IGFBP-1−/− animals. Adult IGFBP-1−/− mice of both sexes were fertile, and litter size was normal. Female IGFBP-1−/− mice were also capable of nursing their young. Minimal inflammatory cell infiltrates were found in histological examination of the pancreas and kidney in some IGFBP-1−/− animals (data not shown). Hepatic lesions of unknown etiology and variable degree of multifocal, mixed inflammatory cell infiltration in the liver were observed in some heterozygous and homozygous mutant animals 13 weeks and older (data not shown). Transgenic mice overexpressing IGFBP-1 demonstrated fasting hyperglycemia, impaired glucose tolerance, and modest insulin resistance (25). In contrast, no significant differences in the blood glucose and serum insulin levels were observed in the homozygous mutant mice before and after glucose challenge (data not shown), suggesting that IGFBP-1 deficiency does not play a major role in glucose regulation.
Histopathology following two-thirds partial hepatectomy.
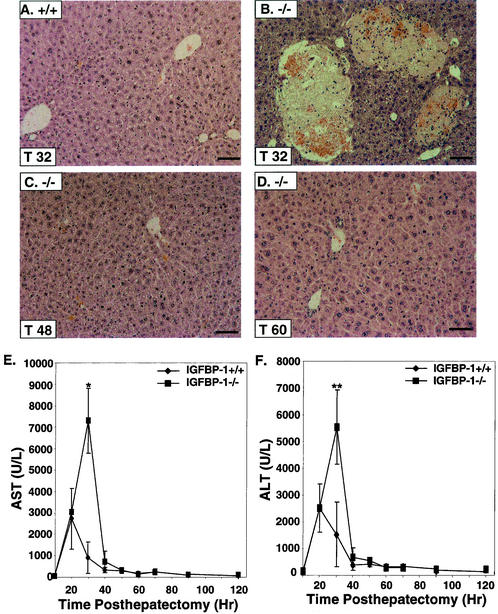
No significant mortality and morbidity were observed in IGFBP-1−/− mice relative to IGFBP-1+/+ mice posthepatectomy. However, well-demarcated areas of liquefaction necrosis, randomly distributed and representing 5 to 10% of the liver parenchyma with necrotic hepatocytes at the periphery, were observed in IGFBP-1−/− livers at 32 and 40 h after hepatectomy (n = 7 and 9) (P < 0.001) (Fig. 2B). Little or no necrosis was seen in the >80% of IGFBP-1−/− livers harvested at 48 h and later posthepatectomy, and regenerative changes were observed, indicating that resolution can occur (Fig. 2C and D). Increased liver damage was further substantiated by a sevenfold increase in aspartate aminotransferase levels (P < 0.006) and a fivefold increase in alanine aminotransferase levels (P < 0.02) in IGFBP-1−/− serum relative to IGFBP-1+/+ livers 32 h after hepatectomy (Fig. 2E and F). Levels of glucose, amylase, creatinine, triglyceride, and cholesterol were similar in IGFBP-1+/+ and IGFBP-1−/− mice (data not shown).
FIG. 2.
Hepatic necrosis in IGFBP-1−/− mice after hepatectomy. (A to D) Hematoxylin- and eosin-stained sections of IGFBP-1+/+ liver at 32 h posthepatectomy (A) and of IGFBP-1−/− livers at 32 h (B), 48 h (C), and 60 h (D) after partial hepatectomy. Bars, 50 μm. (E and F) Aspartate aminotransferase (AST) (E) and alanine aminotransferase (ALT) (F) levels in IGFBP-1+/+ and IGFBP-1−/− livers after partial hepatectomy. For IGFBP-1+/+, n = 4 per time point. For IGFBP-1−/−, n = 6 to 9 per time point. *, P < 0.006. **, P < 0.02. AST normal level = 72 to 288 U/liter. ALT normal level = 24 to 140 U/liter. Error bars indicate standard deviations.
Blunted DNA synthetic response following two-thirds partial hepatectomy.
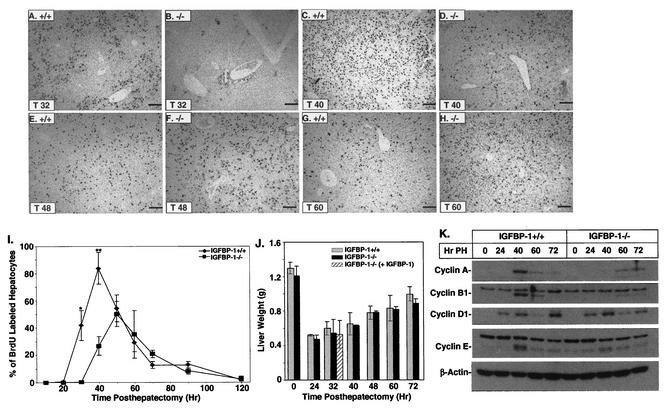
If IGFBP-1 induction is essential to the regenerative response, DNA synthesis should be diminished in IGFBP-1−/− mice posthepatectomy. BrdU incorporation detected by immunohistochemistry was used to measure the number of S phase cells at various times after partial hepatectomy in IGFBP-1+/+ and IGFBP-1−/− animals. No BrdU staining occurred in IGFBP-1+/+ or IGFBP-1−/− livers before surgery (0 h after hepatectomy), consistent with the cells being in the quiescent (G0) stage (data not shown). The level of induced hepatocyte DNA synthesis was abnormal in the IGFBP-1−/− animals, with no detectable DNA synthesis at 32 h (P < 0.00001) and 40% of wild-type levels at 40 h (P < 0.00001) after hepatectomy (Fig. 3A, B, C, D, and I). No significant differences were observed at 48 and 60 h posthepatectomy (Fig. 3E, F, G, H, and I). The peak in DNA synthesis in the IGFBP-1−/− livers was delayed and reduced by 30 to 40% relative to the peak in IGFBP-1+/+ livers. No significant differences in DNA synthesis were observed in nonparenchymal cells, including endothelial, Kupffer, and other sinusoidal cells, between the IGFBP-1+/+ and IGFBP-1−/− livers (data not shown). There was no statistically significant difference in the rate of mass reconstitution in the IGFBP-1−/− and IGFBP-1+/+ livers (Fig. 3J), suggesting that cell size increases were independent of S phase.
FIG. 3.
Delayed and reduced S phase hepatocytes in IGFBP-1−/− livers after partial hepatectomy. (A to H) Lower magnification of IGFBP-1+/+ and IGFBP-1−/− livers at 32, 40, 48, and 60 h posthepatectomy after anti-BrdU immunohistochemical detection. Round, uniformly stained nuclei represent BrdU-positive hepatocytes. Bars, 50 μm. (I) BrdU-positive hepatocytes for each sample were quantitated by counting positively stained cells in three low-power fields. The mean for each time point was expressed as a percentage of total hepatocytes per low-power field. For IGFBP-1+/+, n = 4 per time point. For IGFBP-1−/−, n = 6 to 9 per time point. Statistical significance was calculated versus wild-type littermates. * and **, P < 0.00001. Error bars indicate standard deviations. (J) Normal mass restitution despite reduced DNA synthesis. Liver weights of the indicated animals at the indicated times after partial hepatectomy are shown; n = 6. (K) Immunoblots showing delayed induction of cyclins A and B1 in IGFBP-1−/− posthepatectomy (PH) livers.
Delayed induction of cyclins A and B1 following partial hepatectomy.
Several studies have demonstrated a correlation between cyclin expression and hepatocyte cell cycle progression (2, 7, 11, 21, 34, 35). To assess whether the observed decrease in hepatocyte DNA synthesis in IGFBP-1−/− livers could reflect impaired progression of hepatocytes across the G1/S transition, we measured the steady-state levels of several cell cycle-associated gene products. Peak cyclin B1 and A induction occurred at 40 h posthepatectomy in the IGFBP-1+/+ livers and was threefold greater than that in IGFBP-1−/− livers, whereas a slight induction was noted in the IGFBP-1−/− livers at 60 h after partial hepatectomy (Fig. 3K). An approximately twofold reduction in cyclin E induction was noted in the IGFBP-1−/− livers compared to the IGFBP-1+/+ livers at 40 h posthepatectomy. The expression of cyclin D1 was similar in IGFBP-1+/+ and IGFBP-1−/− livers.
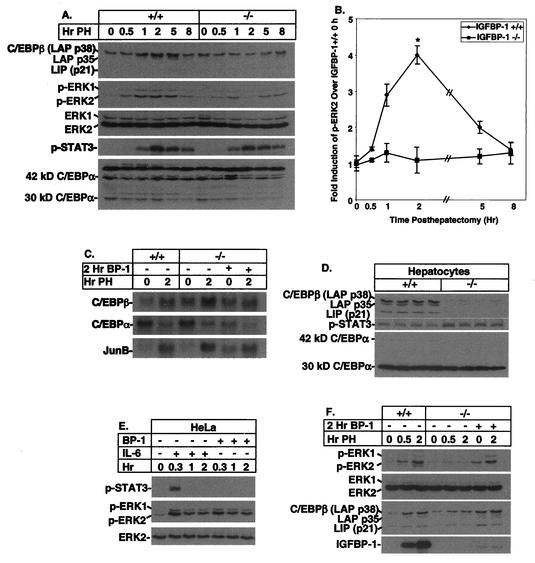
Blunted induction of C/EBPβ and MAPK expression posthepatectomy in IGFBP-1−/− livers restored by IGFBP-1 treatment.
Following partial hepatectomy, mice with a targeted deletion of the C/EBPβ bZip transcription factor gene display impaired hepatic regeneration characterized by a blunted DNA synthetic response and delayed induction of cyclin A, B, and E expression but not of cyclin D1 expression (9). Since the patterns of cyclin dysregulation are similar in IGFBP-1−/− and C/EBPβ−/− livers, we assessed the expression of C/EBPβ in IGFBP-1−/− livers. A highly blunted induction in C/EBPβ expression was observed in the IGFBP-1−/− livers at 1, 2, and 5 h posthepatectomy compared with controls (Fig. 4A). There was a transient twofold elevation of C/EBPα at 1 h posthepatectomy, and the decrease in p30 normally observed after partial hepatectomy (9) appeared to occur more rapidly in the IGFBP-1−/− livers. As previously observed (9), decreased C/EBPβ associated with even small increases in C/EBPα may result in a significant increase in relative levels of C/EBPα and homo- and heterodimers which are known inhibitors of hepatocyte DNA synthesis.
FIG. 4.
Blunted induction of C/EBPβ and p-ERK1/2 in IGFBP-1−/− livers after hepatectomy, corrected by IGFBP-1 treatment. (A) Delayed induction of C/EBPβ and p-EKR1/2 in IGFBP-1−/− livers from 0 to 8 h posthepatectomy (PH) as assessed by immunoblotting using whole-cell extracts. Various forms of C/EBPs and ERKs are indicated. (B) Graphic representation of pERK2 levels (n = 3 for each time point). Error bars indicate standard deviations; P = 0.05 at 2 h. (C) Northern analyses using total RNA isolated from IGFBP-1+/+ and IGFBP-1−/− quiescent and 2-h-posthepatectomy livers, IGFBP-1−/− livers treated with IGFBP-1 for 2 h, and 2-h-posthepatectomy IGFBP-1−/− livers treated with IGFBP-1. BP-1, IGFBP-1. (D) Reduced expression of C/EBPβ in IGFBP-1−/− primary hepatocytes (n = 4; P < 0.00001). Four different preparations of primary hepatocytes were utilized. Immunoblot analyses of C/EBPβ, STAT3, and C/EBPα levels are shown. (E) Western analyses using HeLa whole-cell extracts with or without IL-6 or IGFBP-1 treatment. (F) Western analyses using whole-cell extracts prepared from IGFBP-1+/+ and IGFBP-1−/− quiescent and 2-h-posthepatectomy livers, IGFBP-1−/− livers treated with IGFBP-1 for 2 h, and 2-h-posthepatectomy IGFBP-1−/− livers treated with IGFBP-1.
The MAPK/ERK cascade activation is a key signaling pathway involved in the regulation of G1 phase progression during regeneration (30). Since C/EBPβ could represent a potential downstream target of the MAPK pathway in the regenerating liver (3, 29), we investigated ERK1 (p44 MAPK) and ERK2 (p42 MAPK) activation in IGFBP-1−/− livers by using phospho-ERK-specific antibodies (Fig. 4A). The degree of ERK1 and ERK2 phosphorylation was reduced by as much as fourfold (2 h) in IGFBP-1−/− livers compared with IGFBP-1+/+ livers at 1, 2, and 5 h posthepatectomy (Fig. 4A and B), significantly at 2 h (P = 0.05). The total levels of ERK1 and ERK2 proteins were similar, indicating that signal transduction resulting in ERK phosphorylation was reduced in IGFBP-1−/− livers.
IL-6 is a known activator of C/EBPβ (1, 27), hepatic ERK1/2 phosphorylation (19), and STAT3 activation (4) in regenerating livers. To assess whether reduced IL-6 signaling could contribute to blunted C/EBPβ and ERK1/2 activation in IGFBP-1−/− livers posthepatectomy, we used a phospho-STAT3-specific antibody in Western analyses to examine the induction pattern of STAT3. Similar degrees of STAT3 induction were observed in the wild-type and IGFBP-1−/− livers (Fig. 4A), suggesting that IL-6 signaling was not responsible for the differential kinetics of ERK1/2 and C/EBPβ activation in IGFBP-1−/− livers.
To ascertain whether differences in protein expression may be explained in part by differences in gene expression, we examined C/EBBβ mRNA expression. Northern analyses revealed that there is no significant difference in the expression pattern of C/EBPβ mRNA in IGFBP-1−/− quiescent and 2-h-posthepatectomy livers relative to that in control livers. JunB and C/EBPα mRNA expression patterns were indistinguishable between the quiescent and 2-h-posthepatectomy IGFBP-1+/+ and IGFBP-1−/− livers (Fig. 4C). Treatment with a single dose of IGFBP-1 followed by partial hepatectomy had no effect on the mRNA expression of C/EBPβ, C/EBPα, and JunB. These data suggest that the level of C/EBPβ protein was regulated posttranscriptionally.
To determine if abnormal regulation of C/EBPβ was intrinsic to IGFBP-1−/− hepatocytes, we examined C/EBPβ protein expression in primary hepatocytes isolated from IGFBP-1+/+ and IGFBP-1−/− livers. The amount of C/EBPβ protein in the IGFBP-1+/+ hepatocytes was >7-fold higher than that in the IGFBP-1−/− hepatocytes (P < 0.00001), even though the amount of p-STAT3 in the IGFBP-1−/− hepatocytes was slightly elevated compared to that in the IGFBP-1+/+ hepatocytes (Fig. 4D). On the other hand, the p42 form of C/EBPα was reduced and the less antiproliferative form p30 was relatively elevated in hepatocytes from both IGFBP-1−/− and IGFBP-1+/+ livers, perhaps as a result of relative instability in the p42 form. Levels of C/EBPα were slightly higher in IGFBP-1+/+ hepatocytes, suggesting that in this in vitro system, levels of C/EBPα were not greatly influenced by the presence of IGFBP-1.
Because the IL-6 signaling pathway is constitutively active in both IGFBP-1+/+ and IGFBP-1−/− primary hepatocytes as verified by p-STAT3 Western analyses (Fig. 4D), we employed an alternative cell type to assess whether IGFBP-1 could potentially function as a signaling molecule to activate ERK1/2. In these experiments, IL-6 was used as a positive control because it is a known activator of ERK1/2 phosphorylation (19). HeLa cells treated with IGFBP-1 for 20 min showed a level of ERK activation similar to that of cells stimulated with IL-6 (Fig. 4E). However, activation of STAT3, as shown by p-STAT3, was detected only in extracts prepared from cells treated with IL-6 and not in those from cells treated with IGFBP-1. Taken together, these findings suggested that IGFBP-1 can function upstream of the MAPK/ERK pathway. Even though HeLa cells contain IGF-I receptor (26), which upon stimulation can activate the ERK pathway (6), the studies were conducted under low-serum conditions and no exogenous IGFs were added. Rodent hepatocytes have undetectable IGF-I receptors (31, 32), and IGFs have not been shown to have a growth-regulatory role in isolated hepatocytes. Thus, the stimulatory effect of IGFBP-1 on ERK1/2 activation in regenerating liver may not be dependent on regulation of IGF activity, but this cannot be ruled out.
If the aberrant induction of C/EBPβ and MAPK/ERK activation was a direct effect of IGFBP-1 deficiency in the IGFBP-1−/− mice, then treating IGFBP-1−/− mice with IGFBP-1 should restore the level of C/EBPβ and the level of ERK1/2 induction to near normal. IGFBP-1 treatment led to a level of ERK1/2 activation similar to that seen in IGFBP-1+/+ livers at 30 min posthepatectomy (Fig. 4F). A single preoperative dose of IGFBP-1 returned ERK1/2 activation to near normal following a two-thirds partial hepatectomy. IGFBP-1 treatment was also sufficient to restore C/EBPβ induction. The absolute level of IGFBP-1 achieved in the IGFBP-1−/− livers by injection was significantly less than in wild-type posthepatectomized controls (Fig. 4D). A single preoperative dose of IGFBP-1 was not sufficient to correct the DNA synthetic defect in IGFBP-1−/− livers following 70% partial hepatectomy, suggesting that a more sustained presence of IGFBP-1 may be required to correct the regenerative defect.
DISCUSSION
In this study, we demonstrated that hepatocyte DNA synthesis posthepatectomy was delayed and reduced in animals that lack IGFBP-1. IGFBP-1−/− mice also had increased liver injury in the first 2 days posthepatectomy associated with elevated liver enzymes and areas of necrosis. Liver mass restoration was normal in IGFBP-1−/− livers. Regaining of liver mass despite deficient DNA synthesis is a general phenomenon in animal models that are defective in liver regeneration that most likely results from hypertrophy of hepatocytes despite G1/S arrest (4, 9, 20, 38). Blunted hepatocyte cell cycle progression in the IGFBP-1−/− livers was associated with reduced and delayed induction of cyclin A and cyclin B1 and reduced expression of cyclin E (a model is shown in Fig. 5). In contrast to IL-6−/− livers, which demonstrate reduced cyclin D1 mRNA and protein expression posthepatectomy (4), normal activation of cyclin D1 was observed in IGFBP-1−/− livers. Interestingly, similar changes in the expression of these cell cycle-regulatory gene products were observed in C/EBPβ−/− livers posthepatectomy (9). Here it was shown that the normal induction of C/EBPβ did not occur in IGFBP-1−/− livers, potentially establishing a link between IGFBP-1 levels and the C/EBPβ-regulated mitogenic pathway (Fig. 5).
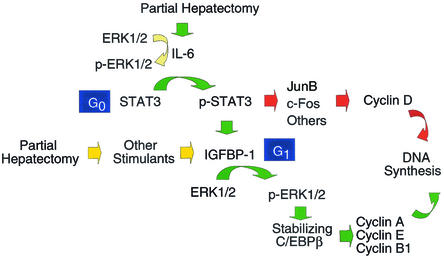
FIG. 5.
Proposed model for IGFBP-1-mediated signal transduction to C/EBPβ posthepatectomy. Upregulation of IGFBP-1, in response to IL-6 and other stimulants posthepatectomy, leads to activation of ERK1/2, which helps to stabilize C/EBPβ, thereby coordinating the progression of hepatocytes into S phase of the cell cycle by regulating the expression of cyclins A, B1, and E.
The C/EBPs (α and β) are leucine-zipper transcription factors that are highly expressed in quiescent livers and are able to heterodimerize with other C/EBP proteins (32). C/EBPα plays a role in specifying mitotic growth arrest, terminal differentiation, or both (37), whereas C/EBPβ expression is required but not sufficient to stimulate cell proliferation (3). Following a two-thirds hepatectomy, an inverse regulation of the C/EBPα and -β proteins results in at least a sevenfold increase in the β-to-α DNA binding ratio between 3 and 24 h posthepatectomy (8, 32), indicating that a coordinated temporal cascade of C/EBPα and -β expression may regulate the balance between cell proliferation and maturational phases of the liver. In hepatectomized IGFBP-1−/− livers, whereas early induction of C/EBPβ did not occur, levels of C/EBPα at the corresponding times were normal or slightly higher (1 h posthepatectomy), suggesting that the α/β ratio was also abnormal. Thus, as in C/EBP−/− livers, the relative elevation of the α/β ratio is likely to contribute to the impaired entry into S phase.
Even though IL-6 has been implicated in playing a role in activating C/EBPβ (1), normal induction of immediate-early genes linked to the IL-6/STAT3 activation pathway in the C/EBPβ−/− livers posthepatectomy suggests that C/EBPβ regulation is independent of the IL-6 pathway (9). This finding is further supported by data that showed normal induction of C/EBPβ expression in IL-6−/− livers following partial hepatectomy (4). In contrast, there was a reduced induction of C/EBPβ protein expression following partial hepatectomy in the IGFBP-1−/− livers, which was corrected with IGFBP-1 treatment. The involvement of IGFBP-1 as a regulator of C/EBPβ protein levels was further indicated by the finding of a greater-than-sevenfold decrease in C/EBPβ expression in IGFBP-1−/− primary hepatocytes. Like for C/EBPβ−/− livers (9), no differences were detected in IL-6 responsive pathways, such as STAT3 activation, and expression of STAT3-responsive genes, such as those for JunB and cyclin D1, in IGFBP-1−/− livers following partial hepatectomy.
Unlike most of the reports which emphasize the establishment of a transcriptional cascade as a means to regulate target gene expression following two-thirds partial hepatectomy, our study suggests that IGFBP-1 functions to help stabilize the C/EBPβ protein and to activate ERK1/2 expression by posttranslational modification (Fig. 5). Normal induction of C/EBPβ mRNA and ERK1/2 protein expression were observed in the IGFBP-1-deficient livers posthepatectomy. Based on our results, treatment of IGFBP-1−/− mice with IGFBP-1 increased the total (phosphorylation state-independent) C/EBPβ protein level in the liver to normal. However, full restoration of ERK1/2 activation occurred only in posthepatectomized IGFBP-1 treated IGFBP-1−/− livers, suggesting that IGFBP-1 cooperates with other hepatectomy-induced growth factors or cytokines to posttranslationally modify MAPK. For instance, activation of ERK1 (p44 MAPK) precedes ERK2 (p42 MAPK) phosphorylation in IL-6−/− livers pretreated with IL-6 (19). However, IGFBP-1 treatment alone led to elevation of the p42 MAPK level in IGFBP-1−/− livers. Since IGFBP-1 mRNA and protein expression are rapidly induced 1 h after IL-6 treatment (18), it is conceivable that IGFBP-1 cooperates with IL-6 or other IL-6 induced growth factors to elicit the activation of ERK1/2 during liver regeneration.
The underlying mechanism leading to the stabilization of C/EBPβ by IGFBP-1 during liver regeneration awaits further elucidation. It is known that in aplysia, phosphorylation by MAPK is required for aplysia CCAAT/enhancer binding protein to act as a transcription activator and to prevent aplysia CCAAT/enhancer binding protein from being degraded through the ubiquitin-proteasome pathway (39). Similarly, inhibition of the ubiquitin/proteasome pathway can lead to the upregulation of Gadd153 and ATF3, both of which belong to the CCAAT/enhancer binding protein (C/EBP) family (40).
Our report showed that an appropriate level of IGFBP-1 is critical for proper control of the hepatocyte cell cycle. It is also the first report to show that a member of the IGFBP family may regulate mitogenic signaling by activating MAPK and C/EBPβ protein levels in vivo. Presently it is not clear if regulation of hepatocyte proliferation by IGFBP-1 is mediated via IGF-dependent or -independent mechanisms. However, it should be noted that IGF-I receptors are expressed at a very low level in hepatocytes, and IGF-I has not been shown to be a hepatocyte mitogen in vitro (24). Moreover, in human hepatoma cell lines, dephosphorylated IGFBP-1 may be mitogenic independent of IGF-I (13, 17). Human conditions in which elevations of IGFBP-1 expression are observed include cirrhosis, in which some degree of liver regeneration may occur (17), and human hepatocellular carcinomas, in which IGFBP-1 has been detected as a highly expressed gene (14). Overexpression of IL-6 and soluble IL-6 receptor in transgenic mice causes massive elevation of IGFBP-1 and the early development of hepatocellular hyperplastic lesions and late formation of liver adenomas (22). These data provide initial insight into a potential role for IGFBP-1 in stimulating growth of the liver in human clinical conditions and malignant transformation.
Acknowledgments
We thank Gary Swain at the University of Pennsylvania School of Medicine's Digestive and Liver Center for help with hematoxylin and eosin staining, BrdU immunohistochemistry, and imaging.
This work was supported in part by Digestive and Liver Center grant P30 DK50306 (technical support) and grants NIH DK 58315 and DK 49629 (to R.T.).
REFERENCES
- 1.Akira, S., H. Isshiki, T. Sugita, O. Tanabe, S. Kinoshita, Y. Nishio, T. Nakajima, T. Hirano, and T. A. Kishimoto. 1990. Nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, J. H., R. Y. Poon, C. L. Ahonen, B. M. Rieland, C. Deng, and G. S. Crary. 1998. Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene 16:2141-2150. [DOI] [PubMed] [Google Scholar]
- 3.Buck, M., V. Poli, T. Hunter, and M. Chojkier. 2001. C/EBPβ phosphorylation by RSK creates functional XEXD caspase inhibitory box critical for cell survival. Mol. Cell 8:807-816. [DOI] [PubMed] [Google Scholar]
- 4.Cressman, D. E., L. E. Greenbaum, R. A. DeAngelis, G. Ciliberto, E. E. Furth, V. Poli, and R. Taub. 1996. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 22:1379-1383. [DOI] [PubMed] [Google Scholar]
- 5.Crissey, M. A,. J. I. Leu, R. A. De Angelis, L. E. Greenbaum, L. M. Scearce, K. Kovalovich, and R. Taub. 1999. Liver specific and proliferation-induced deoxyribonuclease I hypersensitive sites in the mouse insulin-like growth factor binding protein-1 gene. Hepatology 30:1187-1197. [DOI] [PubMed] [Google Scholar]
- 6.Dews, M., M. Prisco, F. Peruzzi, G. Romano, A. Morrione, and R. Baserga. 2000. Domains of the insulin-like growth factor I receptor required for the activation of extracellular signal-regulated kinases. Endocrinology 141:1289-1300. [DOI] [PubMed] [Google Scholar]
- 7.Factor, V. M., and S. S. Thorgeirsson. 1997. Coexpression of c-myc and transforming growth factor alpha in the liver promotes early replicative senescence and diminishes regenerative capacity after partial hepatectomy in transgenic mice. Hepatology 26:1434-1443. [DOI] [PubMed] [Google Scholar]
- 8.Greenbaum, L. E., D. E. Cressman, B. A. Haber, and R. Taub. 1995. Coexistence of C/EBPα, β, growth-induced proteins and DNA synthesis in hepatocytes during liver regeneration. J. Clin. Investig. 96:1351-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenbaum, L. E., W. Li, D. E. Cressman, Y. Peng, G. Ciliberto, V. Poli, and R. Taub. 1998. CCAAT enhancer binding protein β is required for normal hepatocyte proliferation in mice after partial hepatectomy. J. Clin. Investig. 102:996-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haber, B., L. Naji, D. Cressman, and R. Taub. 1995. Coexpression of liver-specific and growth-induced genes in perinatal and regenerating liver: attainment and maintenance of the differentiated state during rapid proliferation. Hepatology 22:906-914. [PubMed] [Google Scholar]
- 11.Hengelin, B., and C. Brechot. 1992. Cyclin A is required in S phase in normal epithelial cells. Biochem. Biophys. Res. Commun. 182:1144-1154. [DOI] [PubMed] [Google Scholar]
- 12.Higgins, G. M., and R. M. Anderson. 1931. Restoration of the liver of the white rat following partial surgical removal. Arch. Pathol. 12:656-665. [Google Scholar]
- 13.Kim, D.-G., D. Lee, C. Baik-Hwan, Y. Kyung-Ran, and K. Mi-Young. 1999. Down regulation of insulin-like growth factor binding proteins and growth modulation in hepatoma cells by retinoic acid. Hepatology 29:1091-1098. [DOI] [PubMed] [Google Scholar]
- 14.Kondoh, N., T. Wakasuki, A. Ryo., A. Hada, T. Aihara, S. Horiuchi, N. Goseki, O. Atsubara, K. Takenaka, M. Shichita, K. Tanaka, M. Shuda, and M. Yamamoto. 1999. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 59:4990-4996. [PubMed] [Google Scholar]
- 15.Kovalovich, K., W. Li, R. DeAngelis, L. E. Greenbaum, G. Ciliberto, and R. Taub. 2001. Interleukin-6 protects against Fas-mediated death by establishing a critical level of anti-apoptotic hepatic proteins FLIP, Bcl-2, and Bcl-xL. J. Biol. Chem. 276:26605-26613. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J., L. Greenbaum, B. A. Haber, D. Nagle, V. Lee, V. Miles, K. L. Mohn, M. Bucan, and R. Taub. 1994. Structure and localization of the IGFBP-1 gene and its expression during liver regeneration. Hepatology 19:656-665. [DOI] [PubMed] [Google Scholar]
- 17.Lee, P. D. K., L. C. Giudice, C. A. Conover, and D. R. Powell. 1997. Insulin-like growth factor binding protein-1: recent findings and new directions. Proc. Soc. Exp. Biol. Med. 216:319-357. [DOI] [PubMed] [Google Scholar]
- 18.Leu, J. I., M. S. Crissey, J. P. Leu, G. Ciliberto, and R. Taub. 2001. Interleukin-6-induced STAT3 and AP-1 amplify hepatocyte nuclear factor-1-mediated transactivation of hepatic genes, an adaptive response to liver injury. Mol. Cell. Biol. 21:414-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, W., X. Liang, J. I. Leu, K. Kovalovich, G. Ciliberto, and R. Taub. 2001. Global changes in interleukin-6-dependent gene expression patterns in mouse livers after partial hepatectomy. Hepatology 33:1377-1386. [DOI] [PubMed] [Google Scholar]
- 20.Li, W., X. Liang, C. Kellendonk, V. Poli, and R. Taub. 2002. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J. Biol. Chem. 277:28411-28417. [DOI] [PubMed] [Google Scholar]
- 21.Loyer, P., S. Cariou, D. Glais, M. Bilodeau, G. Baffet, and C. Guguen-Guillouzo. 1996. Growth factor dependence of progression through G1 and S phases of adult rat hepatocytes in vitro. Evidence of a mitogen restriction point in mid-late G1. J. Biol. Chem. 271:11484-11492. [DOI] [PubMed] [Google Scholar]
- 22.Maione, D., E. Di Carlo, W. Li, P. Musiani, A. Modesti, M. Peters, S. Rose-John, C. Della Rocca, M. Tripodi, D. Lazzaro, R. Taub, R. Savino, and G. Ciliberto. 1998. Coexpression of IL-6 and soluble IL-6 receptor causes nodular regenerative hyperplasia and adenomas of the liver. EMBO J. 17:5588-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michalopoulos, G. K., and M. C. DeFrances 1997. Liver regeneration. Science 276:60-66. [DOI] [PubMed] [Google Scholar]
- 24.Mohn, K. L., A. E. Melby, D. S. Tewari, T. M. Laz, and R. Taub. 1991. The gene encoding rat insulin like growth factor binding protein 1 is rapidly and highly induced in regenerating liver. Mol. Cell. Biol. 11:1393-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, L. J. 2000. Overexpression of insulin-like growth factor binding protein-1 in transgenic mice. Pediatr. Nephrol. 14:567-571. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, K., A. Hongo, J. Kodama, Y. Miyagi, M. Yoshinouchi, and T. Kudo. 2000. Down-regulation of the insulin-like growth factor I receptor by antisense RNA can reverse the transformed phenotype of human cervical cancer cell lines. Cancer Res. 60:760-765. [PubMed] [Google Scholar]
- 27.Poli, V., F. P. Mancini, and R. Cortese. 1990. IL-6DBP, a nuclear protein involved in interleukin-6-signal transduction defines a new family of leucine zipper proteins related to C/EBP. Cell 63:643-653. [DOI] [PubMed] [Google Scholar]
- 28.Scearce, L. M., T. M. Laz, T. G. Hazel, L. F. Lau, and R. Taub. 1996. Rapid activation of latent transcription factor complexes reflects initiating signals in liver regeneration. Death Differ. 3:47-55. [PubMed] [Google Scholar]
- 29.Spector, M. S., K. L. Auer, W. D. Jarvis, E. J. Ishac, B. Gao, G. Kunos, and P. Dent. 1997. Differential regulation of the mitogen-activated protein and stress activated protein kinase cascades by adrenergic agonists in quiescent and regenerating adult rat hepatocytes. Mol. Cell. Biol. 17:3556-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talarmin, H,. C. Rescan, S. Cariou, D. Glaise, G. Zanninelli, M. Bilodeau, P. Loyer, C. Guguen-Guillouzo, and G. Baffet. 1999. The mitogen-activated protein kinase kinase/extracellular signal-regulated kinase cascade activation is a key signaling pathway involved in the regulation of G1 phase progression in proliferating hepatocytes. Mol. Cell. Biol. 19:6003-6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taub, R. 1995. Expression and function of growth induced genes during liver regeneration, p. 71-97. In R. L. Jirtle (ed.), Liver Regenation and Carcinogenesis. Academic Press, Inc., N.Y.
- 32.Taub, R. 1996. Liver regeneration 4: transcriptional control of liver regeneration. FASEB J. 10:413-427. [PubMed] [Google Scholar]
- 33.Taub, R., L. E. Greenbaum, and Y. Peng. 1999. Transcriptional regulatory signals define cytokine-dependent and independent pathways in liver regeneration. Semin. Liver Dis. 19:117-127. [DOI] [PubMed] [Google Scholar]
- 34.Trembley, J. H., J. O. Ebbert, B. T. Kren., and C. J. Steer. 1996. Differential regulation of cyclin B1 RNA and protein expression during hepatocyte growth in vivo. Cell Growth Differ. 7:903-916. [PubMed] [Google Scholar]
- 35.Trembley, J. H., B. T. Kren, and C. J. Steer. 1994. Posttranscriptional regulation of cyclin B messenger RNA expression in the regenerating rat liver. Cell Growth Differ. 5:99-108. [PubMed] [Google Scholar]
- 36.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 37.Wang, N. D., M. J. Finegold, A. Bradley, C. N. Ou, S. V. Abdelsayed, M. D. Wilde, L. R. Taylor, D. R. Wilson, and G. J. Darlington. 1995. Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269:1108-1112. [DOI] [PubMed] [Google Scholar]
- 38.Yamada, Y., I. Kirillova., J. J. Peschon, and N. Fausto. 1997. Initiation of liver growth by tumor necrosis factor: defective liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc. Natl. Acad. Sci. USA 94:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto, N., A. N. Hegde, D. G. Chain, and J. H. Schwartz. 1999. Activation and degradation of the transcription factor C/EBP during long-term facilitation in aplysia. J. Neurochem. 73:2415-2423. [DOI] [PubMed] [Google Scholar]
- 40.Zimmermann, J., D. Erdmann, I. Lalande, R. Grossenbacher, M. Noorani, and P. Furst. 2000. Proteasome inhibitor induced gene expression profiles reveal overexpression of transcriptional regulators ATF3, GADD153 and MAD1. Oncogene 19:2913-2920. [DOI] [PubMed] [Google Scholar]