Abstract
Coactivators constitute a diverse group of proteins that are essential for optimal transcriptional activity of nuclear receptors. In the past few years many coactivators have been identified but it is still unclear whether these proteins interact indiscriminately with all nuclear receptors and whether there is some redundancy in their functions. We have previously cloned and characterized RAP250 (ASC-2/PRIP/TRBP/NRC), an LXXLL-containing coactivator for nuclear receptors. In order to study its biological role, Rap250 null mice were generated by gene targeting. Here we show that genetic disruption of Rap250 results in embryonic lethality at embryonic day (E) 13.5. Histological examination of placentas revealed a dramatically reduced spongiotrophoblast layer, a collapse of blood vessels in the region bordering the spongiotrophoblast, and labyrinthine layers in placentas from Rap250−/− embryos. These findings suggest that the lethality of Rap250−/− embryos is the result of obstructed placental blood circulation. Moreover, the transcriptional activity of PPARγ is reduced in fibroblasts derived from Rap250−/− embryos, suggesting that RAP250 is an essential coactivator for this nuclear receptor in the placenta. Our results demonstrate that RAP250 is necessary for placental development and thus essential for embryonic development.
Nuclear receptors comprise a family of transcription factors that regulate gene expression in a ligand-dependent manner. Upon ligand binding these proteins activate transcription of specific genes involved in the control of diverse physiological processes, such as cellular growth, development, differentiation, and homeostasis (20). Binding of ligands to the ligand-binding domain of nuclear receptors causes conformational changes of the receptor, enabling the recruitment of coactivators harboring LXXLL motifs (see below). A number of coactivators for nuclear receptors have been identified (for a review, see reference 6). Among the most studied is the steroid receptor coactivator 1 (SRC-1) family, which contains three related coactivators, referred to as SRC-1, SRC-2/GRIP1/TIF2, and SRC-3/p/CIP/RAC3/ACTR/AIB1/TRAM1 (1, 4, 7, 17, 23, 28, 30, 33). These factors, together with CBP/p300 and pCAF, bind nuclear receptors and gain access to target promoter regions through histone acetyltransferase-mediated nucleosome remodeling. Another class of coactivators is represented by the TRAP/DRIP/ARC multiprotein complex (5, 22, 26), with one major nuclear receptor interacting subunit, PBP/TRAP220/DRIP205 (25, 31, 41, 45), that appears to act independently of histone acetyltransferase activity.
Nuclear receptor-activating protein 250 (RAP250) (3), also called ASC-2, PRIP, TRBP, and NRC (13, 15, 19, 42), was recently cloned and described as a novel nuclear receptor coactivator which interacts with the ligand-binding domain of ligand-bound nuclear receptors. The interaction is mediated through a short hydrophobic motif called the LXXLL motif (or NR box) which is found in most nuclear receptor coactivators. In contrast to other nuclear receptor coactivators, RAP250 only uses one LXXLL motif when interacting with nuclear receptors. A second LXXLL motif is located in the C-terminal part of the protein, but it appears to be specific for interactions with the liver X receptor β (16). RAP250 has a unique structure and does not belong to a previously defined coactivator family. In addition, no known homologous genes in Saccharomyces cerevisiae or Drosophila melanogaster have yet been reported. RAP250 is expressed in many tissues, and high levels of RAP250 mRNA have been detected in reproductive organs, brains, hearts, and blood (3, 13, 42). During development, RAP250 is widely expressed, with high levels of mRNA in placenta and various parts of the brain (3). The RAP250 gene is also amplified in human breast, colon, and lung cancers (15). The mechanism by which RAP250 activates transcription is not fully understood; however, the protein has an intrinsic activation domain that is likely to contribute to transcriptional activation but does not contain any histone acetyltransferase activity (3, 16, 19). Recently, three different proteins containing RNA-binding domains, termed PIMT, CoAA, and CAPER, were shown to bind to and increase the activity of RAP250 (10, 11, 43), and a role for RAP250 as a link between CBP/p300 and TRAP/DRIP/ARC coactivator complexes has been proposed (21).
The use of knockout technology has helped to elucidate the physiological function of several nuclear receptor coactivators. Mice deficient in SRC-1 are viable and show only minor resistance to steroids and thyroid hormones, indicating redundancy in coactivator functions (24, 35, 38). SRC-3 knockout mice are also viable but show growth retardation and reduced female reproductive function (34, 37). Both Cbp and p300 null mutant mice die in utero, and mice heterozygous for either of these genes have severe abnormalities (29, 40). An embryonically lethal phenotype, resulting from defects in the heart and placenta, was observed when the nuclear receptor interacting component of the TRAP/DRIP/ARC complex was disrupted (9, 44).
To investigate the biological role of RAP250 in vivo, we generated mice lacking Rap250. In this report, we show that RAP250 is required for embryonic development, as disruption of the Rap250 gene in mice results in vascular dysfunction of the placenta and embryonic lethality.
MATERIALS AND METHODS
Construction of RAP250 targeting vector.
To isolate the mouse Rap250 gene, a 129/SvJ genomic phage library (Stratagene) was screened with a mouse Rap250 cDNA probe (EcoRI fragment from pGAD-RAP250 [3]). One of the isolated clones, clone 4, contained an 18-kb genomic region including five exons of the murine Rap250 locus. To construct a targeting vector, a 7-kb BamHI fragment including exons 5 to 7 was subcloned into the plasmid pBluescript KS+ (Stratagene). The TK gene was then subcloned into the SalI site of this plasmid, generating the plasmid p4B1-TK. In a second plasmid, a 7-kb HindIII fragment including exons 8 and 9 was subcloned. The generated plasmid was cut with SalI and XbaI and the neo gene was inserted at those sites. In the next step, this plasmid was cut with KpnI (partial digestion) and a KpnI fragment from p4B1-TK was subcloned into this site, which generated the targeting vector. In the targeting vector, exons 7 and 8 are replaced with the neo gene and the neo gene is flanked by a 5-kb homologous region on each side. The TK gene is located at the 5′ end of the insert (see Fig. 1A for maps).
FIG. 1.
Targeted disruption of the mouse Rap250 gene. (A) Structures of the wild-type Rap250 allele, targeting vector, and targeted allele are shown with the BamHI (B), HindIII (H), KpnI (K), and XbaI (X) restriction sites and primers for PCR screening. The locations of probes used in Southern blot analysis are indicated. Black boxes indicate exons. The exon numbering is adapted from the work of Zhu et al. (42). (B) Homologous recombination in ES cells. Southern blot analysis of DNA from two positive ES clones, clone 1B8 and 2D2, digested with BamHI is shown. The 5-kb band for the wild-type allele (WT) and the 11-kb band for the targeted allele (KO) are indicated. The probes used are indicated in Fig. 1A. (C) Southern blot analysis of mouse yolk sac DNA digested with BamHI. (D) PCR genotyping of mouse embryo DNA. The bottom band, using the U6 and L8 primers, represents the wild-type allele and the top band, using the KOF1 and L3 primers, represents the mutated allele. M, molecular marker (1-kb ladder). (E) Photo of wild-type and Rap250−/− embryos at E13.5. No obvious abnormalities or significant size differences were detected between wild-type and Rap250−/− littermates.
Generation of RAP250 knockout mice.
RW4 embryonic stem (ES) cells were electroporated with the linearized targeting vector and selected with G418 and ganciclovir on embryonic fibroblast feeder cells. Resistant clones were analyzed by Southern blotting after BamHI digestion by use of a 3′ external probe as illustrated in Fig. 1A. Two ES cell lines exhibiting homologous recombination were injected into C57BL/6 blastocysts that were implanted into pseudopregnant females. Chimeric male mice were bred to C57BL/6 females and germ line transmission of the targeted allele was examined in the agouti offspring by Southern blot and PCR analysis. The mice and embryos analyzed are of a mixed 129SvJ-C57BL/6 genetic background. All animal experimentation was conducted in accordance with accepted standards of humane animal care.
Genotyping of mice and embryos.
DNA from embryo tail tips and yolk sacs was genotyped by PCR amplification using primers L8 (5′-TTCCTACTCATGCCCACAT-3′) and U6 (5′-ATTCCCCTCAGTATGCTAGA-3′), which detect intron 7 of the wild-type allele, whereas primers L3 (5′-AGCAGTCTAGTTAGTAGACA-3′) from intron 8 and KOF1 (5′-TAGGTCTCTTAAGCAGAGGTACCGG-3′) overlapping the neomycin cassette were used to detect the targeted allele. Southern blot analysis of BamHI-digested genomic DNA using a 3′ external probe (HindIII/SacI fragment from exon 9 and downstream sequence) was done to confirm PCR genotyping results.
Histology, immunohistochemistry, and TUNEL assay.
Placentas and embryos were collected and fixed in 4% paraformaldehyde for 14 h, dehydrated, and embedded in paraffin. Serial sections were cut to 4 μm and stained with hematoxylin and eosin or periodic acid-Schiff stain (PAS) for microscopic examination. For immunohistochemistry, deparaffinized and rehydrated sections were pretreated with 0.3% H2O2 in phosphate-buffered saline (PBS) for 15 min and then washed in PBS. The sections were blocked for 30 min in 5% horse serum in PBS and then incubated with anti-von Willebrand polyclonal antibody (Dako) diluted 1:500 or anti-prolactin (C-17) polyclonal antibody (Santa Cruz Biotechnology) diluted 1:2,000 in 5% horse serum in PBS for analysis of placenta or with anti-MAP-2B (Becton Dickinson) for analysis of embryo. Antibody binding was detected by sequential incubation of the sections with biotinylated goat anti-rabbit serum or anti-mouse serum and streptavidin-peroxidase complex (Vector). Positive staining was detected by a substrate reaction with diaminobenzidine. Sections were counterstained with Gill's hematoxylin and mounted in Permount (Fisher). To detect DNA fragmentation in situ, sections were analyzed by the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) method using an in situ Cell Death Detection kit (Roche) according to the manufacturer's protocol.
Isolation of fibroblasts from embryos, RT-PCR, Western blot analysis, and transient transfections.
Mouse embryonic fibroblasts (MEFs) were isolated from E13.5 embryos. After removal of the head and inner organs and digestion with collagenase, the cells were cultured in Dulbecco's modified Eagle's high-glucose medium with 10% fetal calf serum and sodium pyruvate. Total RNA was prepared by using a kit (Qiagen). Reverse transcription (RT)-PCR was carried out using poly(dT) as a primer for cDNA synthesis and Superscript II reverse transcriptase (Life Technologies). The PCR primers used to detect Rap250 expression were ex5F (5′-TCTCCAGGTCGGAATCCTAT) and ex7R (5′-TTTGTCGACTGGACGATTATCTGGGGTGT). Preparation of nuclear extracts and Western blot analysis were performed as described previously (3). The rabbit anti-RAP250 antibody was derived against amino acids 818 to 931 of human RAP250 fused to glutathione S-transferase. Transient transfections were done with Lipofectin (Life Technologies) according to the manufacturer's instructions using the following plasmids: pUAS-tk-Luc, pGAL4-PPARγ, pGAL4-RXRα (3), pGAL4-EERβ (generous gift from S. Sanyal), and pCMVβ (Clontech). Cell extracts were prepared 24 h after transfection and assayed for luciferase and β-galactosidase activities.
RESULTS
Targeted disruption of Rap250 gene in mice.
To investigate the biological role of RAP250 in vivo, we used homologous recombination in ES cells for the generation of mice in which the Rap250 gene was disrupted. Genomic mouse Rap250 clones were isolated from libraries using a cDNA probe. To understand the genomic organization of the gene, one of the isolated clones was sequenced, and the corresponding intron-exon structure for that part of the gene was determined (Fig. 1A). RAP250 contains one functional NR box and an intrinsic transactivation domain that are considered necessary for enhancement of transcription activation by nuclear receptors. Therefore, we expected that removal of these regions of the Rap250 gene would eliminate the function of RAP250 as a nuclear receptor coactivator. These two functional domains were localized to exon 7. The targeting vector was designed to delete exons 7 and 8, including the region encoding amino acids 563 to 975 of RAP250, and to replace this region with the neomycin gene (Fig. 1A). ES cells were electroporated with targeting vectors and subjected to positive and negative selection. Ganciclovir- and G418-resistant clones were isolated and analyzed for homologous recombination by Southern blot analysis using both internal and 3′-flanking probes (Fig. 1B). Two of these targeted ES cell lines, 1B8 and 2D2, were injected into C57BL/6 blastocysts to produce chimeric mice. Both clones contributed to the germ line.
Disruption of the Rap250 gene results in embryonic lethality.
Mice heterozygous for the Rap250 mutation were viable and fertile and appeared phenotypically indistinguishable from their wild-type littermates. Genotype analysis (Fig. 1C and D) of progenies obtained from crosses between F1 Rap250+/− mice revealed that 39% were wild type and 61% were heterozygous, whereas no homozygous mutated mice were born (Table 1). To determine the time point of embryonic lethality, heterozygous females from timed matings with heterozygous males were sacrificed at various days postconception, and the genotype of the embryos was analyzed (Table 1). No homozygous mutant embryos remained alive, except for two which were severely growth retarded, at E14.5 or later. At E13.5 Rap250−/− embryos showed no gross abnormalities or great differences in size compared to their wild-type littermates (Fig. 1E). To verify that the Rap250 gene was not expressed in mutant embryos we used RT-PCR on RNA extracted from fibroblasts derived from Rap250−/− embryos. Rap250 mRNA was present in fibroblasts derived from wild-type embryos but not from Rap250−/− embryos (Fig. 2A). This result was further confirmed by Western blot analysis of nuclear extracts from MEFs (Fig. 2B).
TABLE 1.
Genotypes of progeny from crosses of F1 Rap250+/− mice
| Stage | No. of offspring with genotype
|
Total no. of offspring | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| P21a (clone 1B8) | 62 | 96 | 0 | 158 |
| P21 (clone 2D2) | 16 | 24 | 0 | 40 |
| E16.5-E14.5 | 27 | 48 | 2b | 77 |
| E13.5 | 11 | 29 | 9 | 49 |
P, postnatal day.
Severely retarded in growth and development.
FIG. 2.
Absence of Rap250 mRNA and protein in Rap250−/− embryonic fibroblasts. (A) RT-PCR analysis of total RNA from wild-type (+/+) and Rap250−/− (−/−) mouse embryonic fibroblasts. Expression of Rap250 is detected in wild-type but not null cells, and actin mRNA is detected in both cell types. M, molecular marker (1-kb ladder); 0, negative control without DNA. (B) Western blot analysis of nuclear extracts from wild-type (WT) and null (KO) mouse embryonic fibroblasts using RAP250 polyclonal antibodies. RAP250 protein, indicated by an arrow, is detected in wild-type but not null fibroblasts.
Developmental defects in placentas of Rap250−/− mice.
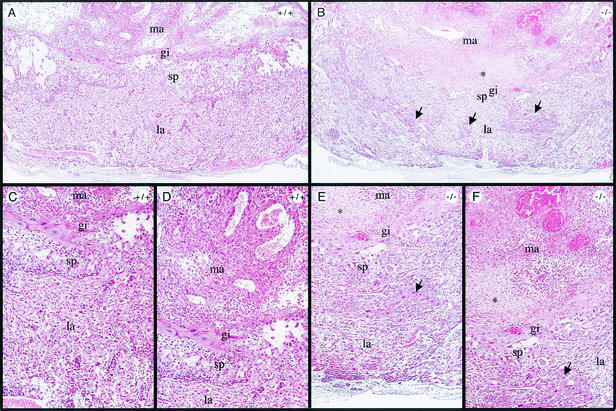
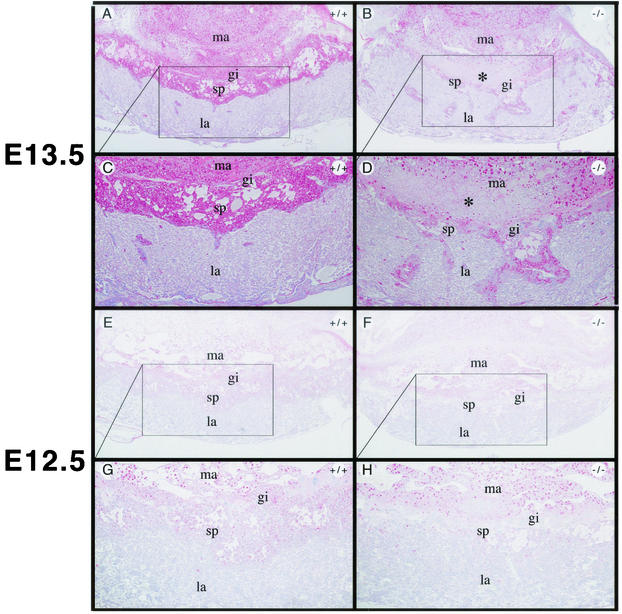
Histological analysis of placentas from Rap250 null embryos at E13.5 revealed that the overall structure was disorganized (Fig. 3A and B). In wild-type placentas, the labyrinthine layer, spongiotrophoblast layer, and giant cell layer are localized in distinct layers (Fig. 3A, C, and D). However, the spongiotrophoblast layer in Rap250 null placentas is markedly reduced and the labyrinthine layer contains areas of spongiotrophoblast-like cells (Fig. 3B, E, and F). In addition, the labyrinthine layer of Rap250−/− placentas is less vascularized than that of wild-type placentas, and an area with necrosis at the maternal-embryonal interface was observed. Placentas from Rap250−/− embryos at E12.5 did not show any signs of necrosis. Sections of placenta stained with PAS, which stains glycogen-containing cells, showed that the spongiotrophoblast layer of Rap250 null placentas is markedly reduced at both E13.5 (Fig. 4A to D) and E12.5 (Fig. 4E to H). We also observed that the labyrinthine layer of Rap250−/− placentas contained islands of spongiotrophoblast-like cells that were not detected in the wild type. We next visualized spongiotrophoblast and giant cells in placentas with an anti-prolactin antibody that recognizes prolactin and prolactin-related proteins. As shown in Fig. 5, placentas from Rap250−/− embryos showed much less staining both at E13.5 and E12.5. The reduced staining is mainly due to the reduced number of spongiotrophoblast cells because the number of giant cells was approximately the same in placentas from wild-type and Rap250−/− embryos. These results suggest a progressive developmental defect in Rap250 null embryos.
FIG. 3.
Developmental defects in Rap250−/− placentas, as shown by hematoxylin and eosin staining of Rap250+/+ and Rap250−/− placentas at E13.5. Overview pictures of wild-type (A) and Rap250−/− (B) placentas at E13.5 are shown. (C and D) Higher-power views of panel A. (E and F) Higher-power views of panel B. The spongiotrophoblast layer is markedly reduced in null placentas compared to the wild type, and the labyrinthine layer contains islands of spongiotrophoblast-like cells, indicated by arrows. The labyrinthine layer is less dense in null placentas than in wild-type placentas. Note an area of cell death in null placentas in the maternal decidua bordering the giant cell layer indicated by an asterisk. gi, giant cells; la, labyrinthine layer; ma, maternal decidua; sp, spongiotrophoblast layer.
FIG. 4.
Reduced spongiotrophoblast layer in Rap250−/− placentas, as shown by PAS staining, which stains glycogen-containing cells, of sections of wild-type and Rap250−/− placenta. Glycogen-containing cells appear in pink. (A and B) Sections of PAS staining of wild-type and Rap250−/− placentas, respectively, at E13.5. (C and D) Higher-power views of panels A and B, respectively. The number of stained cells in the spongiotrophoblast layer is markedly reduced in Rap250−/− placentas compared to wild-type placentas. While the border between the labyrinth and spongiotrophoblast layers is distinct in wild-type placentas, it is not as sharp in null placentas. The labyrinthine layer of Rap250−/− placentas contains several islands of glycogen-containing spongiotrophoblast-like cells that are not seen in wild-type placentas. An area with cell death in the maternal decidua of Rap250−/− placentas is indicated by an asterisk. (E and F) PAS staining of sections of wild-type and Rap250−/− placentas, respectively, at E12.5. (G and H) Higher-power views of panels E and F. The number of stained cells in the spongiotrophoblast layer is markedly reduced in Rap250−/− placentas compared to wild-type placentas. gi, giant cells; la, labyrinthine layer; ma, maternal decidua; sp, spongiotrophoblast layer.
FIG. 5.
Reduced expression of prolactin-like proteins in Rap250−/− placentas. Staining of wild-type and Rap250−/− placentas with antibodies against prolactin. Immunohistochemical analysis of wild-type (A and E) and Rap250−/− (B and F) placentas at E13.5 and E12.5 with antibodies against prolactin is shown. (C, D, G, and H) Higher-power views of panels A, B, E, and F, respectively. The number of stained cells in the spongiotrophoblast layer is markedly reduced in Rap250−/− placentas compared to wild-type placentas. gi, giant cells; la, labyrinthine layer; ma, maternal decidua; sp, spongiotrophoblast layer.
Vascular defects in placentas of Rap250−/− mice.
The necrotic area detected in Rap250 null placentas at E13.5 (Fig. 3) suggested the presence of vascular alterations and we next visualized endothelial cells of the placentas by use of antibodies against von Willebrand's factor. We observed that blood vessels in the region bordering the spongiotrophoblast and labyrinthine layers had collapsed in Rap250−/− placentas (Fig. 6, compare panels A and C with panels E and G). The collapse of blood vessels is restricted to the vessels at the border of the spongiotrophoblast and labyrinthine domains of the placenta, as the morphology of vessels of the maternal decidua and labyrinthine layer appeared normal. The region in proximity of the collapsed vessels showed signs of necrosis. We therefore analyzed the placental sections for DNA degradation using the TUNEL technique. The areas close to the collapsed vessels of the Rap250−/− placentas were positive for TUNEL staining, displaying a staining pattern characteristic of necrotic tissue, thus confirming that these areas were necrotic (Fig. 6B and D). However, no DNA degradation was apparent in null placentas at E12.5 (data not shown), and the corresponding tissue of Rap250+/− placentas did not show any signs of DNA degradation (Fig. 6F and H). The severe defects detected in the blood vessels bordering the labyrinth and spongiotrophoblast layers of knockout mouse placentas suggest that blood circulation was obstructed, resulting in local necrosis. We argue that these vascular defects cause placental ischemia and subsequent embryonic death.
FIG. 6.
Vascular defects in Rap250−/− placentas. The collapse of vessels bordering the labyrinth and the spongiotrophoblast region of Rap250−/− placentas at E13.5 is shown. Immunohistochemical analysis was performed for Rap250−/− (A and C) and Rap250+/− (E and G) extra-embryonic tissues. Panels A, C, E, and G show staining with antibodies against von Willebrand's factor. Note the release of von Willebrand's factor in the collapsed vessel of the labyrinth and the spongiotrophoblast region of the Rap250−/− placenta. In contrast, the placental vessels in the labyrinth region were intact and positive staining was only detected in the endothelial cells. Similar vessels of Rap250+/− placentas were unaffected. (B, D, F, and H) TUNEL staining to detect DNA fragmentation. An area juxtaposed to the collapsed vessels shown in panels B and D stained positive with TUNEL staining (marked with an asterisk). Bars: A, B, E, and F, 125 μm; C, D, G, and H, 50 μm.
Deficiency in heart and neural development in Rap250−/− mice.
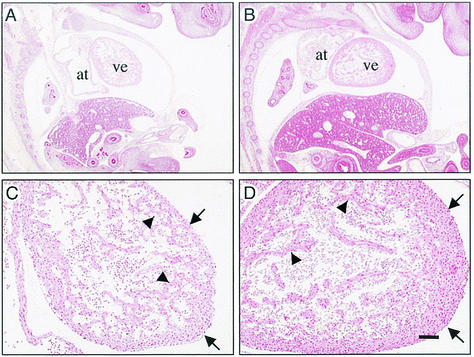
Examination of hematoxylin- and eosin-stained sagittal sections of Rap250−/− embryos revealed no gross developmental defects. However, myocardial walls of both atriums and ventricles of Rap250−/− embryos were thinner than those of wild-type embryos at E13.5 (Fig. 7A and B). At a higher magnification, Rap250−/− embryos showed hypoplasia of ventricular walls and the trabecular zone (Fig. 7C and D). Histological examination of the E13.5 embryos showed retarded development in most areas of the brain. The neopallial cortex was significantly thinner in Rap250−/− embryos than in wild-type littermates (Fig. 8A and B). Immunohistological staining with anti-MAP2B monoclonal antibody, an antibody against differentiated neurons, indicated a reduced number of differentiated neurons in the neopallial cortex in Rap250−/− embryos (Fig. 8C and D). The olfactory lobes, which can be easily distinguished in wild-type embryos at this stage, were not seen in Rap250−/− mice (Fig. 8E and F). Differentiation and enlargement of the thalamus and hypothalamus regions of Rap250−/− embryos were not as well developed as in wild-type littermates, and the volume of the third ventricle was correspondingly enlarged in the mutant embryos (Fig. 8E and F). These morphological findings suggest that neuroepithelial cells of the Rap250−/− embryos are defective in cell proliferation.
FIG. 7.
Histological abnormalities of hearts from Rap250−/− embryos at E13.5, as shown by hematoxylin and eosin staining of sagittal sections of Rap250−/− and wild-type embryos. (A and B) Hematoxylin and eosin staining of sagittal sections of Rap250−/− and wild-type embryos, respectively. Myocardial walls are thinner in Rap250−/− embryos than in wild-type littermates. at, atrium; ve, ventricle. Bar = 400 μm. (C and D) Hypoplasia in ventricular walls (arrows) and degeneration of trabecular zone (arrowheads) in Rap250−/− embryos and wild-type littermates, respectively. Bar = 100 μm.
FIG. 8.
Defective neural development in Rap250−/− mice. (A and B) Neopallial cortex of E13.5 embryos. Note the significantly thinner layer (arrows) of the Rap250−/− embryo (A) compared to that of the wild-type embryo (B). Bar = 50 μm. (C and D) Immunohistochemical analysis of neopallial cortex from Rap250−/− and wild-type embryos, respectively, at E13.5 with antibodies against MAP-2B. Less-differentiated neurons are seen in the neopallial cortex of Rap250−/− embryos than in the wild type. Bar = 25 μm. (E and F) Overview of E13.5 brains, as shown by hematoxylin and eosin staining of sagittal sections. Note the olfactory lobe in the wild type (arrows) (F) and the enlarged third ventricle (stars) in the Rap250−/− embryo (E). Bar = 400 μm.
Transcriptional activities of PPARγ are impaired in Rap250−/− MEFs.
In order to investigate the role of RAP250 in regulating the activity of nuclear receptors important for placental development, we isolated fibroblasts from wild-type and Rap250−/− embryos and analyzed transcriptional activities using transient transfections. Plasmids containing the PPARγ, EERβ, and RXRα ligand-binding domains fused to the GAL4 DNA binding domain were cotransfected with a GAL4 responsive reporter gene into MEFs, whereupon these cells were cultured in the presence or absence of ligand. PPARγ and RXRα showed ligand-dependent transcriptional activity in both wild-type and Rap250−/− cells. However, the activity of PPARγ was markedly reduced in Rap250−/− cells, by 50% compared to wild-type cells (Fig. 9). The activity of RXRα was not significantly changed, whereas ERRβ had no activity compared to that of the GAL4 DNA binding domain alone. These results suggest that RAP250 is necessary for optimal function of PPARγ.
FIG. 9.
Reduced PPARγ-mediated transcriptional activation in Rap250−/− MEFs. Fibroblasts isolated from wild-type and Rap250−/− embryos were cotransfected with 0.5 μg of Gal4-luciferase reporter plasmid, 0.5 μg of pGAL4-DBD, pGal4-PPARγ, pGal4-ERRβ, or pGal4-RXRα, and 0.2 μg of pCMVβ and cultured in the absence (−) or presence (+) of ligand (10 μM BRL49653 for PPARγ or 10 μM 9-cis-retinoic acid for RXRα). Cells were harvested 24 h after transfection and assayed for luciferase and β-galactosidase activities. The luciferase activity was adjusted with β-galactosidase activity for each cellular extract to correct for transfection efficiency. The results represent means ± standard deviations of a representative experiment performed in duplicate and are represented as fold increases compared to the activity of pGAL4-DBD.
DISCUSSION
Previous results suggest an important role for RAP250 as a coactivator for several nuclear receptors (3, 13, 15, 19, 42). In this report, we provide genetic evidence that RAP250 is required for embryonic development. Embryos lacking this coactivator die around E13.5 and our results indicate that the embryonic lethality is most likely due to placental dysfunction, since morphological examination of placentas from Rap250−/− embryos revealed severe developmental and vascular defects. We found that the spongiotrophoblast layer was markedly reduced and that maternal blood vessels bordering the spongiotrophoblast and labyrinthine layers had collapsed at E13.5 in Rap250 null placentas. In addition, a necrotic area was found in proximity to the collapsed blood vessels. These findings suggest that blood circulation has been obstructed in placentas from Rap250−/− embryos, leading to placental ischemia followed by embryonic death. Interestingly, the collapse of blood vessels is specific for those vessels found at the border of the spongiotrophoblast and labyrinthine layers, since normal blood vessels were found outside this region. The mechanism that causes the collapse of blood vessels bordering the spongiotrophoblast and labyrinthine layers in placentas from Rap250−/− embryos is unclear. One explanation could be a lack of local production of factors, such as vascular endothelial growth factor, needed for proper blood vessel function. This explanation is supported by our finding that the spongiotrophoblast layer, which is known to have endocrine functions, is markedly reduced in placentas from Rap250−/− embryos as early as E12.5.
The severe morphological changes we observed in placentas from Rap250−/− embryos affected both the labyrinth and spongiotrophoblast layers, suggesting that RAP250 is involved in multiple signaling pathways in the placenta. The striking reduction of the spongiotrophoblast layer is similar to what was found in the null mutation of nuclear receptor ERR-β in mice. ERR-β plays an important role in trophoblast differentiation and the null mutation results in embryonic lethality at E10.5 (18). The labyrinthine layer was also less vascularized in placentas from Rap250−/− embryos than in the wild type. This abnormality might reflect impaired function of the nuclear receptor PPARγ since genetic studies have shown that this receptor and its dimerization partner RXR are essential for placental development and vasculature (2, 14, 27, 36). It is also interesting that the nuclear receptor coactivator TRAP220, which is a coactivator for PPARγ, also has an essential role in the labyrinthine layer (44). RAP250 is a coactivator for PPARγ (3, 16, 42) in vitro and our transfection studies with cultured MEFs derived from Rap250−/− embryos further indicate that RAP250 is a coactivator for PPARγ in vivo since the activity of the PPARγ AF2 function was only 50% in Rap250−/− MEFs compared to wild-type cells. Examination of Rap250−/− embryos revealed that hearts had thin cardiac ventricular walls and trabecular hypoplasia. This heart phenotype is comparable to that described for PPARγ−/−, RXRα−/−, and TRAP220−/− embryos (2, 9, 12), and the severity of these defects suggests that they might contribute to the lethality of mutant embryos. The existence of a placenta-heart axis has been demonstrated (2), and therefore it is likely that the compromised heart development observed in Rap250−/− embryos is a result of placental dysfunction. In addition, the central nervous system was generally underdeveloped at E13.5; however, whether the observed defects in the central nervous system are primary or secondary to those in the placenta is unclear.
Our observation that Rap250−/− embryos die rather late during development suggests a selective role for RAP250 in the regulation of transcription which is in contrast to the function of the more general coactivators CBP, p300, and the mammalian mediator. Targeted disruption of SRB7, a component of the mammalian mediator complex, showed that this factor is essential for cell viability and consequently null embryos die at the blastocyst stage (32). Moreover, mice lacking TRAP220/PBP, the nuclear receptor interacting component of the TRAP/DRIP/ARC subcomplex of the mediator complex die in utero at E11.5 (8, 44). The two related proteins CBP and p300, which are coactivators for a large number of transcription factors, are both required for embryonic development, and null embryos die at E9.5 (29, 40). However, there are also examples in which the lack of a coactivator can be compensated for by related factors. In mice lacking SRC-1, which are viable but display partial hormone resistance, elevated levels of SRC-2/TIF2 were detected and proposed to compensate for the lack of SRC-1 (38). Targeted disruption of PCAF resulted in mice without a distinct phenotype; however, elevated levels of PCAF-B/GCN5 were suggested to compensate for the loss of PCAF (39). The lethal phenotype of Rap250−/− embryos might in part be explained by the fact that RAP250 does not have any related homologues that could compensate for its loss. Although mice lacking RAP250 die in utero, we believe that RAP250 has important functions after birth since it is expressed in many tissues in adults. We are presently investigating this possibility using conditional knockout technology.
In summary, we have shown that targeted disruption of the RAP250 gene results in embryonic lethality. Our results demonstrate that RAP250 is essential for normal function of the placenta and indicate a nonredundant but selective function of RAP250 in vivo.
Acknowledgments
We thank Stefan Teglund, Britt-Marie Skoog, and Gunnel Brolin for ES cell work; Lars-Ährlund-Richter and Jose Insunsa for blastocyst injections; Mikael Zmarzlak, Sara Selenius, and Behnosh Esni for technical assistance; and Urban Lendahl for analyzing sections of knockout embryo. We also thank Pauline Flodby and Jason Matthews for critically reading the manuscript.
This work was supported by the Swedish Cancer Fund and by KaroBio AB. P.A. was supported by a fellowship from Svenska Sällskapet för Medicinsk Forskning and by funds from Lars Hiertas Minne and Magnus Bergvalls Stiftelse. G.S. was supported by a grant from the Swedish Cancer Fund and B.R. was supported by a grant from the Wallenberg Consortium North for Functional Genomics.
REFERENCES
- 1.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 2.Barak, Y., M. C. Nelson, E. S. Ong, Y. Z. Jones, P. Ruiz-Lozano, K. R. Chien, A. Koder, and R. M. Evans. 1999. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol. Cell 4:585-595. [DOI] [PubMed] [Google Scholar]
- 3.Caira, F., P. Antonson, M. Pelto-Huikko, E. Treuter, and J.-Å. Gustafsson. 2000. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem. 275:5308-5317. [DOI] [PubMed] [Google Scholar]
- 4.Chen, H. W., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 5.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 7.Hong, H., K. Kohli, A. Trivedi, D. L. Johnson, and M. R. Stallcup. 1996. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. USA 93:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, M., and R. G. Roeder. 2001. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab. 12:127-134. [DOI] [PubMed] [Google Scholar]
- 9.Ito, M., C.-X. Yuan, H. J. Okano, R. B. Darnell, and R. G. Roeder. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5:683-693. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki, T., W. W. Chin, and L. Ko. 2001. Identification and characterization of a RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276:33375-33383. [DOI] [PubMed] [Google Scholar]
- 11.Jung, D. J., S. Y. Na, D. S. Na, and J. W. Lee. 2002. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J. Biol. Chem. 277:1229-1234. [DOI] [PubMed] [Google Scholar]
- 12.Kastner, P., J. M. Grondona, M. Mark, A. Gansmuller, M. LeMeur, D. Decimo, J. L. Vonesch, P. Dolle, and P. Chambon. 1994. Genetic analysis of RXR alpha developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell 78:987-1003. [DOI] [PubMed] [Google Scholar]
- 13.Ko, L., G. R. Cardona, and W. W. Chin. 2000. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. USA 97:6212-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubota, N., Y. Terauchi, H. Miki, H. Tamemoto, T. Yamauchi, K. Komeda, S. Satoh, R. Nakano, C. Ishii, T. Sugiyama, K. Eto, Y. Tsubamoto, A. Okuno, K. Murakami, H. Sekihara, G. Hasegawa, M. Naito, Y. Toyoshima, S. Tanaka, K. Shiota, T. Kitamura, T. Fujita, O. Ezaki, S. Aizawa, T. Kadowaki, et al. 1999. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell 4:597-609. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. K., S. L. Anzick, J. E. Choi, L. Bubendorf, X. Y. Guan, Y. K. Jung, O. P. Kallioniemi, J. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. C. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283-34293. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S. K., S. Y. Jung, Y. S. Kim, S. Y. Na, Y. C. Lee, and J. W. Lee. 2001. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol. Endocrinol. 15:241-254. [DOI] [PubMed] [Google Scholar]
- 17.Li, H., P. J. Gomes, and J. D. Chen. 1997. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. USA 94:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, J., R. Sladek, J. A. Bader, A. Matthyssen, J. Rossant, and V. Giguere. 1997. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-beta. Nature 388:778-782. [DOI] [PubMed] [Google Scholar]
- 19.Mahajan, M. A., and H. H. Samuels. 2000. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol. Cell. Biol. 20:5048-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 21.Misra, P., C. Qi, S. Yu, S. H. Shah, W. Q. Cao, M. S. Rao, B. Thimmapaya, Y. Zhu, and J. K. Reddy. 2002. Interaction of PIMT with transcriptional coactivators CBP, p300, and PBP differential role in transcriptional regulation. J. Biol. Chem. 277:20011-20019. [DOI] [PubMed] [Google Scholar]
- 22.Näär, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 23.Oñate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354-1357. [DOI] [PubMed] [Google Scholar]
- 24.Qi, C., Y. Zhu, J. Pan, A. V. Yeldandi, M. S. Rao, N. Maeda, V. Subbarao, S. Pulikuri, T. Hashimoto, and J. K. Reddy. 1999. Mouse steroid receptor coactivator-1 is not essential for peroxisome proliferator-activated receptor alpha-regulated gene expression. Proc. Natl. Acad. Sci. USA 96:1585-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rachez, C., B. D. Lemon, Z. Suldan, V. Bromleigh, M. Gamble, A. M. Näär, H. Erdjument-Bromage, T. Paul, and L. P. Freedman. 1999. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824-828. [DOI] [PubMed] [Google Scholar]
- 26.Rachez, C., Z. Suldan, J. Ward, C. P. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sapin, V., P. Dolle, C. Hindelang, P. Kastner, and P. Chambon. 1997. Defects of the chorioallantoic placenta in mouse RXRalpha null fetuses. Dev. Biol. 191:29-41. [DOI] [PubMed] [Google Scholar]
- 28.Takeshita, A., G. R. Cardona, N. Koibuchi, C. S. Suen, and W. W. Chin. 1997. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 272:27629-27634. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, Y., I. Naruse, T. Maekawa, H. Masuya, T. Shiroishi, and S. Ishii. 1997. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. USA 94:10215-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 31.Treuter, E., L. Johansson, J. S. Thomsen, A. Wärnmark, J. Leers, M. Pelto-Huikko, M. Sjöberg, A. P. H. Wright, G. Spyrou, and J.-Å. Gustafsson. 1999. Competition between TRAP220 and TIF2 for binding to nuclear receptors. J. Biol. Chem. 274:6667-6677. [DOI] [PubMed] [Google Scholar]
- 32.Tudor, M., P. J. Murray, C. Onufryk, R. Jaenisch, and R. A. Young. 1999. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 13:2365-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voegel, J. J., M. J. Heine, C. Zechel, P. Chambon, and H. Gronemeyer. 1996. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 15:3667-3675. [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, Z., D. W. Rose, O. Hermanson, F. Liu, T. Herman, W. Wu, D. Szeto, A. Gleiberman, A. Krones, K. Pratt, R. Rosenfeld, C. K. Glass, and M. G. Rosenfeld. 2000. Regulation of somatic growth by the p160 coactivator p/CIP. Proc. Natl. Acad. Sci. USA 97:13549-13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss, R. E., J. Xu, G. Ning, J. Pohlenz, B. W. O'Malley, and S. Refetoff. 1999. Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J. 18:1900-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wendling, O., P. Chambon, and M. Mark. 1999. Retinoid X receptors are essential for early mouse development and placentogenesis. Proc. Natl. Acad. Sci. USA 96:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, J., L. Liao, G. Ning, H. Yoshida-Komiya, C. Deng, and B. W. O'Malley. 2000. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. USA 97:6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, J., Y. Qiu, F. J. DeMayo, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1998. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922-1925. [DOI] [PubMed] [Google Scholar]
- 39.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97:11303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 41.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, Y., L. Kan, C. Qi, Y. S. Kanwar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2000. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 275:13510-13516. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, Y., C. Qi, W. Q. Cao, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2001. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc. Natl. Acad. Sci. USA 98:10380-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, Y., C. Qi, Y. Jia, J. S. Nye, M. S. Rao, and J. K. Reddy. 2000. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J. Biol. Chem. 275:14779-14782. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, Y. J., C. Qi, S. Jain, M. S. Rao, and J. K. Reddy. 1997. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 272:25500-25506. [DOI] [PubMed] [Google Scholar]