Abstract
The ubiquitin-conjugating enzymes HR6A and HR6B are the two mammalian homologs of Saccharomyces cerevisiae RAD6. In yeast, RAD6 plays an important role in postreplication DNA repair and in sporulation. HR6B knockout mice are viable, but spermatogenesis is markedly affected during postmeiotic steps, leading to male infertility. In the present study, increased apoptosis of HR6B knockout primary spermatocytes was detected during the first wave of spermatogenesis, indicating that HR6B performs a primary role during the meiotic prophase. Detailed analysis of HR6B knockout pachytene nuclei showed major changes in the synaptonemal complexes. These complexes were found to be longer. In addition, we often found depletion of synaptonemal complex proteins from near telomeric regions in the HR6B knockout pachytene nuclei. Finally, we detected an increased number of foci containing the mismatch DNA repair protein MLH1 in these nuclei, reflecting a remarkable and consistent increase (20 to 25%) in crossing-over frequency. The present findings reveal a specific requirement for the ubiquitin-conjugating activity of HR6B in relation to dynamic aspects of the synaptonemal complex and meiotic recombination in spermatocytes.
Ubiquitin is present in all cells, and the ubiquitin system is involved in different essential cellular processes such as cell division, responses to stress, and apoptosis. Protein ubiquitination occurs through the activities of ubiquitin activating-enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin-ligating enzymes (E3) (52). Polyubiquitination usually targets a substrate for degradation by a multisubunit structure called the proteasome. Ubiquitination, and in particular monoubiquitination, may also serve other purposes, such as activation or inactivation of transcription factors (12), internalization of transmembrane receptors (22, 52), and alteration of chromatin structure through stable ubiquitination of histones (13, 36). The genome encodes only very few E1 enzymes (1 or 2), and the diverse functions of the ubiquitin system are brought about by 10 to 20 different E2s and an even greater variation of E3 enzymes (52). The ubiquitin-conjugating E2 enzyme HR6B is essential for male fertility in the mouse (41).
HR6B is one of the two mammalian homologs of the Saccharomyces cerevisiae E2 enzyme named RAD6/UBC2 (24). The other mammalian RAD6 homolog, HR6A, shows 96% amino acid identity to HR6B. Between mouse and human enzymes, the identity is 100%. The mouse and human HR6B genes are autosomal, whereas HR6A is located on the X chromosome in both species (24, 41).
RAD6 in yeast is essential for sporulation but is also involved in many other processes, as illustrated by the pleiotropic phenotype of RAD6-null mutants (27). These mutants are defective in a specific DNA damage response pathway named postreplication repair. Furthermore, there are defects in gene silencing and changes in mitotic homologous recombination frequency (10, 23, 44). The precise mechanism of the role of RAD6 in yeast meiosis and sporulation is unclear. In Rad6 mutants, premeiotic DNA replication takes place, but there is a very early block in sporulation, precluding analysis of meiotic recombination in haploid spores (15). However, after withdrawal from sporulation medium at different time points after induction, intragenic recombinants have been recovered, albeit in lower numbers than in the wild type (8, 32). Also, analysis of DNA has shown that recombined DNA products appear in RAD6 mutants, but the timing was delayed compared to the wild type (8).
Recent data show that RAD6 is required for the ubiquitination of histone H2B, and this modification is essential for sporulation and gene silencing (13, 40, 46). Ubiquitination of histone H2A was not detected, and no effect of mutation of the ubiquitination sites in H2A was found. During the preparation of this study, it was reported that ubiquitination of H2B by RAD6 is a prerequisite for methylation of lysine 4 and lysine 79 of histone H3 (9, 13, 46), and these modifications are essential for silencing of telomeres and ribosomal DNA (11, 13, 26, 46). This phenomenon is referred to as “trans-tail” regulation of histone modification (46).
During postreplication DNA repair in yeast, RAD6 interacts with the E3 enzyme RAD18. The human homolog of this E3 enzyme, hRAD18Sc, has also been shown to interact with HR6A and HR6B (55). We have recently identified the gene encoding the mouse homolog mRAD18Sc, and the expression of this gene was shown to be highly elevated in the testis, in particular in spermatocytes (49).
In HR6B knockout mice, spermatogenesis appears to start up normally. The first prominent morphological signs of defective spermatogenesis become visible when the spermatids differentiate during the first wave of spermatogenesis. The number of elongating and condensing spermatids is reduced in HR6B knockout versus wild-type mouse testes, and the nuclear morphology is highly abnormal (41). In contrast to the male infertility phenotype of the HR6B knockout mice, the phenotype of HR6A knockout mice involves normal somatic development associated with maternal-factor infertility (18; H. P. Roest et al., unpublished data). The HR6A knockout males are fertile, showing normal spermatogenesis. An important role of HR6A/HR6B in somatic cells, in addition to the role of these proteins in gametogenesis, is evident from the fact that HR6A and HR6B double-knockout animals are not viable (Roest et al., unpublished).
In mammalian somatic cells, ca. 10% of H2A and 1 to 1.5% of H2B are ubiquitinated. In a previous report we described ubiquitination of histone H2A during spermatogenesis in the mouse (3). Marked ubiquitination of H2A was seen, in particular for the sex body in pachytene spermatocytes. The sex body is a heterochromatic region in the periphery of the pachytene spermatocyte nucleus that contains the X and Y chromosomes. After completion of the meiotic divisions, little H2A ubiquitination was found in round spermatids, but H2A ubiquitination is increased in elongating spermatids, just prior to replacement of the histones by protamines. No abnormalities in the pattern of histone H2A ubiquitination were observed in HR6B knockout testes (3). Since the level of ubiquitinated H2B in spermatocytes and spermatids was below the level of detection on Western blots (3) and since no antibodies specifically recognizing ubiquitinated H2B on cell preparations are available, it is not known whether mutation of HR6B affects H2B ubiquitination in mammals.
In the present study we describe how HR6B plays a primary role during the meiotic prophase in spermatocytes.
MATERIALS AND METHODS
Mice.
HR6A and HR6B knockout mouse strains were maintained on a FVB/N background and, as mentioned below, for one experiment we used an HR6B knockout mouse strain on a C57BL/6 background. Heterozygote (for HR6A or HR6B) or wild-type control animals were either littermates of the knockouts or progeny from breedings with heterozygotes from the same backcross generation.
Isolation of different cell types from mouse testis.
Spermatocytes and round spermatids were purified from 30-day-old and adult mice by using collagenase and trypsin treatment, followed by sedimentation at unit gravity (StaPut procedure) and density gradient centrifugation through Percoll (19). The purity of the cell fractions was >90% as determined by microscopic analysis of an aliquot of the purified cells fixed in Bouins' fixative on glass slides.
Another protocol for purification of spermatocytes or spermatids involved inclusion of 10 mM iodoacetamide throughout the cell isolation procedure to inhibit protein deubiquitination (29). Iodoacetamide exerts a negative effect on cell viability, and therefore the cell isolation procedure was shortened as follows. Decapsulated testes were shaken (90 cycles/min, amplitude 10 mm) at 32 to 34°C in 20 ml of phosphate-buffered saline (PBS) with Ca2+ and Mg2+ (PBS+Ca/Mg; 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 6.5 mM Na2HPO4, 1.1 mM CaCl2, 0.5 mM MgCl2), containing 10 mM iodoacetamide, 1 mg of trypsin (40 to 110 U/mg; Boehringer Mannheim, Mannheim, Germany)/ml, 1 mg of collagenase (0.435 U/mg; Boehringer Mannheim)/ml, and 0.5 mg of hyaluronidase (1,000 U/mg; Boehringer Mannheim)/ml in a siliconized 100-ml Erlenmeyer flask for 25 min. The tubule fragments obtained by this enzyme treatment were shaken (120 cycles/min) at 32 to 34°C in 20 ml of PBS without Ca2+ and Mg2+ (PBS−Ca/Mg; 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4) containing 10 mM iodoacetamide for 10 min. The larger cell clumps were removed by using a Pasteur pipette, and the cell suspension was filtered through a 60-μm (pore-size) nylon filter. The obtained cell preparation was centrifuged at 62 × g (spermatocyte-enriched preparation) or 249 × g (round spermatid-enriched preparation) for 3 min.
Polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Proteins were run on sodium dodecyl sulfate (SDS)-polyacrylamide gels and blotted onto nitrocellulose by using the Bio-Rad Mini-Protean II electrophoresis and blot cells (Bio-Rad, Hercules, Calif.). Blotting was performed for 1 h at room temperature in 25 mM Tris-192 mM glycine-20% (vol/vol) methanol (pH 8.3). Two-dimensional gels were run as previously described (25).
Ubiquitinated histones were detected with anti-ubiquitin (α-ubi; polyclonal antibody; Dako Diagnostics BV, Amsterdam, The Netherlands). After a blocking step for 1 h in buffer A (0.5 M NaCl, 20 mM Tris-HCl [pH 7.5], 0.1% [vol/vol] Tween 20), the blot was incubated with α-ubi (at 1:200) in buffer A, followed by three 10-min washings in buffer A. A second antibody incubation (peroxidase-labeled goat anti-rabbit; Sigma-Aldrich, Zwijndrecht, The Netherlands) and a subsequent washing were also done in buffer A. Antigen-antibody complexes were detected by using a chemiluminescence kit (Du Pont/NEN, Bad Homburg, Germany) according to the instructions provided by the manufacturer. HR6A and HR6B were detected with a polyclonal antibody (α-HR6A/B) raised against a peptide representing the N terminus of HR6A and HR6B, which are identical. After the blocking of nonspecific sites with 2.5% (wt/vol) nonfat milk in 20 mM Tris-HCl (pH 7.5)-0.15 M NaCl (buffer B) for 1 h at room temperature, α-HR6A/B was diluted 1:500 in buffer B containing 0.2% nonfat milk (wt/vol) and then incubated with the immunoblot for 90 min. Washings were performed in buffer B with 0.05% (vol/vol) Tween 20. For the final washing step after the second antibody incubation (peroxidase-labeled goat anti-rabbit), 10 mM Tris-HCl (pH 8.0)-0.15 M NaCl-0.05% Tween 20 was used. Antigen-antibody complexes were detected as described for α-ubi.
Immunocytochemistry.
Testes were isolated from 30-day-old wild-type or HR6B knockout mice. For irradiation experiments, mice were exposed to ionizing radiation from a 137Cs source. A single dose of 4 Gy was used, and testes were isolated 3 or 7 days after exposure; two animals of each genotype were used for each time point.
Embryonal ovaries were isolated at E18 from wild-type, HR6A knockout, HR6A heterozygote, HR6B knockout, and HR6B heterozygote embryos. The upper half of each embryo was used to genotype the animals.
Testis and ovary tissues were processed for immunocytochemistry as described by Peters et al. (35). Spreads of spermatocytes and oocytes were double stained with rabbit polyclonal or mouse monoclonal anti-SCP3 (gifts from C. Heyting, Wageningen, The Netherlands) in combination with one of the following antibodies: mouse monoclonal anti-uH2A (a gift from J. Celis, Aarhus, Denmark), and anti-Mut L homolog 1 mismatch repair protein (anti-MLH1; BD Pharmingen, Lexington, Ky.), rabbit polyclonal anti-SCP2, anti-SCP1 (a gift from C. Heyting), and anti-RAD51 (a gift from R. Kanaar, Rotterdam, The Netherlands). For polyclonal first antibodies, the secondary antibodies were fluorescein isothiocyanate- or TRITC (tetramethyl rhodamine isothiocyanate)-labeled goat anti-rabbit immunoglobulin G (IgG) antibodies (Sigma-Aldrich, Zwijndrecht, The Netherlands); fluorescein isothiocyanate-labeled goat anti-mouse IgG and IgM (Sigma) were used as secondary antibodies for anti-MLH1 and anti-uH2A, respectively. Before incubation with antibodies, slides were washed in PBS (three times, each for 10 min), and nonspecific sites were blocked with 0.5% (wt/vol) bovine serum albumin (BSA) and 0.5% (wt/vol) milk powder in PBS. First, antibodies were diluted in 10% (wt/vol) BSA in PBS, and incubations were carried out overnight at room temperature in a humid chamber. Slides were then subjected to three 10-min washes in PBS, blocked in 10% (vol/vol) normal goat serum (Sigma) in blocking buffer (supernatant of 5% [wt/vol] milk powder in PBS centrifuged at 14,000 rpm for 10 min), and incubated with secondary antibodies in 10% normal goat serum in blocking buffer at room temperature for 2 h. Finally, slides were then subjected to three 10-min washes in PBS (in the dark) and embedded in Vectashield containing DAPI (4′,6′-diamidino-2-phenylindole to counterstain the DNA (Vector Laboratories, Burlingame, Calif.). Fluorescent images were observed by using a fluorescence microscope (Axioplan 2; Carl Zeiss, Jena, Germany) equipped with a digital camera (Coolsnap-Pro; Photometrics, Waterloo, Canada). Digital images were processed by using Adobe Photoshop software (Adobe Systems). The number of loose synaptonemal complex (SC) beads was counted in 38 nuclei (at least 9 nuclei per animal were chosen at random) from both wild-type and HR6B knockout mice (four animals in each group). The total length of the SC was determined in 18 wild-type pachytene nuclei (6 per animal), 15 wild-type diplotene nuclei (5 per animal), and 18 HR6B knockout pachytene nuclei (6 per animal) by using Image J software analysis (National Institutes of Health). Each group contained three animals, and values were not significantly different between animals within a single group. Statistical differences in total SC length per nucleus were calculated by using the Mann-Whitney U test. The number of MLH1 spots was counted in at least 10 spermatocyte or oocyte nuclei of each animal. For each animal, the average number of MLH1 foci was calculated, and statistical comparison was made between five wild-type, eight HR6B knockout, and two HR6A knockout males and between four HR6A heterozygote and four HR6A knockout female E18 embryos by using the Mann-Whitney U test. MLH1 foci in pachytene oocytes from a single wild-type E18 embryo were counted to compare with the HR6A heterozygotes. In a separate experiment, MLH1 spots were counted in at least 10 oocyte nuclei from one wild-type, one HR6B heterozygote, and one HR6B knockout E18 embryo(s) on a C57BL/6 background. To validate the use of this strain, MLH1 foci in pachytene spermatocytes of one wild-type male and one HR6B knockout male were counted. The results were similar to those obtained with HR6B knockout males on a FVB/N background (not shown).
For the irradiation experiments, at least 20 nuclei were analyzed for each animal, with two animals per group.
Meiotic chromosome preparation.
Meiotic preparations of two 6-week-old wild-type and two 6-week-old HR6B knockout males were made as described elsewhere (14). After overnight aging of the slides at 37°C, they were incubated at 60°C in 2× SSC (0.3 M sodium chloride containing 0.03 M trisodium citrate; pH 7.0) for 2 h, followed by a rinse in distilled water. Subsequently, the slides were embedded in Vectashield containing DAPI to stain the DNA (Vector Laboratories). Spread diakinesis/metaphase I nuclei with a complete set of identifiable chromosome pair configurations were selected. Fluorescent images were captured and processed as described above. The number of chiasmata was counted in at least 15 nuclei per animal. Each group contained two animals, and values were not significantly different between animals within a single group. Statistical differences in the number of chiasmata per nucleus were calculated by using the Mann-Whitney U test.
TUNEL assay.
Testes were isolated from wild-type, HR6B heterozygote, and HR6B knockout mice that were 2, 3, 4, and 6 weeks old and also from adult HR6A knockout mice. Tissues were formaldehyde fixed and embedded in paraffin according to standard procedures. Sections were mounted on aminoalkylsilane-coated glass slides, dewaxed, and pretreated with proteinase K (Sigma) and peroxidase as described elsewhere (16). Slides were subsequently washed in terminal deoxynucleotidyl transferase (TdT) buffer for 5 min (17) and then incubated for at least 30 min in TdT buffer containing 0.01 mM Biotin-16-dUTP (Roche Diagnostics, Almere, The Netherlands) and 0.4 U of TdT enzyme (Promega, Leiden, The Netherlands)/μl. The enzymatic reaction was stopped by incubation in TB buffer, and the sections were washed (16). Slides were then incubated with StreptABComplex-horseradish peroxidase conjugate (Dako) for 30 min and washed in PBS. dUTP-biotin labeled cells were visualized with 3,3′-diaminobenzidine tetrahydrochloride-metal concentrate (Pierce, Rockford, Ill.). Tissue sections were counterstained with nuclear fast red-5% (wt/vol) Al2(SO4)3. For each animal, the number of TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling)-positive cells was counted in at least 200 tubule sections, and the average number of positive cells per 100 cross sections was calculated. Data were analyzed by using the Mann-Whitney U test.
Fluorescence in situ hybridization analysis.
A TRITC-labeled PNA probe directed against mouse telomeres (a gift from M. Zijlmans, Rotterdam, The Netherlands) was used to identify the telomeres in spread spermatocytes. Meiotic spread preparations were denatured in 70% (vol/vol) deionized formamide in 2× SSC at 72°C for 2.5 min, followed by dehydration with ethanol. Slides were air dried, and 20 μl of denatured probe (0.3 μg/ml) in hybridization mix (70% [vol/vol] deionized formamide, 10 mM Tris-HCl [pH 7.0], 0.25% [wt/vol] blocking reagent [Du Pont/NEN, Boston, Mass.]; stock 1% [wt/vol] in 40 mM Tris-HCl [pH 7.0]) was applied. Hybridization was carried out under a coverslip in a humid chamber for 2 h in the dark at room temperature, followed by sequential washing in 70% (vol/vol) formamide, 10 mM Tris-HCl (pH 7.0), and 0.1% (wt/vol) BSA (two times, 15 min each time) and in 0.1 M Tris-HCl (pH 7.0), 0.15 M NaCl and 0.08% (vol/vol) Tween 20 (three times, 5 min each time). Subsequently, immunocytochemistry with the anti-SCP3 polyclonal antibody was carried out as described above.
RESULTS
HR6A/B protein expression is strongly decreased in spermatogenic cells from HR6B−/− mice compared to wild-type and HR6AY/− mice.
Some functional redundancy between HR6A and HR6B must exist, since HR6A and HR6B double-knockout mice are inviable, whereas the single HR6A and HR6B knockout mice show mild somatic phenotypes. In most cell types isolated from rat tissues, HR6A and HR6B protein levels appear to be comparable (25). Only in postmeiotic spermatids does the level of HR6B exceed that of HR6A (25).
The male infertility of HR6B knockout mice may signify that HR6B performs a specific function during spermatogenesis. Alternatively, the quantity of HR6A protein in the HR6B knockout spermatogenic cells may be insufficient to allow full compensation for the lack of HR6B.
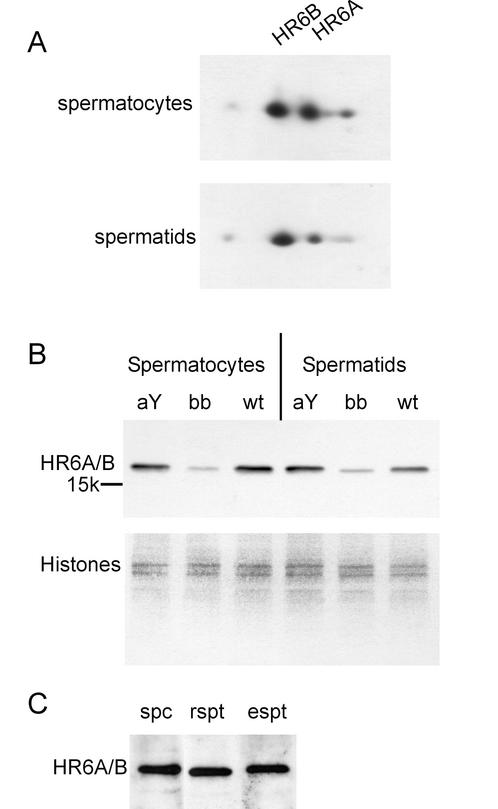
On two-dimensional gels the amounts of HR6A and HR6B in wild-type spermatocytes is approximately equal, showing a tendency toward more HR6B, which becomes evident in spermatids (Fig. 1A). We estimated the total HR6A and/or HR6B protein levels in spermatocytes and spermatids from wild-type mice compared to those in HR6AY/− and HR6B−/− mice by using an anti-HR6A/B antibody that recognizes both HR6A and HR6B (Fig. 1B). In the cells from the HR6B knockout mice, the intensity of the HR6A/B band is much lower than in the wild-type cells. When spermatocytes and spermatids were isolated from the HR6A knockout, the intensity of the HR6A/B band was found to be approximately equal to that in the wild type. Thus, in HR6B knockout spermatocytes, as well as in spermatids, the level of HR6A/B protein expression is markedly reduced, whereas in HR6A knockout spermatocytes and spermatids no reduction in the amount of HR6A/B is observed. Comparison of the total level of HR6A/B protein in different testicular cell types of wild-type mice indicates that this level is equally high in spermatocytes, round spermatids, and elongating spermatids (Fig. 1C).
FIG. 1.
Immunoblot analysis of HR6A and HR6B expression in germ cells from wild-type and HR6B knockout mouse testis. Highly purified spermatocytes and spermatids were isolated from wild-type (wt), HR6A knockout (aY), and HR6B knockout (bb) mouse testes. (A) Total protein extracts from wild-type cells were run on two-dimensional gels and subjected to isoelectric focusing in the first dimension and SDS-PAGE in the second dimension. HR6A and HR6B were detected with anti-HR6A/B. The positions of the two proteins are indicated; the pH increases from left to right in the gel. (B) Total protein extracts were run on SDS-PAGE gels and then analyzed by using anti-HR6A/B. The position of the 15K molecular weight marker is indicated. The lower panel in B shows part of the Ponceau red-stained blot, with several histone bands, as a control for equal protein loading of the gel. (C) Abundant presence of HR6A and/or HR6B proteins in highly purified spermatocytes (spc), round spermatids (rspt), and elongating spermatids (espt) isolated from wild-type mice.
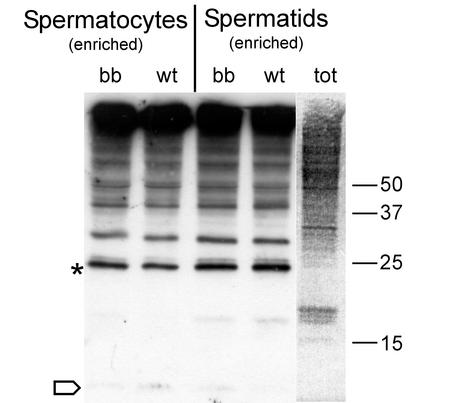
To determine whether HR6B is important for quantitative maintenance of overall protein ubiquitination in spermatocytes and spermatids, we studied protein ubiquitination by using one-dimensional SDS-PAGE gels. It was observed that the overall ubiquitination capacity and the global pattern of protein ubiquitination were similar in the spermatocytes and spermatids of wild-type mice compared to that of HR6B knockout mice (Fig. 2). This result indicates that HR6B most likely is not required for the maintenance of bulk protein ubiquitination but is involved in the ubiquitination of one or a few specific and possibly low-abundance substrates whose function is essential to spermatogeis.
FIG. 2.
Immunoblot of ubiquitinated proteins in germ cells from wild-type and HR6B knockout mouse testes. Preparations enriched in spermatocytes and spermatids were isolated in the presence of iodoacetamide by using a rapid procedure to try to minimize deubiquitination during isolation (see Materials and Methods). Immunoblots of total protein samples from isolated cell fractions of wild-type (wt) and HR6B knockout (bb) mice were analyzed by using an anti-ubiquitin antibody. For comparison, the total (tot) protein pattern is shown in the right lane, which is a Ponceau red-stained representative lane of the immunoblot. The asterisk indicates the position of ubiquitinated H2A/H2B, and the open arrow indicates the position of free ubiquitin. The positions of different molecular weight markers (in thousands) are also indicated.
Knockout of HR6B, but not of HR6A, results in an increased death rate of spermatocytes in immature mouse testis.
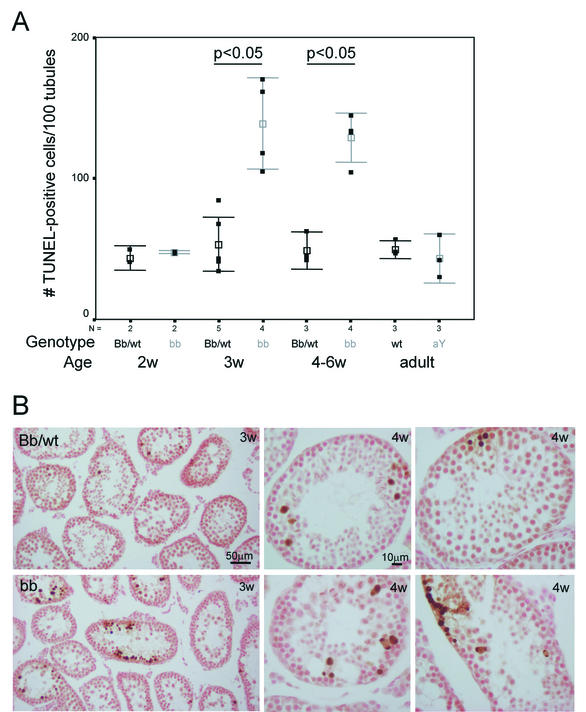
During the first wave of spermatogenesis in HR6B knockout mice, derailment of spermatogenesis becomes visible during postmeiotic spermatid elongation and condensation. Previously, we have shown that in 6-week-old animals, apoptosis of spermatonesTcytes is increased (41). This may be an indirect effect caused by impaired spermatid development, exerting a detrimental feedback on earlier steps in spermatogenesis. Since we found that the level of HR6A/B protein was already greatly affected in HR6B−/− spermatocytes, we studied a possible role of HR6B in spermatocytes through TUNEL analysis of wild-type and HR6B knockout testis of immature animals, before the first appearance of elongating spermatids. At the age of 2 weeks, the first wave of spermatogenesis had progressed up to the meiotic prophase, and early pachytene spermatocytes are present in most testis tubules of wild-type and HR6B knockout mice. In the testes of 3-week-old mice, late pachytene cells are present, and meiotic divisions have occurred in several tubules, resulting in the formation of round spermatids. Elongating spermatids have not yet formed at this stage of postnatal development (not shown). Figure 3A shows that the frequency of germ cell death is significantly increased in testis sections from HR6B−/− mice aged 3 weeks and onward compared to age-matched wild types and heterozygotes. In wild-type and in HR6B knockout testes tubuli, the TUNEL-positive cells are found grouped together in a relatively small percentage of tubule cross sections. The location within the tubule and the identification of the surrounding cells indicate that the TUNEL-positive cells are mainly primary spermatocytes in mid-pachytene and metaphase/anaphase I. In testes from 3-week-old HR6B knockout animals, the average number of apoptotic spermatocytes per positive tubule cross section is higher, and also the number of tubule cross sections containing apoptotic spermatocytes is increased by ca. 50% (Fig. 3B). In 4-week-old animals, large compact clusters of apoptotic cells occasionally are present, but not all cell types present within these clusters can be determined, and the clusters may include a few spermatogonia in addition to spermatocytes (Fig. 3B). The results strongly indicate that spermatocyte development in HR6B knockout animals is impaired already during the first wave of spermatogenesis, before the appearance of elongating spermatids. This eliminates the possibility that apoptotic loss of HR6B−/− spermatocytes is an indirect consequence of impaired postmeiotic differentiation of spermatids. In contrast, in HR6A knockout males, which show normal fertility, no increase in the number of TUNEL-positive cells per tubule cross section was observed (Fig. 3A).
FIG. 3.
Analysis of TUNEL-positive cells in HR6A and HR6B knockout mouse testes. (A) The numbers of TUNEL-positive cells per 100 tubules were determined in testis sections from wild-type (wt), HR6AY/− (aY), HR6B+/− (Bb), and HR6B−/− (bb) mice of different ages. The results for wild-type and HR6B+/− immature animals were pooled. The number of animals used in each group is indicated below the figure (N). The actual data are indicated as black squares, and the mean and standard error of the mean (SEM) values are indicated in black (wt and Bb) and gray (bb and aY). A significant increase (P < 0.05) in the amount of apoptotic spermatocytes per tubule cross section was found only for HR6B knockout mice that were 3 weeks old or older. (B) Representative photomicrographs of TUNEL staining in testis sections from wild-type or HR6B+/− (Bb/wt, upper panels) and HR6B−/− (bb, lower panels) mice that were 3 (3w) or 4 (4w) weeks old. Note the larger clusters of apoptotic cells in tubules of HR6B knockout mice.
In HR6B knockout pachytene and diplotene spermatocytes, SC structure and telomere localization are disturbed.
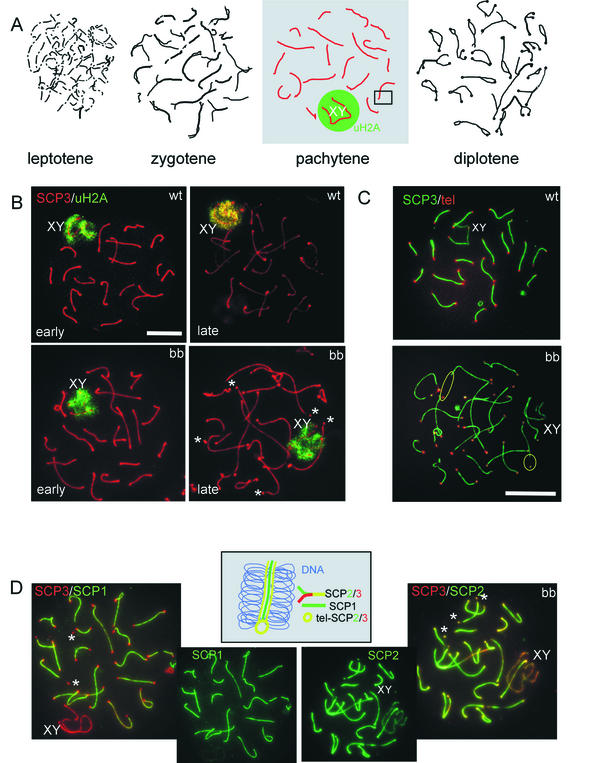
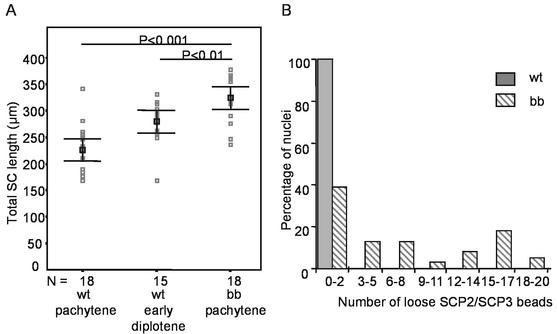
Based on the above-described findings, a detailed study was performed on the progression of the meiotic prophase in HR6B knockout spermatocytes. Meiotic spread preparations were stained with antibodies against SC protein 3 (SCP3), SCP2, and SCP1, which visualize the (paired) chromosomal axes during late zygotene, throughout pachytene, and during diplotene (21) (Fig. 4A). Sex body chromatin in pachytene and early diplotene cells was stained by using a monoclonal antibody against ubiquitinated H2A (anti-uH2A) (3). In accordance with previous immunohistochemical studies (3), no difference in the staining pattern of anti-uH2A was observed between wild-type and HR6B knockout spermatocytes (Fig. 4B). Using the anti-SCP antibodies, it was found that formation of the SC appeared to occur normally in HR6B−/− cells. However, during late pachytene the SC appeared to be thinner, and the average total length of the SC was significantly increased compared to that in wild-type pachytene and early diplotene cells (Fig. 4B and D and 5A). Also, there was loss of SC staining in the near telomeric regions at the end of the pachytene stage in HR6B−/− spermatocytes, when the ends of the SC show a thickened bead-like appearance (Fig. 4B and D and 5B). The seemingly detached ends, or beads, were found to contain SCP3 and SCP2, which are components of the lateral elements of the SC. The protein SCP1, which is a constituent of the transverse filaments that connect the lateral elements, was absent from the SCP2 and SCP3 beads, irrespective of whether they where still observed as attached to the rest of the paired axes (Fig. 4D). Fluorescent in situ hybridization with a mouse telomere PNA probe, followed by immunocytochemistry with anti-SCP3 polyclonal antibody, confirmed that the SCP3 and SCP2 beads colocalized with the telomeres (Fig. 4C). In wild-type cells, a similar loss of SCPs from near telomeric regions occurs in diplotene spermatocytes, but not during pachytene, when the chromosomes are still fully paired. Hence, loss of SC proteins from near telomeric regions may be associated with changes in chromatin structure in this region and/or may be due to premature SC degradation.
FIG. 4.
SCs and the sex body in wild-type and HR6B knockout spermatocytes. (A) Schematic representation of the appearance of lateral elements of the SC during the different stages of the meiotic prophase. SC fragments are formed during leptotene. During zygotene, homologous chromosome pairing is initiated; this is visualized by the appearance of longer and thicker (in paired regions) axial and/or lateral elements of the SC. During pachytene, all autosomal chromosomes are fully paired, and each visible SC axis has a chromosome that consists of two chromatids on either side (the number of visible axes corresponds to the haploid number of chromosomes). The X and Y chromosomes pair only in the pseudoautosomal region. These chromosomes are heterochromatic and form the so-called sex body (indicated by XY), located in the nuclear periphery. During diplotene, the chromosomal axes start to separate but remain attached in the regions where crossing-over has occurred. Differences between wild-type (wt) and HR6B knockout (bb) spermatocytes were detected mainly during pachytene. Therefore, this stage is highlighted, and the photomicrographs in panels B, C, and D represent pachytene spermatocytes. XY indicates the sex body, and the green circleindicates that this area is enriched in ubiquitinated H2A (uH2A). (B) Spermatocyte spreads from wild-type and HR6B knockout mice stained with a polyclonal antibody against SCP3 (red) and a monoclonal antibody against ubiquitinated H2A (uH2A, green). Early and late pachytene spermatocytes are shown. The SC axes in many bb spermatocytes are longer and apparently thinner, and this effect is most pronounced during late pachytene. Also, loose-appearing SCP2/SCP3 beads at the ends of some SC axes can be observed. Sex body chromatin and uH2A staining appear normal. Upon longer exposure, uH2A also localizes to the telomeres of late pachytene and early diplotene cells in wild-type and in HR6B knockout spermatocytes (not shown). Asterisks indicate loose SCP2/SCP3 beads; XY indicates the sex body. Scale bar, 20 μm. (C) Meiotic spread preparations from wild-type (wt) and HR6B knockout (bb) mouse testis were hybridized to a mouse telomere probe (tel; red signal) and stained with anti-SCP3 (SCP3; green signal). In the HR6B knockout, a large number of the telomeres is not associated with the ends of the axes of the SCs but with loose bead-shaped SC fragments. For two of these telomeres, a yellow circle indicates the large distance between the telomere and the presumed beginning of the respective SC axis. XY indicates the X and Y chromosome pair. Scale bar, 20 μm. (D) HR6B knockout pachytene spermatocytes double stained with anti-SCP3 (red) and anti-SCP1 (green) (SCP3/SCP1) or with anti-SCP3 (red) and anti-SCP2 (green) (SCP3/SCP2). Single stains with anti-SCP1 and anti-SCP2 are also shown. Note that SCP2, but not SCP1, colocalizes to the loose SCP2/SCP3 beads stained with SCP3. In the middle, a schematic interpretation is shown of a fragment from the SC complex, enlarged from the box indicated in the pachytene nucleus in panel A. It visualizes localization of SCP3 and SCP2 on the lateral elements that have formed on the chromosomal axes, whereas SCP1 is a component of the transverse filaments that connect the lateral elements of the SC during chromosome pairing (no staining of SCP1 is observed on unpaired X and Y chromosomal axes). SCP1 is also absent from near telomeric DNA. In these regions SCP2 and SCP3 are both present (tel-SCP2/3).
FIG. 5.
Numbers of detached SCP2/SCP3 beads, and the length of the SC per cell in spermatocytes from wild-type and HR6B knockout mice. (A) Total length of the SC (in micrometers) per nucleus of wild-type (wt) pachytene and early diplotene spermatocytes and of HR6B knockout (bb) pachytene spermatocytes. Individual measurements are indicated in gray. Mean and SEM values (black squares and error bars, respectively) are indicated. P values are indicated above the lines connecting the two compared groups. N indicates the number of nuclei that were investigated. Nuclei were from three different animals in each group. (B) Frequency distribution of the number of loose SCP2/SCP3 beads per pachytene nucleus of wild-type (wt; black bars) and HR6B knockout (bb; hatched bars) mice. Thirty-eight nuclei from four different animals were analyzed for each group. For the wild type, all nuclei fell in the first category of zero to two loose SCP2/SCP3 beads; of these nuclei, 76% had no loose beads at all, and there was only 1 nucleus that showed 2 loose SCP2/SCP3 beads. For the HR6B knockout animals, only 24% of all analyzed nuclei had no loose SCP2/SCP3 beads.
Mutation of HR6B specifically results in a marked increase in the number of MLH1 foci in pachytene spermatocytes.
Meiotic recombination frequency is generally lower in heterochromatic regions and thus appears to be influenced by chromatin structure (53). In yeast cells, RAD6 is involved in the regulation of several aspects of chromatin structure and influences mitotic homologous recombination. The observed lengthening of SC axes in HR6B−/− pachytene spermatocytes is an indicator of aberrant (less-condensed) chromatin structure. Hence, we hypothesized that in HR6B knockout spermatocytes meiotic recombination might be affected. Therefore, we studied the nuclear localization of RAD51 and MLH1 in meiotic spread preparations of wild-type and HR6B knockout spermatocytes.
RAD51 is similar to Escherichia coli RecA and functions as a single-stranded DNA-binding protein that has DNA-dependent ATPase activity and stimulates strand exchange. It localizes in foci to the axial elements of unpaired chromosomal axes during leptotene, zygotene, and early pachytene of the prophase of the first meiotic division in mouse and rat spermatocytes (1, 6, 30, 31, 37, 47). The number of RAD51 foci increases during leptotene or early zygotene from 0 to ∼250 foci and decreases again during zygotene or early pachytene. In later pachytene stages, the foci disappear from the autosomal SCs but persist longer at the X chromosome (30, 31). This localization pattern is in accordance with its proposed role in recombination complexes during the early phases of meiotic recombination. We observed no abnormalities in the pattern of RAD51 localization in HR6B−/− spermatocytes (not shown). Since the number of RAD51 foci is dynamically changing during leptotene or zygotene, it was not possible to determine whether there is an absolute difference in the number of RAD51 foci between wild-type and HR6B knockout spermatocytes.
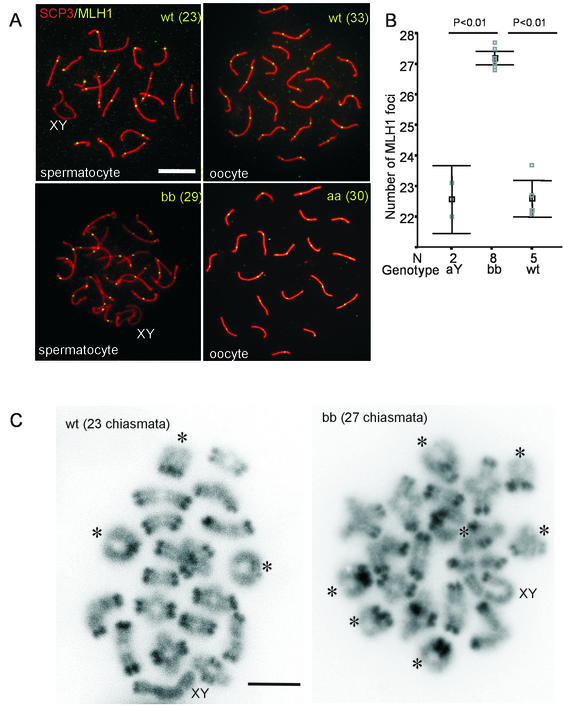
MLH1 localizes to the sites of crossing-over in mid-to-late pachytene spermatocytes and oocytes (2, 5). Antibodies against this protein can be used to determine the number and localization of crossing-overs. We detected 22.5 ± 1.5 and 22.4 ± 2.2 MLH1 foci in wild-type and HR6AY/− pachytene spermatocytes, respectively. However, 27.1 ± 1.9 MLH1 foci were found in HR6B−/− pachytene spermatocytes (Fig. 6A and B). In these HR6B−/− spermatocytes, the general distribution of MLH1 foci was not visibly different from the wild-type pattern (Fig. 6A). To confirm that the increase in the number of MLH1 foci in HR6B knockout spermatocytes actually leads to an increase in the number of crossing-overs, we counted the number of chiasmata in diakinesis/metaphase I nuclei of wild-type and HR6B knockout mice. We found 22.8 ± 1.2 and 25.6 ± 1.9 chiasmata in wild-type spermatocytes and HR6B knockout spermatocytes, respectively (Fig. 6C). This difference is statistically significant (P < 0.001). In HR6B knockout spermatocytes, the number of detected chiasmata is slighly lower then the number of MLH1 foci (this is discussed below).
FIG. 6.
Increased frequency of meiotic recombination during pachytene in HR6B−/− spermatocytes. (A) Meiotic spread preparations from wild-type (wt) and HR6B knockout (bb) mouse testes and from wild-type and HR6A knockout (aa) mouse fetal ovaries (E18) were stained with anti-SCP3 (SCP3, red signal) and anti-MLH1 (MLH1, green signal). In the HR6B knockout (bb), the number of MLH1 foci is increased (29 foci in this nucleus) compared to the wild type (23 foci). The average number of foci between wild-type and HR6A knockout oocytes does not differ (the number of foci for these two individual nuclei are 33 and 30, respectively). The SC structure of the knockout pachytene spermatocyte nucleus shown here has relatively few abnormalities. An increased number of MLH1 foci is found in bb nuclei that appear relatively normal and also in bb nuclei that show long and thin SC axes and/or loose SCP2/SCP3 beads (the latter is not shown). XY indicates the sex body. Scale bar, 20 μm. (B) The number of MLH1 foci per spermatocyte nucleus is indicated on the y axis. Please note that this axis starts at 21 foci. The gray squares indicate the average number of foci per nucleus per animal (N = the number of animals tested). For each animal at least 10 nuclei were counted. In black, the mean value and SEM are indicated (black square and error bars, respectively). MLH1 foci were counted in wild-type (wt), HR6A knockout (aY), and HR6B knockout (bb) spermatocytes. (C) Diakinesis/metaphase I spread preparations of wild-type (wt) and HR6B knockout (bb) spermatocyte nuclei. Bivalents showing two crossover events are indicated by and asterix. The bivalents containing the X and Y chromosome are indicated by XY. Note that the centromeric regions of the chromosomes are more heavily stained by DAPI. Scale bar, 10 μm.
An increase in meiotic recombination may be due to an increase in the number of double-strand DNA breaks. During the meiotic prophase, these breaks are normally induced by a meiosis-specific topoisomerase variant named SPO11 (7, 42, 43). In somatic cells, homologous recombination can occur to repair damage-induced double-strand DNA breaks. There are indications that such damage-induced double-strand DNA breaks can be converted to sites of meiotic recombination (20). To investigate whether irradiation of spermatocytes at or around the time of meiotic double-strand break formation by SPO11 would result in an increase in the number of MLH1 foci in mid-pachytene spermatocytes isolated from wild-type and HR6B−/− several days later, mice were exposed to 4 Gy (total body gamma irradiation), and the number of MLH1 foci was studied 3 and 7 days postirradiation. We found no effect of irradiation on the number of MLH1 spots in HR6B knockout spermatocytes (the average numbers of MLH1 spots were 27.8 and 27.2 at 3 and 7 days postirradiation, respectively), and in the wild type there was an average increase of 1.4 and 1.6 spots per nucleus at 3 and 7 days postirradiation, respectively.
The average number of crossing-overs differs between male and female gametogenesis: ca. 22 in mouse spermatocytes and 30 in mouse oocytes (2). Mutation of HR6A, but not of HR6B, leads to maternal-factor infertility (Roest et al., unpublished). To investigate whether mutation of HR6A might affect female meiotic recombination frequency, we studied the number of MLH1 spots in wild type, HR6A+/−, and HR6A−/− pachytene oocytes. In female gametogenesis, meiotic crossing-over occurs during fetal development. We isolated ovaries at E18 from embryos of HR6A+/− females crossed with HR6AY/− males and from wild-type embryos. The embryos were genotyped, and oocytes were spread on slides. No significant difference in the number of MLH1 foci (heterozygote, 32.1 ± 0.9; HR6A knockout, 32.1 ± 0.4; wild type [n = 1], 32.0) between wild type/heterozygote and HR6A knockout pachytene oocytes was detected (Fig. 6A). In a separate experiment, MLH1 foci in pachytene oocytes from one wild type, one HR6B heterozygote and one HR6B knockout female E18 embryo were also counted (10 oocytes per E18 embryo), and we found averages of 29.6, 28.9, and 28.8, respectively, indicating that mutation of HR6B does not affect the number of MLH1 foci during female meiosis. The somewhat lower numbers of MLH1 foci at E18 in this experiment may be due to a small difference in timing of collection of the oocytes and/or strain differences (C57BL/6 versus FVB/N). No SC abnormalities were observed in HR6A or HR6B knockout pachytene oocytes.
DISCUSSION
Partial redundancy between HR6A and HR6B.
In yeast, RAD6 is involved in postreplication DNA repair and sporulation (54). In addition, RAD6 plays a role in a number of other processes, mostly involving chromatin structure regulation. The elucidation of possible similar functions of RAD6 homologs in higher eukaryotes is complicated by several factors. First, two very similar homologs, HR6A and HR6B, exist in mice and humans. Thus, to obtain a cell that has no functioning RAD6 homologs, the alleles of both genes would have to be mutated. Second, it appears that mutational inactivation of both HR6A and HR6B genes is incompatible with cell survival (Roest et al., unpublished). This contrasts with yeast, since RAD6 mutant cells survive under normal circumstances, although they do show general signs of decreased fitness. Thus, functions of mammalian RAD6 homologs can be studied only in mutant cells or in animal models that still possess at least one functional copy of either gene. The HR6B knockout mouse is such a model. Although male HR6B knockout cells have only a single HR6A gene (located on the X chromosome), no postreplication DNA repair phenotype is present (41). Combining this finding with the lethality of the double HR6A and HR6B mutation in mouse, it can be concluded that HR6A and HR6B are redundant with respect to functions that involve normal cell survival and postreplication DNA repair. This leads to the third and last factor: the uncertainty with regard to the degree of redundancy of all functions between HR6A and HR6B. The differences between the HR6A and HR6B proteins (7 amino acid residues among a total of 152 amino acids) are located at nonconserved sites, or the different amino acids, when located at conserved sites, are similar. The relevance of these differences between HR6A and HR6B is unknown. Differential effects of HR6A versus HR6B mutation, such as the increased rate of cell death of HR6B−/− but not HR6AY/− spermatocytes described here, may result from functional divergence of the two proteins and/or from differential expression patterns. We have previously shown that the level of HR6B protein in wild-type spermatids is higher than that of HR6A (25). In the present study, we have demonstrated that spermatocytes as well as spermatids of HR6B−/− mice have a very low level of HR6A protein, compared to the level of HR6B protein in HR6AY/− spermatocytes and spermatids. These findings lead to the suggestion that the impairment of functions in HR6B−/− spermatocytes might be explained by a degree of compensation by HR6A that is too low. At present we cannot exclude that HR6A and/or HR6B also exert some differential functions. However, if the male infertility phenotype of HR6B−/− mice is caused by differential expression, the quantitative defect in HR6A and HR6B functioning may lead to a quantitative decrease in the ubiquitination of critical substrates rather than an absolute absence of such ubiquitination. This complicates the search for substrates.
Approximately 25 mammalian E2 or E2-like enzymes have been identified (52). Several of these enzymes show specific or enhanced expression in testis (4). Immunodepletion studies by Rajapurohitam et al. (38) have shown that a significant fraction of protein ubiquitination in testis extracts depends upon UBC4, and little or no effect was seen when HR6A and HR6B were immunodepleted (38). Results with the mouse HR6B knockout model, as presented here, confirm that HR6A and HR6B are not involved in maintaining the overall pattern and quantity of protein ubiquitination. Thus, the infertility phenotype of male HR6B knockout mice must be due to lack of ubiquitination of only one or a few selected HR6A- and/or HR6B-specific substrates.
HR6B is important for control of chromatin structure during the male meiotic prophase.
During the first wave of spermatogenesis in HR6B knockout mice, we found an approximately twofold increase in the number of TUNEL-positive spermatocytes. This result strongly indicates a role for HR6B in normal male meiosis.
During the meiotic prophase, the pairing of homologous chromosomes and meiotic recombination take place. To get more insight in the possible role of HR6B during meiosis, we studied SC structure in wild-type and HR6B knockout spermatocytes. Longer and apparently thinner SCs, with loose SCP2/SCP3 beads at the telomeres, as observed in HR6B−/− pachytene spermatocytes, are features also seen in wild-type diplotene cells. It appears that the chromatin structure that is associated with the paired chromosome axis in HR6B−/− pachytene spermatocytes is somewhat less compact and that there is premature breakdown of near telomeric SC structures. In addition, or alternatively, there might be a delay in the desynapsis of paired homologous chromosomes, whereas other diplotene-associated processes, such as lengthening of the SC and removal of SC proteins, proceed according to a normal schedule.
The SC, which ensures interaction between homologous chromosomes, is functionally associated with the cohesion complex, which keeps the sister chromatids associated (50). SCC1, a component of the cohesion complex, is cleaved during the metaphase-to-anaphase transition in mitotic cells (48), and RAD6 is required for the degradation of one of the cleavage products (39). During meiosis, SCC1 is replaced by a meiosis-specific variant named Rec8 (45). In yeast Rec8 mutants, the sister chromatids separate precociously at meiosis I, resulting in an equational instead of a reductional division (51). It is not known whether RAD6 plays a role in the degradation of Rec8 during yeast meiosis. In view of the present results, it will be of interest to study the metabolism of the mouse meiotic cohesion component Rec8 (34) in wild-type and HR6B knockout spermatocytes.
HR6B mutation increases the frequency of meiotic recombination in mouse spermatocytes.
The distribution of meiotic recombination sites and the frequency of meiotic recombination are strictly regulated in normal pachytene cells. In HR6B−/− spermatocytes, there is a marked increase in the number of MLH1 foci and also a significant increase in the number of chiasmata, providing evidence that the frequency of meiotic recombination is increased. The number of detected chiasmata is slightly lower than the number of MLH1 foci in HR6B knockout spermatocytes. In wild-type spermatocytes, most autosomal chromosome pairs show only one MLH1 focus and crossing-over event. When the crossing-over frequency is increased, in HR6B knockout spermatocytes, an increased number of crossover events will result in an increase in the number of bivalents with two chiasmata. Two chiasmata per bivalent are much more difficult to identify compared to bivalents with a single crossover, and we expect a systematic underestimation of the number of chiasmata in HR6B knockout spermatocytes. Still, we cannot exclude the possibility that not all MLH1 foci are converted into actual sites of crossover when there are two MLH1 foci per bivalent.
Lynn et al. (28) have shown that there appears to be a positive correlation between normal length of the SC and the number of meiotic recombination sites, indicating that the length of the SC reflects genetic rather than physical distance. Although we also find an increased SC length in association with an increased recombination frequency, we question whether in our case the two parameters are directly related. The SC length per number of recombination sites is constant between different mouse strains (28) but is increased by 20% in HR6B−/− spermatocytes. It is more likely that the increased length of the SC represents a decrease in chromatin compaction. This phenomenon has also been described for SCP3 knockout pachytene oocytes (56). In these SCP3−/− oocytes, a doubling of SC length was found, and there were many axial gaps. However, no clear increase in meiotic recombination frequency was observed (56).
Knockout of HR6A results in maternal-factor infertility, as evidenced by a block at the two-cell stage of embryonic development when HR6A−/− oocytes are fertilized by wild-type sperm (Roest et al., unpublished). In analogy to the meiotic defects in the male HR6B knockout, meiotic recombination frequency might be affected in HR6A−/− pachytene oocytes. However, we found no difference in the number of MLH1 foci between wild-type and HR6A knockout pachytene oocytes and also not between wild-type and HR6B knockout pachytene oocytes. In addition, HR6A knockout spermatocytes contained a normal number of MLH1 foci. These data indicate that, in pachytene oocytes, the level of HR6A in the absence of HR6B and vice versa may be sufficiently high to allow meiotic recombination at normal frequency and to maintain normal SC structure. Alternatively, the finding that mutation of HR6B and not HR6A influences meiotic recombination may reflect a functional difference between HR6A and HR6B. The observation of this effect in males only may be due to differences between mechanisms that regulate meiotic recombination during the male and female meiotic prophase.
Little is known about the mechanisms that control the processing of the early recombination nodules, which are in 10-fold excess of the actual meiotic recombination sites formed later. One such a mechanism is meiotic interference; this ensures that two crossover sites rarely occur in close proximity to one another. However, the distribution of MLH1 spots in HR6B knockout spermatocytes appeared to be normal. The increased crossing-over frequency may also result from an increase in the number of double-strand DNA breaks. Since RAD6 is involved in postreplication DNA repair, there may be an inability to correctly process DNA lesions during the last round of spermatogenic DNA replication, in HR6B−/− preleptotene spermatocytes. This could then lead to an increase in the number of double-strand DNA breaks, which could subsequently be converted to sites of meiotic recombination.
Exposure of HR6B knockout mice to ionizing irradiation did not result in a further increase of the recombination frequency by 3 or 7 days after irradiation. Although the results show that gamma irradiation may slightly increase the level of meiotic recombination in wild-type spermatocytes, there are no indications that HR6B−/− spermatocytes have an increased activity to convert this type of damage to sites of meiotic recombination. However, we do not exclude that impairment of DNA repair, due to the absence of HR6B, is involved in causing the meiotic phenotype.
In yeast cells there is evidence that suggests that meiotic SPO11-induced double-strand breaks occur in particular in regions containing an open chromatin structure and that changes in chromatin structure produce parallel changes in the occurrence of meiotic double-strand breaks (33, 53). Thus, based on the observations described here, certain changes in chromatin structure in spermatocytes may occur in the absence of HR6B that allow SPO11 to create DNA breaks in genomic regions that normally are not accessible to this specialized topoisomerase. This may then lead to the observed increase in meiotic recombination frequency in HR6B knockout spermatocytes.
Acknowledgments
We thank Julio E. Celis (Institute of Cancer Biology, Copenhagen, Denmark) for providing the anti-uH2A antibody (monoclonal antibody E6C5); Christa Heyting (Wageningen University, Wageningen, The Netherlands) for the anti-SCP1, -SCP2, and -SCP3 antibodies; and Roland Kanaar (Erasmus MC Rotterdam, Rotterdam, The Netherlands) for the anti-RAD51 antibody. In addition, we thank Mark Zijlmans (Erasmus MC Rotterdam) for the telomere probe and P. de Boer (UMC St. Radboud, Nijmegen, The Netherlands) for technical advice.
This work was supported by the Dutch Science Foundation (NWO) through GB-MW (Medical Sciences) and by the Dutch Cancer Society (EUR 99-2003).
REFERENCES
- 1.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. Baxter, G. Helt, C. R. Nelson, G. L. Gabor, J. F. Abril, A. Agbayani, H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, L. K., A. Reeves, L. M. Webb, and T. Ashley. 1999. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151:1569-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baarends, W. M., J. W. Hoogerbrugge, H. P. Roest, M. Ooms, J. Vreeburg, J. H. J. Hoeijmakers, and J. A. Grootegoed. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 207:322-333. [DOI] [PubMed] [Google Scholar]
- 4.Baarends, W. M., R. van der Laan, and J. A. Grootegoed. 2000. Specific aspects of the ubiquitin system in spermatogenesis. J. Endocrinol. Investig. 23:597-604. [DOI] [PubMed] [Google Scholar]
- 5.Baker, S. M., A. W. Plug, T. A. Prolla, C. E. Bronner, A. C. Harris, X. Yao, D. M. Christie, C. Monell, N. Arnheim, A. Bradley, T. Ashley, and R. M. Liskay. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13:336-342. [DOI] [PubMed] [Google Scholar]
- 6.Barlow, A. L., F. E. Benson, S. C. West, and M. A. Hulten. 1997. Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 16:5207-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking spo11. Mol. Cell 6:989-998. [DOI] [PubMed] [Google Scholar]
- 8.Borts, R. H., M. Lichten, and J. E. Haber. 1986. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics 113:551-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briggs, S. D., T. Xiao, Z. W. Sun, J. A. Caldwell, J. Shabanowitz, D. F. Hunt, C. D. Allis, and B. D. Strahl. 2002. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498. [DOI] [PubMed] [Google Scholar]
- 10.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255-269. [DOI] [PubMed] [Google Scholar]
- 11.Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis, and F. Winston. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in Saccharomyces cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12:165-170. [DOI] [PubMed] [Google Scholar]
- 12.Conaway, R. C., C. S. Brower, and J. W. Conaway. 2002. Emerging roles of ubiquitin in transcription regulation. Science 296:1254-1258. [DOI] [PubMed] [Google Scholar]
- 13.Dover, J., J. Schneider, M. A. Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by RAD6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 14.Evans, E. P., G. Breckon, and C. E. Ford. 1964. An air-drying method for meiotic preparations from mammalian testes. Cytogenetics 3:289-294. [DOI] [PubMed] [Google Scholar]
- 15.Game, J. C., T. J. Zamb, R. J. Braun, M. Resnick, and R. M. Roth. 1980. The role of radiation (rad) genes in meiotic recombination in yeast. Genetics 94:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavrieli, Y., Y. Sherman, and S. A. Ben-Sasson. 1992. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119:493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorczyca, W., J. Gong, and Z. Darzynkiewicz. 1993. Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res. 53:1945-1951. [PubMed] [Google Scholar]
- 18.Grootegoed, J. A., W. M. Baarends, H. P. Roest, and J. H. Hoeijmakers. 1998. Knockout mouse models and gametogenic failure. Mol. Cell. Endocrinol. 145:161-166. [DOI] [PubMed] [Google Scholar]
- 19.Grootegoed, J. A., R. Jansen, and H. J. van der Molen. 1986. Effect of glucose on ATP dephosphorylation in rat spermatids. J. Reprod. Fertil. 77:99-107. [DOI] [PubMed] [Google Scholar]
- 20.Hanneman, W. H., M. E. Legare, S. Sweeney, and J. C. Schimenti. 1997. Cisplatin increases meiotic crossing-over in mice. Proc. Natl. Acad. Sci. USA 94:8681-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heyting, C. 1996. Synaptonemal complexes: structure and function. Curr. Opin. Cell Biol. 8:389-396. [DOI] [PubMed] [Google Scholar]
- 22.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell. Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 23.Huang, H., A. Kahana, D. E. Gottschling, L. Prakash, and S. W. Liebman. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koken, M. H., P. Reynolds, I. Jaspers-Dekker, L. Prakash, S. Prakash, D. Bootsma, and J. H. Hoeijmakers. 1991. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. USA 88:8865-8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koken, M. H. M., J. W. Hoogerbrugge, I. Jaspers-Dekker, J. de Wit, R. Willemsen, H. P. Roest, J. A. Grootegoed, and J. H. J. Hoeijmakers. 1996. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 173:119-132. [DOI] [PubMed] [Google Scholar]
- 26.Krogan, N. J., J. Dover, S. Khorrami, J. F. Greenblatt, J. Schneider, M. Johnston, and A. Shilatifard. 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 277:10753-10755. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, C. 1994. The RAD6 repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays 16:253-258. [DOI] [PubMed] [Google Scholar]
- 28.Lynn, A., K. E. Koehler, L. Judis, E. R. Chan, J. P. Cherry, S. Schwartz, A. Seftel, P. A. Hunt, and T. J. Hassold. 2002. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science 296:2222-2225. [DOI] [PubMed] [Google Scholar]
- 29.Matsui, S., A. A. Sandberg, S. Negoro, B. K. Seon, and G. Goldstein. 1982. Isopeptidase: a novel eukaryotic enzyme that cleaves isopeptide bonds. Proc. Natl. Acad. Sci. USA 79:1535-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moens, P. B., D. J. Chen, Z. Shen, N. Kolas, M. Tarsounas, and H. H. Q. Heng. 1997. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106:207-215. [DOI] [PubMed] [Google Scholar]
- 31.Moens, P. B., N. K. Kolas, M. Tarsounas, E. Marcon, P. E. Cohen, and B. Spyropoulos. 2002. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115:1611-1622. [DOI] [PubMed] [Google Scholar]
- 32.Montelone, B. A., S. Prakash, and L. Prakash. 1981. Recombination and mutagenesis in rad6 mutants of Saccharomyces cerevisiae: evidence for multiple functions of the RAD6 gene. Mol. Gen. Genet. 184:410-415. [DOI] [PubMed] [Google Scholar]
- 33.Nicolas, A. 1998. Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc. Natl. Acad. Sci. USA 95:87-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisi, S., M. J. McKay, M. Molnar, M. A. Thompson, P. J. van der Spek, E. van Drunen-Schoenmaker, R. Kanaar, E. Lehmann, J. H. Hoeijmakers, and J. Kohli. 1999. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol. Cell. Biol. 19:3515-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters, A. H., A. W. Plug, M. J. van Vugt, and P. de Boer. 1997. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5:66-68. [DOI] [PubMed] [Google Scholar]
- 36.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 37.Plug, A. W., J. Xu, G. Reddy, E. I. Golub, and T. Ashley. 1996. Presynaptic association of Rad51 protein with selected sites in meiotic chromatin. Proc. Natl. Acad. Sci. USA 11:5920-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajapurohitam, V., C. R. Morales, M. El-Alfy, S. Lefrancois, N. Bedard, and S. S. Wing. 1999. Activation of a UBC4-dependent pathway of ubiquitin conjugation during postnatal development of the rat testis. Dev. Biol. 212:217-228. [DOI] [PubMed] [Google Scholar]
- 39.Rao, H., F. Uhlmann, K. Nasmyth, and A. Varshavsky. 2001. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature 410:955-959. [DOI] [PubMed] [Google Scholar]
- 40.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 41.Roest, H. P., J. van Klaveren, J. de Wit, C. G. van Gurp, M. H. M. Koken, M. Vermey, J. H. van Roijen, J. T. M. Vreeburg, W. M. Baarends, D. Bootsma, J. A. Grootegoed, and J. H. J. Hoeijmakers. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes a defect in spermatogenesis associated with chromatin modification. Cell 86:799-810. [DOI] [PubMed] [Google Scholar]
- 42.Romanienko, P. J., and R. D. Camerini-Otero. 1999. Cloning, characterization, and localization of mouse and human SPO11. Genomics 61:156-169. [DOI] [PubMed] [Google Scholar]
- 43.Romanienko, P. J., and R. D. Camerini-Otero. 2000. The mouse spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6:975-987. [DOI] [PubMed] [Google Scholar]
- 44.Singh, J., V. Goel, and A. J. Klar. 1998. A novel function of the DNA repair gene rhp6 in mating-type silencing by chromatin remodeling in fission yeast. Mol. Cell. Biol. 18:5511-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stoop-Myer, C., and A. Amon. 1999. Meiosis: Rec8 is the reason for cohesion. Nat. Cell. Biol. 1:E125-E127. [DOI] [PubMed] [Google Scholar]
- 46.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 47.Tarsounas, M., T. Morita, R. E. Pearlman, and P. B. Moens. 1999. RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J. Cell Biol. 147:207-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37-42. [DOI] [PubMed] [Google Scholar]
- 49.van der Laan, R., H. Roest, J. Hoogerbrugge, E. Smit, R. Slater, W. Baarends, J. Hoeijmakers, and J. Grootegoed. 2000. Characterization of mRAD18Sc, a mouse homolog of the yeast post-replication repair gen RAD18. Genomics 69:86-94. [DOI] [PubMed] [Google Scholar]
- 50.van Heemst, D., and C. Heyting. 2000. Sister chromatid cohesion and recombination in meiosis. Chromosoma 109:10-26. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe, Y., and P. Nurse. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400:461-464. [DOI] [PubMed] [Google Scholar]
- 52.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 53.Wu, T. C., and M. Lichten. 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263:515-518. [DOI] [PubMed] [Google Scholar]
- 54.Xiao, W., B. L. Chow, S. Broomfield, and M. Hanna. 2000. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics 155:1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xin, H., W. Lin, W. Sumanasekera, Y. Zhang, X. Wu, and Z. Wang. 2000. The human RAD18 gene product interacts with HHR6A and HHR6B. Nucleic Acids Res. 28:2847-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan, L., J. G. Liu, M. R. Hoja, J. Wilbertz, K. Nordqvist, and C. Hoog. 2002. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296:1115-1118. [DOI] [PubMed] [Google Scholar]