Abstract
FLI-1 is an ETS family transcription factor which is overexpressed in Friend erythroleukemia and contributes to the blockage of differentiation of erythroleukemic cells. We show here that FLI-1 represses the transcriptional activity of the β-globin gene promoter in MEL cells and interacts with two of its critical transactivators, GATA-1 and EKLF. Unexpectedly, FLI-1 enhances the stimulating activity of GATA-1 on a GATA-1-responsive promoter but represses that of EKLF on β-globin and an EKLF-responsive artificial promoters. This repressive effect of FLI-1 requires the ETS DNA binding domain and its association with either the N- or C-terminal domain, which themselves interact with EKLF but not with GATA-1. Furthermore, the FLI-1 ETS domain alone behaves as an autonomous repression domain when linked to the Gal4 DNA binding domain. Taken together, these data indicate that FLI-1 represses EKLF-dependent transcription due to the repression activity of its ETS domain and its indirect recruitment to erythroid promoters by protein-protein interaction with EKLF. Reciprocally, we also show that EKLF itself represses the FLI-1-dependent megakaryocytic GPIX gene promoter, thus further suggesting that functional cross-antagonism between FLI-1 and EKLF might be involved in the control of the erythrocytic versus megakaryocytic differentiation of bipotential progenitors.
The ETS gene family encodes transcriptional regulators which are involved in multiple cellular and developmental processes. ETS proteins are characterized by a conserved ETS domain, which is responsible for their DNA binding to the core motif GGA. Other domains are involved in either activation or repression of transcription. Specificity is conferred both at the level of DNA binding and through interactions of ETS proteins with other transcriptional regulators. SPI-1/PU.1 and FLI-1 proteins are two divergent members of the ETS family initially identified as transactivators which are both involved in hematopoiesis and in erythroleukemia induced by the Friend viral complex in mice.
SPI-1/PU.1 or FLI-1 overexpression by proviral insertion mutagenesis is a recurrent event observed in murine clonal erythroleukemia that is induced, respectively, by the spleen focus-forming virus (SFFV) or the Friend murine leukemia virus component of the Friend viral complex (8, 45, 68). Direct evidence for deregulated expression of SPI-1/PU.1 in leukemogenesis was provided by the spontaneous development of multistep erythroleukemia in SPI-1 transgenic mice (4, 5, 44). SPI-1/PU.1 overexpression is involved in both the survival and the inhibition of differentiation of erythroid cells (47). Indeed, the chemical induction of terminal differentiation of SFFV erythroleukemic cell lines is associated with the downregulation of SPI-1/PU.1, and this differentiation is blocked by enforced expression of SPI-1/PU.1 (54, 69). In avian primary erythroblasts, SPI-1/PU.1 cooperates with an activated erythropoietin receptor (EPO-R) to inhibit apoptosis and Epo-dependent differentiation (52, 53). Importantly, this transforming property of SPI-1/PU.1 evidenced in avian erythroblasts is strictly dependent upon specifically activated forms of EPO-R (52). Cooperation between SPI-1/PU.1 and EPO-R activation by the gp55env protein encoded by SFFV has also been recently demonstrated with mice (1). One of the known contributions of SPI-1/PU.1 in erythroleukemia is to inhibit the activity of GATA-1, which is one of the most critical DNA binding factors required for erythroid cell differentiation and survival. Indeed, several studies have shown that the very N-terminal region and the ETS domain of SPI-1/PU.1 interact with the Zn fingers region of GATA-1 (40, 46, 47, 55, 70, 78). Some of these studies have shown that the N-terminal region of SPI-1/PU.1 is involved in the inhibition of the DNA-binding of GATA-1 both in vitro and in vivo. However, other studies have shown that although the ETS domain of SPI-1/PU.1 is not required to inhibit the DNA binding of GATA-1, it is nevertheless required to fully inhibit erythroid differentiation. Very recently, SPI-1/PU.1 has been shown to interact with the acetyltransferase domain of CBP (71, 29) and to inhibit the CBP-mediated acetylation of several nuclear proteins including GATA-1 itself. SPI-1/PU.1 has also been shown to interact and to interfere with the splicing activity of p54rnb (26) and TLS (27) and to interact with histone deacetylase 1 (HDAC1) (32). However, the significance of these other interactions of SPI-1/PU.1 in the induction of erythroleukemia is still unclear. Furthermore, we have shown recently that SPI-1/PU.1 is a transcriptional activator of the fli-1 gene (63), thus further suggesting that SPI-1/PU.1 overexpression might also contribute to erythroleukemia through the induction of FLI-1 overexpression (see below). Outside erythroleukemic contexts, SPI-1/PU.1 is normally expressed in myeloid and B-lymphoid cell lineages, and targeted inactivation of the mouse Spi-1/PU.1 gene has shown that it is critically required for myeloid and B-lymphoid cell differentiation (10, 48, 60, 61). Interestingly enough, GATA-1 behaves as a functional antagonist of SPI-1/PU.1 by impeding its interaction with one of its cofactors, c-JUN, which is required for the transcriptional activation of myeloid genes (77). Further studies have shown that due to this functional cross-antagonism between GATA-1 and SPI-1/PU.1, the relative levels of the two proteins constitute a critical determinant controlling the balance between myeloid and erythrocytic differentiation (17, 29, 55).
Unexpectedly, FLI-1 transgenic mice do not spontaneously develop erythroleukemia but develop an autoimmune renal disease associated with enhanced survival of lymphoid cells (76). Nevertheless, several concordant results strongly argue that FLI-1 overexpression undoubtedly contributes to erythroleukemia (41). For example, members of our group and others have shown that the forced expression of FLI-1 inhibits erythroid differentiation in several mouse or human erythroleukemic cell lines (2, 3, 56, 63, 66, 68, 80). Likewise, FLI-1 inhibits the differentiation and induces the proliferation of avian primary erythroblasts in response to EPO (36, 51). Unlike SPI-1/PU.1, the inhibition of erythroid differentiation and apoptosis by FLI-1 in avian primary erythroblasts does not require constitutive activation of the EPO-R signaling (36, 51). Increased resistance to apoptosis has also been observed in FLI-1-transfected 3T3 cells (75), and it has been shown recently that FLI-1 is directly involved in the transcriptional activation of the BCL-2 gene in avian erythroblasts (36). However, enforced expression of BCL-2 alone is unable to inhibit erythroid differentiation of avian erythroblasts, thus implying that FLI-1 deregulates other pathways distinct from BCL-2 up-regulation to block differentiation (36). Among other effects, FLI-1 has been described to contribute partially to the transcriptional repression of the Rb gene (66), to interfere with the splicing function of EWS (33), and to interfere with nuclear hormone receptor response (15), but the significance of these other effects in the context of erythroleukemia remain to be established. Outside erythroleukemic contexts, targeted inactivation of the fli-1 gene has shown that FLI-1 is required for embryonic vasculogenesis and megakaryopoiesis (28, 31, 62). Consistent with the role of FLI-1 in megakaryopoiesis, FLI-1 can activate the promoters of several megakaryocyte-specific genes (GPIX and cMPL) (6, 7, 19, 23), and the forced expression of FLI-1 in K562 cells stimulates their megakaryocytic differentiation at the expense of their erythroid differentiation (2, 3).
The aim of this study was to investigate the molecular mechanisms responsible for the inhibition of erythroid differentiation by FLI-1. We show here, by transient expression assays, that FLI-1 is involved in the specific inhibition of EKLF-dependent transcription of β-globin gene and artificial promoters. By using deletion mutants of FLI-1, we determined that this repression by FLI-1 is critically dependent on its ETS domain, which behaves itself as an autonomous repression domain when it is recruited indirectly to promoters through its association either with the Gal4 DNA binding domain or with the N- or C-terminal domains of FLI-1, themselves interacting with EKLF. Interestingly, we also show that EKLF itself inhibits FLI-1-dependent transcription of the megakaryocytic GPIX gene and artificial FLI-1-responsive promoters, thus further indicating that functional antagonism between FLI-1 and EKLF is reciprocal. Together, these results suggest that the functional cross-antagonism between FLI-1 and EKLF not only is involved in the inhibition of erythroleukemic cell differentiation but also might be involved in the control of erythrocytic versus megakaryocytic differentiation of bipotential progenitors in vivo.
MATERIALS AND METHODS
Cell culture transfection and reporter gene assays.
COS cells, 3T3 cells, and MEL cells (clone 745-A) (63) were grown in Iscove modified Dulbecco’s medium supplemented with 10% fetal calf serum and antibiotics. The differentiation of MEL cells was induced by the addition of 5 mM hexamethylene-bisacetamide (HMBA). Transfections were done in six-well culture plates using DAC-30 (Eurogentec) according to the manufacturer's recommendations. In all experiments, each well was transfected by a constant amount of total DNA (usually 3 μg) and a fixed DAC-30/DNA ratio of 5 (wt/wt). The total amount of transfected DNA was kept constant by adding appropriate amounts of empty vector. Except for MEL cells, each transfection mix also included a fixed amount of pCMV-βGal DNA used to control transfection efficiency. Green fluorescent protein (GFP) fluorescence was determined by fluorescence-activated cell sorter (FACS) analysis of the transfected MEL cells, and the total fluorescence activity was estimated by the product of the number of GFP-positive cells and their mean fluorescence level. Luciferase, β-galactosidase (β-Gal), and chloramphenicol acetyltransferase (CAT) activities were determined on cell lysates, respectively, by luminometry, chemiluminescence, and enzyme-linked immunosorbent assay (ELISA) using appropriate kits (Roche) and according to the manufacturer's recommendations. Growth hormone activities were determined on the supernatants by immunoassay using the hGH ELISA kit (Roche). CAT, LUC, or hGH activities were standardized on β-Gal activities, and final results were expressed as relative values.
Plasmid constructs.
The following plasmids were used as reporters. pEV3-GFP was constructed by first subcloning a HindIII/XbaIII GFP cassette taken from the pGFPemd-P plasmid (Packard) into pBSIISK+ plasmid (Stratagene). This GFP cassette was then isolated by XbaIII/SalI digestion and subcloned into the NotI/SalI sites of the pEV3-EKLF vector (67) in place of the EKLF cDNA. The pC1G3CAT (EKLF responsive; a gift from J. Bieker) (43), pM1αGH (GATA-1 responsive; a gift from J. Crispino) (39), and pTORU-TK-LUC (ETS and AP1 responsive; a gift from M Duterque-Coquillaud) (25) reporters have been already described. Reporter plasmids harboring Gal4 binding sites (pGal4-TK-LUC and its control, pTK-LUC, devoid of Gal4 binding sites) (35) or Gal4 and LexA binding sites (L8G5-LUC and its control, L8-LUC, devoid of Gal4 binding sites) (35) were obtained from S. Khochbin. The pGPIX5′-203LUC reporter harboring the minimal promoter of the human GPIX gene (6) was kindly provided by G. Roth.
The following plasmids were used to produce glutathione-S-transferase (GST) fusion proteins in bacteria. pGEX2T-GATA1 (amino acids [aa] 1 to 413) and pGEX2T-GATA-N-C (aa 200 to 318) (a gift from M. Crossley), pGST-EKLF-C′2 (aa 57 to 376), pGST-EKLF PRO (aa 20 to 291), and pGST-EKLF-ZnF (aa 287 to 376) (9) (a gift from J. Bieker) and pGEX2T-FLI (12) (a gift from M. Duterque-Coquillaud) have already been described.
The following plasmids were used to produce EKLF proteins either by transfection or by in vitro translation. pSG5-EKLF (a gift from J. Bieker) (43) and pSG5-HA-EKLF (encoding a hemagglutinin [HA]-tagged version of EKLF kindly provided by T. Townes) (50) have already been described (Fig. 1). pSG5-HA-EKLF PRO was constructed by cutting pSG5-HA-EKLF by SacI and BamHI, filling ends by Klenow DNA polymerase and religation. pSG5-HA-EKLF-Zn was constructed similarly by cutting pSG5-HA-EKLF by Bpu10I and XbaI, filling ends by Kleenow DNA polymerase and religation.
FIG. 1.
Schematic structure of the FLI-1 and EKLF proteins and deletion or fusion mutants derived thereof. Proteins encoded by the expression plasmids used in this study are depicted. The ETS DNA binding domain of FLI-1 (aa 276 to 360) and the three-zinc-fingers DNA binding domain of EKLF (aa 293 to 376) are shown as black boxes. GAL4 (aa 1 to 147) and HA (21 aa) encoded sequences are shown as hatched boxes, and ER (aa 281 to 599) encoded sequences are shown as a grey box. Other domains are shown as white boxes. Details of the constructions are given in Material and Methods.
The following plasmids were used to produce FLI-1 proteins either by transfection or by in vitro translation. pEF-FLI WT (56, 63) and pSG5-FLI WT (12) have already been described. pcDNA3-FLI WT was obtained by subcloning the NotI FLI-1 cDNA cassette taken from pEF FLI WT into pcDNA3 (Invitrogen). pSG5 FLI 276-403, pSG5 FLI 276-452, pSG5 FLI 1-403, and pSG5 FLI Δ276-361 (Fig. 1) were generated by PCR from pSG5 FLI 1WT. pcDNA3 FLI 345-452 and pcDNA3 FLI 262-452 were generated by PCR from pcDNA3 FLI. pcDNA Gal4-FLI 276-370, pcDNA Gal4-FLI WT, and pcDNA Gal4-FLI Δ276-361 (Fig. 1) were obtained by PCR amplification of the corresponding FLI-1 sequences followed by subcloning them in frame to the Gal4 1-147 coding sequence into pcDNA3 Gal4 (a gift from S. Khochbin). pcDNA3 FLI-I347E carrying the mutation I347E in the ETS domain of FLI-1 (30) was generated from pcDNA3 FLI WT using the QuickChange site-directed mutagenesis kit (Stratagene) and the following oligonucleotide: CTATGACAAAAACGAGATGACCAAAGTGCATG. pEF FLI ER was generated from pEF FLI WT by first destroying the stop codon of FLI-1 using PCR (generating a BclI site) and further subcloning in frame the BamHI/EcoRI fragment encoding the hormone binding domain of the human estrogen receptor (aa 281 to 599) isolated from pBpuroMycER (a gift from T. Littlewood) (38). pCMV Gal4 Elk 1-205 encoding a fusion protein between the ETS domain of Elk1 and the DNA binding domain of Gal4 (a gift from A. Sharrocks) has already been described (73).
Protein expression and purification.
GST fusion proteins were expressed in Escherichia coli BL21 and purified using a standard protocol. A colony was inoculated into 5 ml of Luria-Bertani broth and incubated for 8 to 10 h at 37°C. The 5-ml subculture was then diluted in 45 ml of Luria-Bertani broth and incubated for 1 h at 37°C. Isopropyl-d-galactopyranoside was added to a 0.1 mM final concentration, and the culture was incubated at 37°C for 3 h. After centrifugation, the bacterial pellet was resuspended in 1.5 ml of PBS-NaK (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 1% Triton X-100, supplemented with 2 mM orthovanadate and with 50-μl/ml protease inhibitor cocktail [Complete; Roche]) and lysed by sonication on ice. Lysates were centrifuged at 15,000 × g for 20 min at 4°C, and GST fusion proteins present in the supernatant were purified on glutatnione-Sepharose beads (Amersham). Beads were washed five times in PBS-NaK and resuspended at 50% (vol/vol) in PBS-NaK. The protein concentration was determined by the Bradford assay (Bio-Rad), and the quality of GST fusion proteins was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by Coomassie blue staining.
The synthesis of the FLI-1, EKLF, or GATA-1 protein by in vitro transcription and translation was carried out with the TNT-coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine (Amersham) according to the manufacturer's recommendations.
In vitro protein-protein interaction assays.
Interactions between each GST fusion and in vitro-translated proteins were investigated using pull-down assays. Briefly, 10 μg of GST or GST fusion protein bound to glutathione-Sepharose beads was incubated for 1 h at 4°C with 100,000 cpm of 35S-labeled in vitro-translated proteins in 50 μl of binding buffer (50 mM HEPES [pH 7.6], 1 mM EDTA, 150 mM NaCl, 5 mM KCl, 0.1 mM ZnCl2, 0.5 mM dithiothreitol, 10% glycerol, 10% Triton X-100 supplemented with 2 mM orthovanadate, and 50-μl/ml protease inhibitor cocktail [Complete; Roche]). Beads were then centrifuged for 3 min at 400 × g and washed five times with 250 μl of binding buffer. The final pellet was resuspended in SDS sample buffer and analyzed by SDS-PAGE followed by autoradiography.
Gel shift analyses.
Double-stranded, synthetic oligonucleotides that contain either a CAC binding site derived from the mouse β-Major globin gene (5′-TCGAGTCCGTAGAGCCACACCCTGCAGAGCATATAAGGTGAGGTAGTGCA-3′) or a GGAA binding site derived from the human GPIIb gene (5′-GGGAAGCTTCAGCATCCTCCACAGGAAGTCTTTGGCTCCCG-3′) were used as probes. Purified GST-EKLF (∼1 μg) was incubated with 200 pg of the labeled CAC probe in the presence or absence of increasing excess (1-, 5-, or 10-fold) of purified GST-FLI-1. Reciprocally, GST-FLI-1 (∼1 μg) was incubated with 200 pg of the labeled GGAA probe in the presence of increasing excess of GST-EKLF. Binding reactions were left for 60 min on ice before they were loaded on a nondenaturing 5% polyacrylamide gel as previously described (63). After electrophoresis, the gel was dried and exposed to film.
Immunoprecipitation and Western blot analyses.
The in vivo interaction between FLI-1 and GATA-1 was addressed by coimmunoprecipitation of endogenous proteins present in MEL cell extracts. MEL cells (8 × 107) were resuspended in 2 ml of lysis buffer (50 mM HEPES [pH 7.6], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.125% NP-40, 0.875% Brij-96, and protease inhibitor cocktail [Complete; Roche]). The suspension was incubated for 1 h on ice. The cell extract was precleared by centrifugation at 18,000 × g for 30 min at 4°C and stored in 1-ml aliquots at −80°C until use. Six micrograms of specific (mouse monoclonal anti-FLI-1 [42]) or control (mouse monoclonal anti-PML [PMG3; Santa Cruz Biotechnology]) antibodies was added together with 100 μl of protein G-Sepharose (50% slurry) to 1 ml of cell extract and left in an orbital shaker for 12 h at 4°C. The Sepharose beads were washed eight times in lysis buffer, resuspended in 50 μl of SDS sample buffer, and loaded for SDS-10% PAGE, followed by Western blot analysis. Western blot analysis was performed as previously described (56, 63) using rabbit polyclonal anti-GATA-1 primary antibody and horseradish peroxidase-conjugated secondary antibody, followed by ECL (Amersham). Western blot analyses of HA-EKLF and FLI-1 proteins produced in 3T3 or COS transfected cells were performed on total cells lysates and using commercial FLI-1 (Sc356; Santa Cruz) or HA (Sc-805; Santa Cruz) antibodies as previously described (56).
RESULTS
Transient transfection mimics the inhibition of HMBA-induced endogenous β-globin gene transcription in MEL cells overexpressing FLI-1.
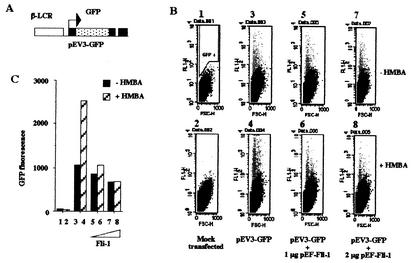
We have shown previously that enforced expression of FLI-1 protein inhibits HMBA-induced differentiation of MEL cells, including HMBA-induced β-globin gene transcription (63; also data not shown). To understand the molecular mechanisms involved in this inhibition, we decided to investigate the effect of FLI-1 on the transcriptional activity of the β-globin gene promoter in transient expression assays performed in HMBA-induced or uninduced MEL cells. For that purpose, we designed the pEV3-GFP reporter construct (Fig. 2A), which contains EGFP cDNA placed under the control of the human β-globin gene promoter and β-locus control region. MEL cells were transfected with pEV3-GFP, and the number and mean fluorescence of GFP-positive cells were determined by FACS analyses (Fig. 2B). The products of these two parameters were taken as an indication of the relative fluorescence activities of the construct in the different conditions (Fig. 2C). We found that the activity of the pEV3-GFP reporter is minimally affected by the cotransfection of the FLI-1 expression vector in the absence of HMBA (Fig. 2C, compare lanes 3, 5, and 7). In the absence of FLI-1, this activity increases by 2.5-fold in response to HMBA (Fig. 2C, lanes 3 and 4). In marked contrast, this HMBA-induced increase is progressively inhibited by the cotransfection of increasing amounts of FLI-1 (Fig. 2C, lanes 3 to 8). These data indicated that enforced expression of FLI-1 inhibits the HMBA-induced activation of a transfected β-globin gene promoter in MEL cells, thus mimicking the inhibition of endogenous β-globin gene transcription.
FIG. 2.
Enforced expression of FLI-1 inhibits β-globin gene promoter activity in MEL cells. (A) Schematic structure of the pEV3-GFP reporter construct. (B) MEL cells were transfected with a fixed amount of the pEV3-GFP plasmid in the presence or absence of increasing doses of the FLI-1 expression vector pEF-FLI-1 as indicated. Immediately following transfection, each set of transfected cells was cultured for 72 h in the absence (upper panels) or in the presence (lower panels) of 5 mM HMBA. The mean fluorescence and the number of GFP-positive cells were then determined by FACS analysis on forward-scattering (FSC) versus fluorescence (FL1) records. The gate used to identify positive cells (GFP+: grey spots) is shown on panel 1. (C) Quantitative analysis of the data presented in panel B. Total fluorescence activities were quantified by determining the product of the percentage of GFP-positive cells and their mean fluorescence value. Each lane corresponds, respectively, to the different panels presented in panel B. Panels B and C show typical results obtained from at least three different transfection experiments.
Transcriptional activity of FLI-1 is not required to inhibit β-globin gene transcription.
Since FLI-1 is known to be a transactivator, its negative effect on β-globin gene transcription could be mediated through the transactivation of gene(s) encoding repressor(s). To facilitate the identification of such putative repressor target gene(s) of FLI-1, we thought to design a tamoxifen-inducible FLI-1 protein. For that purpose, the cDNA sequence encoding the mouse wild-type FLI-1 protein was fused in frame to the cDNA sequence encoding the hormone binding domain (aa 281 to 599) of the tamoxifen-inducible mutant version of the human estrogen receptor (38) (Fig. 1). The resulting coding sequence was subcloned under the control of the human EF1α gene promoter into the pEF-LAC plasmid, thus leading to the pEF-FLI-ER expression vector. MEL cells were then cotransfected with equal amounts of either pEF-FLI-1 or pEF-FLI-ER expression vectors and the two reporter constructs, pEV3-GFP and TORU-TK-LUC, which are, respectively, repressed or induced by wild-type FLI-1. As expected, we confirmed that the HMBA-induced expression of the pEV3-GFP reporter (Fig. 3A, compare lanes 1 and 2) is inhibited by wild-type FLI-1 (Fig. 3A, compare lanes 1 and 2 to 3 and 4) independently of the presence of hydroxy-tamoxifen (Fig. 3A, compare lanes 3 and 4 to 5 and 6). We found also that the HMBA-induced expression of the pEV3-GFP reporter is not inhibited by FLI-ER alone (Fig. 3A, compare lanes 7 and 8 to 1 and 2) but is progressively inhibited by increasing doses of hydroxy-tamoxifen (Fig. 3A, lanes 9 and 10). These results thus indicated that the FLI-ER protein behaves as a conditional inhibitor of pEV3-GFP reporter activity depending on the presence of hydroxy-tamoxifen. Symmetrically, we found that the activity of the TORU-TK-LUC reporter is stimulated by wild-type FLI-1 (Fig. 3B, compare lanes 1 and 3), the extent of the stimulation being much more pronounced in the presence (lanes 4, 5, and 6) than in the absence (lane 3) of HMBA. This difference in the extent of the stimulation can be explained by the known reduction of endogenous FLI-1 levels in HMBA-treated cells (63). In marked contrast, FLI-ER is completely unable to stimulate the TORU-TK-LUC reporter even in the presence of both hydroxy-tamoxifen and HMBA (Fig. 3B, lanes 9 and 10). Taken together, these data demonstrated that at least in the FLI-ER context, the repressing effect of FLI-1 on β-globin gene transcription can be uncoupled from its transactivating activity, thus arguing against the possibility that FLI-1 acts through the activation of gene(s) encoding repressor(s). This led us to suspect that FLI-1 might act directly through protein-protein interaction with transcription factors involved in the positive regulation of β-globin gene transcription.
FIG. 3.
Transactivation activity of FLI-1 is not required to inhibit β-globin gene transcription. MEL cells were transiently cotransfected with the pEV3-GFP and FLI-responsive pTORU-TK-LUC reporter plasmids in the presence of the same amount of expression vectors encoding wild-type FLI-1 or FLI-ER proteins. Immediately following transfection, transfected cells were cultured for 72 h in the presence or absence of HMBA and in the presence of increasing doses of hydroxy-tamoxifen (OHT) (150 or 750 nM). The figure shows typical results of GFP (A) and luciferase (B) activities obtained from two different experiments.
FLI-1 interacts with GATA-1 and EKLF.
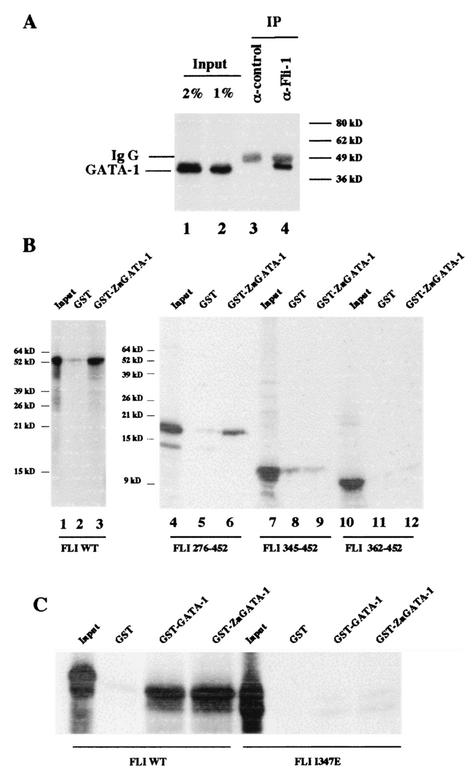
Among several putative FLI-1 partners in MEL cells, we decided to focus our investigation on GATA-1 and EKLF, which correspond to the two most important DNA binding proteins involved in the positive control of the β-globin gene promoter (10, 11, 21, 24, 37, 67). Preliminary experiments showed that recombinant GATA-1 protein (22) coupled to Sepharose 4B, but not Sepharose 4B alone, is able to retain FLI-1 protein present in 745-A cell nuclear extracts (data not shown). Reciprocally, we found by GST pull-down assays that GATA-1 protein present in 745-A cell nuclear extracts can interact with a GST-FLI-1 fusion protein but not with GST alone (data not shown). Further evidence for physical interaction between FLI-1 and GATA-1 was obtained by coimmunoprecipitation experiments showing that GATA-1 protein present in 745-A cell extracts can be coimmunoprecipitated with FLI-1 (Fig. 4A). By GST pull-down assay, we found that the full-length FLI-1 protein interacts with a GST protein fused to the zinc finger region of GATA-1 (aa 200 to 318) but not with GST alone (Fig. 4B, lanes 2 and 3). This interaction still occurs after deletion of the N-terminal part of FLI-1 (Fig. 4B, lanes 4 to 6), but it is completely lost as soon as the N-terminal deletion reaches position 345 in the ETS domain (Fig. 4B, lanes 7 to 12). We also determined that a full-length FLI-1 protein carrying a point mutation (I347E) in the ETS domain (30) fails to interact with GST-Zn-GATA-1, thus confirming that the ETS domain is the only domain of FLI-1 interacting with the Zn finger domain of GATA-1 (Fig. 4C).
FIG. 4.
Interaction between FLI-1 and GATA-1 proteins. (A) MEL cell nuclear extracts were immunoprecipitated with an irrelevant antibody (lane 3) or a FLI-1 specific monoclonal antibody (lane 4). Immunoprecipitated proteins were then analyzed by Western blotting using a GATA-1-specific antibody. Lanes 1 and 2 correspond, respectively, to 2 and 1% of the total cell extract used for immunoprecipitation. The positions of immunoglobulin G (Ig G) and GATA-1 proteins are indicated to the left. (B and C) Interaction of 35S-labeled deletion mutants (B), point mutant (C), or full-length FLI-1 protein (B and C) with GST, GST-GATA-1, and GST-Zn-GATA-1 was investigated by GST pull-down assays. Equal amounts of 35S-labeled proteins were used in each reaction, and 5% of the input proteins are shown in the input lanes.
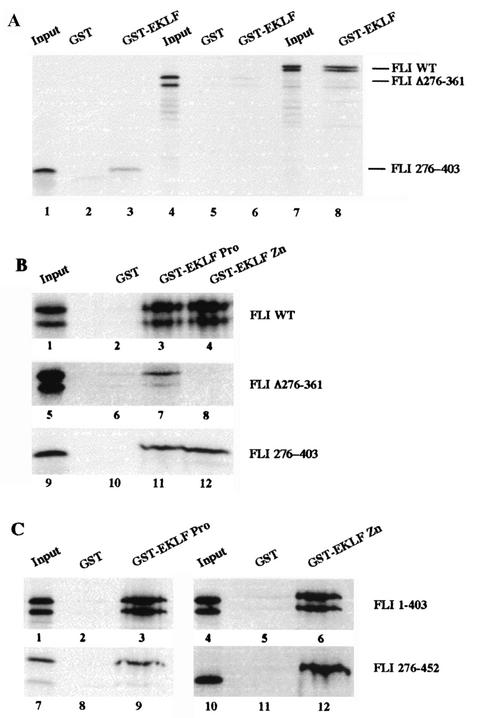
Similarly, we found that a GST-EKLF fusion protein is able to capture FLI-1 protein present in 745-A cell nuclear extracts, thus indicating physical interaction between FLI-1 and EKLF (data not shown). Further GST-pull down assays were performed to identify the domains of EKLF and FLI-1 involved in the interaction (Fig. 5). The FLI 276-403 protein corresponding to the FLI-1 ETS domain alone interacts with all three GST-EKLF fusion proteins tested (GST-EKLF [Fig. 5A, lane 3], GST-EKLF Pro [Fig. 5B, lane 11], and GST-EKLF Zn [Fig. 5B, lane 12]). FLI Δ276-361 carrying a deletion of the ETS domain still interacts with GST-EKLF (Fig. 5A, lane 6), thus indicating that the N- and/or C-terminal domains of FLI-1 also interact with EKLF. However, in contrast to the ETS domain alone, the N- and/or C-terminal domains of FLI-1 can interact only with GST-EKLF PRO (Fig. 5B, lane 7) but not with GST-EKLF Zn (Fig. 5B, lane 8). In agreement with these results, FLI 276-452 and FLI 1-403 carrying, respectively, a deletion of the N- or the C-terminal domain interacts both with GST-EKLF PRO (aa 20 to 291) (Fig. 5C, lanes 3 and 9) and GST-EKLF Zn (aa 287 to 376) (Fig. 5C, lanes 6 and 12). Together, these results thus indicated that the ETS domain of FLI-1 interacts with both the zinc finger DNA binding domain and the proline-rich transactivating domain of EKLF, whereas its N- and/or C-terminal domains interact only with the transactivating domain of EKLF.
FIG. 5.
Interaction between FLI-1 and EKLF proteins. Interaction between 35S-labeled deletion mutants or full-length FLI-1 proteins and GST, GST-EKLF, GST-EKLF PRO, or GST-EKLF-Zn was investigated by GST-pull down assays. Equal amounts of 35S-labeled proteins were used in each reaction, and 5% of the input proteins are shown in the input lanes. The nature of the 35S-labeled FLI-1 protein and that of the GST or GST fusion protein used in the reactions are indicated to the right and on the top of each panel, respectively. Note that due to the presence of an alternative initiation codon at position 33 (56), all the in vitro-translated FLI-1 proteins including the N-terminal domain appear as doublets.
FLI-1 represses EKLF but stimulates GATA-1 activities.
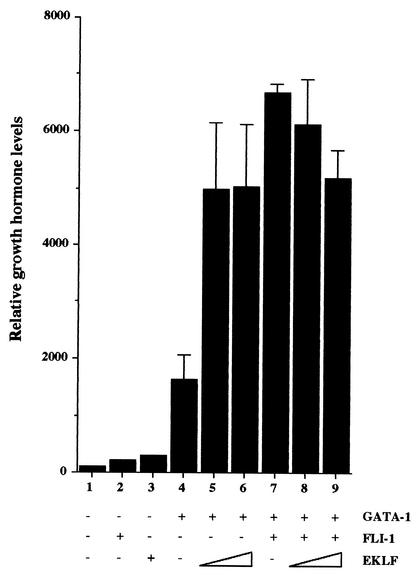
The effect of FLI-1 on GATA-1 activity was addressed by transient expression assay with 3T3 cells using the M1α-GH reporter (39) expressing human growth hormone under the control of an artificial GATA-1-responsive promoter (Fig. 6). As expected, the expression of this M1α-GH reporter is strongly stimulated by GATA-1 (Fig. 6, compare lanes 1 and 4) but is not affected by FLI-1 alone (Fig. 6, lane 2). Unexpectedly, although FLI-1 does not affect the level of GATA-1 protein produced (data not shown), FLI-1 increases by at least fourfold the stimulating effect of GATA-1 on the M1α-GH reporter (Fig. 6, lane 7). This stimulating effect of FLI-1 on GATA-1 activity is slightly higher than the stimulating effect of EKLF (24) (Fig. 6, compare lanes 5 and 6 to lane 7). Furthermore, the addition of increasing doses of EKLF in the presence of GATA-1 and FLI-1 progressively restores an expression level of the M1α-GH reporter equivalent to that observed in the presence of GATA-1 and EKLF alone (Fig. 6, compare lanes 5 and 6 and 7 to 9), suggesting a competition between FLI-1 and EKLF for cooperating with GATA-1. These results thus established that, like EKLF (24), FLI-1 is able to cooperate with GATA-1 to activate the transcription of the M1α-GH reporter in nonerythroid cells.
FIG. 6.
FLI-1 and EKLF stimulate the transcriptional activity of GATA-1 in nonerythroid cells. 3T3 cells were cotransfected with a fixed amount (0.25 μg) of the GATA-1-responsive pM1Gα-GH reporter plasmid and with expression vectors encoding GATA-1 (0.5 μg in lanes 4 to 9), FLI-1 (0.75 μg in lanes 2 and 7 to 9), or EKLF (0.25 or 0.5 μg in lanes 5 and 8 or 6 and 9, respectively) as indicated. Growth hormone levels accumulated in the supernatants of transfected cells 24 h posttransfection were determined by ELISA. Results are expressed as percentages of the level obtained in lane 1 (mean and standard deviation from three different experiments).
The effect of FLI-1 on EKLF activity was first addressed by transfection assays with MEL cells using the pEV3-GFP reporter. We found that the repressing effect of FLI-1 on the expression level of pEV3-GFP can be reversed by the cotransfection of increasing doses of EKLF (Fig. 7A, lanes 1 to 10). Reciprocally, the expression level of pEV3-GFP is stimulated by increasing doses of EKLF (Fig. 7A, lanes 13 to 16), and this stimulation by EKLF is repressed by increasing doses of FLI-1 (Fig. 7A, compare lanes 13 to 14, 11 to 12, and 7 to 8). Furthermore, we verified that the endogenous level of EKLF protein present in 745-A MEL cells is not affected by FLI-1 overexpression (63; also data not shown). These results indicated that the expression level of pEV3-GFP is strongly dependent on the relative levels between the FLI-1 and EKLF proteins. Interestingly enough, the stimulating effect of HMBA is also progressively decreased by increasing doses of transfected FLI-1 (Fig. 7A, compare black bars and hatched bars). Knowing that HMBA induces a reduction of the endogenous FLI-1 protein level (63), the latter observation further suggests that the increased expression level of pEV3-GFP in response to HMBA may be due to the increase in the EKLF/FLI-1 ratio.
FIG. 7.
FLI-1 represses the transcriptional activity of EKLF. (A) MEL cells were cotransfected with a fixed amount of the pEV3-GFP reporter plasmid and various amounts of expression plasmids encoding EKLF or FLI-1 as indicated. Immediately following transfection, each set of transfected cells was cultured for 72 h in the absence (black bars) or in the presence (hatched bars) of 5 mM HMBA. Quantification of GFP fluorescence was performed by FACS analysis as described in Fig. 2. (B) 3T3 cells were cotransfected with fixed amounts of the EKLF-responsive pC1G3-CAT and the CMV-β-Gal reporter plasmids and various amounts of expression plasmids encoding either FLI-1 or a HA-tagged version of EKLF as indicated. CAT and β-Gal activities were determined 24 h posttransfection. Results are expressed as relative values of CAT activities after standardization on β-Gal activities.
Since the pEV3-GFP reporter includes a large sequence derived from the β-locus control region which can bind several other transcription factors in addition to GATA-1 and EKLF, it could not be firmly established from the above-described results that EKLF is indeed a direct target of FLI-1. To ensure that FLI-1 acts directly on EKLF activity, we performed transient expression assays with 3T3 cells using a CAT reporter gene placed under the control of a minimal TK promoter and four EKLF DNA binding sites (pC1G3-CAT) (Fig. 7B). As previously reported (43), this construct is strongly stimulated by HA-EKLF, which corresponds to a HA-tagged version of the wild-type EKLF (50) (Fig. 7B, compare lanes 1 and 2). Whereas the expression level of this construct is unaffected by FLI-1 alone (Fig. 7B, lane 3), its stimulation by HA-EKLF is progressively repressed by increasing doses of FLI-1 (Fig. 7B, lanes 4 to 6). Western blot analyses were used to verify that the level of HA-EKLF protein is not affected by the cotransfection of FLI-1 in these experiments (data not shown). Taken together, these results established that FLI-1 behaves as a specific antagonist of EKLF activity.
Mapping FLI-1 domains required to repress EKLF activity.
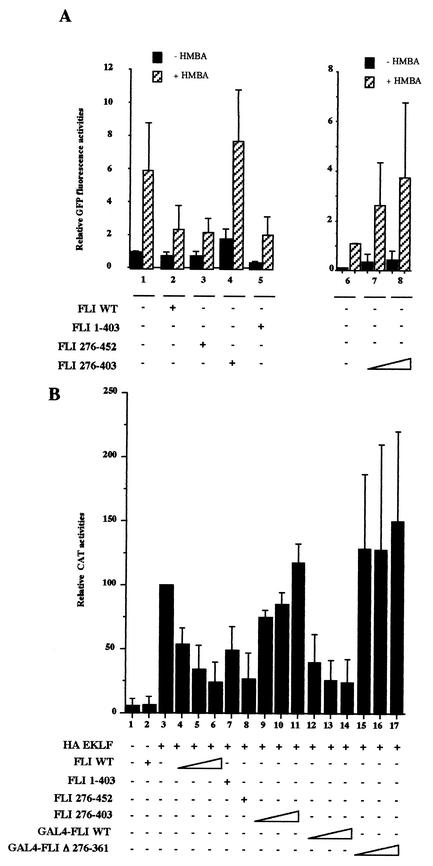
To understand the mechanism by which FLI-1 represses EKLF activity, we determined the minimal domains of FLI-1 required for repression. For that purpose, we compared the effect of various deletion mutants of FLI-1 on the expression of the pEV3-GFP reporter in MEL cells (Fig. 8A) and on the expression of the pC1G3-CAT reporter in 3T3 cells (Fig. 8B). FLI-1 mutants harboring deletion of either the C- or N-terminal domain (FLI 1-403 and FLI 276-452) display a repressing effect similar to that of wild-type FLI-1 on pEV3-GFP (Fig. 8A, compare lanes 2, 3, and 5). Intriguingly, the ETS domain of FLI-1 alone does not repress but instead stimulates the expression of pEV3-GFP in a dose-dependent manner (Fig. 8A, lanes 4 and 6 to 8). The FLI-1 ETS domain thus behaves like a dominant-negative protein against the repressing activity of endogenous FLI-1 in MEL cells. Likewise, the same FLI-1 mutants harboring deletion of either the N- or the C-terminal domain display a repressing effect similar to that of wild-type FLI-1 on the EKLF-dependent activity of the pC1G3-CAT reporter (Fig. 8B, compare lanes 3 and 6 to 8), whereas the ETS domain alone displays a very limited dose-dependent stimulating effect (Fig. 8B, compare lanes 3 and 9 to 11). Since all of the ETS family proteins analyzed to date have been shown to contain a nuclear localization signal in the conserved ETS domain, the possibility that the lack of repressing activity of the FLI-1 ETS domain might be due to its failure to enter the nucleus appeared very unlikely. Nevertheless, we excluded this possibility by verifying that the GAL4 FLI 276-370 protein (Fig. 1), in which the FLI-1 ETS domain is fused to GAL4 DNA binding domain containing its own nuclear localization signal, is still unable to repress the EKLF-dependent activity of the pC1G3-CAT reporter (data not shown). Taken together, these results tend to suggest that the repressing effect of FLI-1 on EKLF activity requires the ETS domain and either the N- or C-terminal domain. In the next experiment, we fused the GAL4 DNA binding domain in the N-terminal position to the wild-type full-length FLI-1 (Fig. 1, GAL4-FLI WT) and then introduced internal deletion of the ETS domain (Fig. 1, GAL4-FLI Δ276-361). The GAL4 FLI WT protein is still able to repress the EKLF-dependent expression of pC1G3-CAT in a dose-dependent manner (Fig. 8B, compare lanes 4 to 6 to lanes 12 to 14). In contrast, GAL4-FLI Δ276-361 carrying a deletion of the ETS domain is unable to repress the expression of the pC1G3-CAT reporter even at the highest dose tested (Fig. 8B, compare lanes 4 to 6 to 15 to 17). The latter result thus established that the repressing effect of FLI-1 on EKLF activity is critically dependent on the ETS domain.
FIG. 8.
Mapping of FLI-1 domains involved in repression of EKLF activity. (A) MEL cells were cotransfected in the same conditions as for Fig. 7A with a fixed amount of the pEV3-GFP reporter plasmid and expression vectors encoding the indicated deletion mutants of FLI-1. Results are expressed as relative GFP fluorescence activities (mean and standard deviation from three different experiments). (B) 3T3 cells were cotransfected in the same conditions as for Fig. 7B with a fixed amount of pC1G3-CAT and the CMV-β-Gal reporters and HA-EKLF expression plasmid with or without expression plasmids encoding the indicated FLI-1 proteins. CAT activities were standardized on β-Gal activities, and results are expressed as percentages of the standardized CAT activity obtained for the pC1G3-CAT reporter transfected with HA-EKLF alone (lane 3) (mean and standard deviation from three different experiments).
The ETS domain of FLI-1 behaves as an autonomous repression domain when recruited indirectly to promoters.
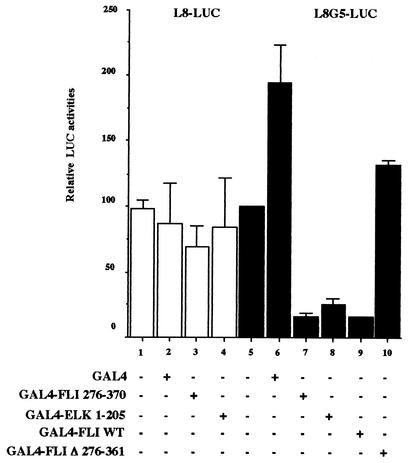
It has been shown recently that the ETS domain of Elk-1 behaves as an autonomous repression domain when recruited indirectly to DNA (73). In the light of our finding that the ETS domain is involved in the repressing activity of FLI-1, this prompted us to investigate if the FLI-1 ETS domain could also behaves as a repressor when recruited indirectly to promoters. To address this question, we fused the ETS domain of FLI-1 (aa 276 to 370) to the DNA binding domain of Gal4 and tested its effect on the expression of a composite Lex-Gal4-driven promoter reporter (L8G5-LUC) in the presence of the strong LexA-VP16 activator. As expected, the GAL4-FLI 276-370 protein displays the same strong repressing effect as the Gal4-ELK 1-205 protein on L8G5-LUC activity (Fig. 9, compare lanes 5, 7, and 8). In the same conditions, even the GAL4 FLI WT protein displays a similar strong repressing effect (Fig. 9, lane 9), and the repressing effect is completely abolished by the deletion of the ETS domain (Fig. 9, lane 10). Importantly, all these GAL4 fusion proteins display very limited effect on the L8-LUC control reporter devoid of GAL4 binding sites (Fig. 9, lanes 3 and 4), thus ensuring that the repressing activity of the FLI-1 ETS domain is truly dependent on its indirect recruitment to the promoter.
FIG. 9.
Autonomous repression activity of the FLI-1 ETS domain. COS cells were cotransfected with a fixed amount of the CMV-β-Gal and either L8-LUC or L8G5-LUC reporter plasmids as well as a fixed amount of expression plasmid encoding the LexA-VP16 fusion protein and a fixed amount of expression plasmids encoding the indicated Gal4 fusion proteins. Luciferase and β-Gal activities in transfected cells were determined 24 h posttransfection. Luciferase activities were standardized on β-Gal activities, and final results are expressed as percentages of the standardized luciferase activity obtained for L8G5-LUC reporter transfected in the presence of LexA-VP16 alone (lane 5) (mean and standard deviation from three different experiments).
EKLF represses FLI-1 target genes.
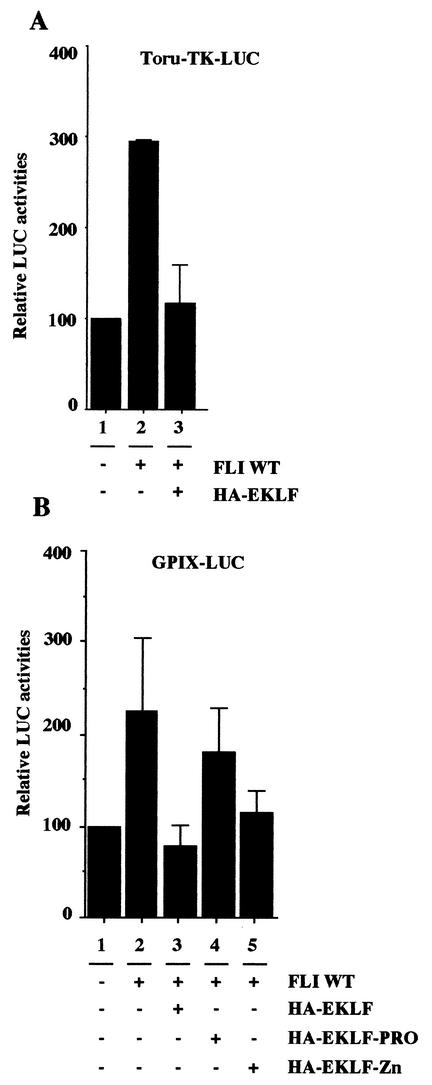
The above results thus established the unanticipated duality of FLI-1 behaving as either an activator or a repressor depending on its mode of recruitment to promoters. It has been shown recently that EKLF also activates or represses transcription depending on a very similar mechanism due to the autonomous repressing activity of its DNA binding domain (13). This prompted us to investigate if, reciprocally, EKLF itself could be involved in the repression of FLI-1 target genes. For that purpose, we used a luciferase reporter gene driven by the GPIX gene promoter which has been shown recently to be a natural target of FLI-1 in vivo (6, 7, 23, 28). As expected, we found that this GPIX-LUC reporter is stimulated about twofold by FLI-1 in cotransfection experiments performed with COS cells (Fig. 10B, lanes 1 and 2). Interestingly enough, this twofold stimulation of the GPIX-LUC reporter by FLI-1 is completely inhibited in the presence of HA-EKLF (Fig. 10B, compare lanes 2 and 3). Similar results were obtained using the FLI-1-responsive TORU-TK-LUC reporter, the stimulation of which is also repressed by HA-EKLF (Fig. 10A, lanes 1 to 3). Furthermore, we found that the Zn finger DNA binding domain of EKLF, but not its proline-rich domain, is sufficient to reproduce most of the repressing activity observed on both of these FLI-1-responsive reporters (Fig. 10B, lanes 4 and 5, and data not shown). These results thus indicate that the reciprocal inhibition of FLI-1 activity by EKLF is also dependent on its DNA binding domain.
FIG. 10.
EKLF represses the transcriptional activity of FLI-1. COS cells were cotransfected with a fixed amount of the TORU-TK-LUC (A) or GPIX-LUC (B) and CMV-β-Gal reporter plasmids with or without expression vectors encoding wild-type FLI-1, wild-type HA-EKLF, or deletion mutants of HA-EKLF as indicated. Luciferase activities were standardized on β-Gal activities, and final results are expressed as percentages of the standardized luciferase activity obtained for the luciferase reporter alone (lane 1) (mean and standard deviation from two different experiments).
GST-EKLF and GST-FLI-1 do not cross-inhibit their DNA binding activities in vitro.
One eventual mechanism responsible for the functional cross-antagonism evidenced between FLI-1 and EKLF could be the reciprocal inhibition of their specific DNA binding activities. We addressed this possibility by gel shift assays using purified GST-EKLF and GST-FLI-1 proteins and two labeled oligonucleotides carrying a specific binding motif for either FLI-1 (GGAA motif) or EKLF (CACCC motif) (Fig. 11). As expected, GST-FLI-1 forms a stable complex with the GGAA probe (Fig. 11A, lane 3), whereas GST alone does not (Fig. 11A, lane 2). However, the formation of this complex by GST-FLI-1 remains unaffected even in the presence of a 10-fold excess of GST-EKLF (Fig. 11A, compare lanes 3 and 6). Reciprocally, the specific complex formed by GST-EKLF with the CAC probe remains unaffected even in the presence of a 10-fold excess of GST-FLI-1 (Fig. 11B, compare lanes 3 to 6). Taken together, these data demonstrate that FLI-1 and EKLF do not cross-inhibit their DNA binding activities, at least in vitro.
FIG. 11.
GST-EKLF and GST-FLI-1 do not cross-inhibit their DNA binding activities. (A) GST-FLI-1 DNA binding activity was analyzed by gel shift assay using the GGAA probe either alone (lane 3) or in the presence of increasing excess of GST-EKLF (1-, 5-, or 10-fold excess, respectively, in lanes 4, 5, and 6). Control lanes correspond, respectively, to the probe incubated without protein (lane 1), with GST alone (lane 2), or with GST-FLI-1 and a 10-fold excess of GST (lane 7). The free probe was allowed to run out of the gel in order to facilitate the detection of putative tripartite complexes between GST-EKLF, GST-FLI-1, and DNA and for that reason is not visible on the autoradiograph. (B) Reciprocal experiment performed as for panel A but using the CAC probe.
DISCUSSION
FLI-1 is a functional antagonist of EKLF.
The present study presents several evidences that FLI-1 is a functional antagonist of the erythroid-specific transcription factor EKLF. The first evidence is given by the results of our transient expression assays performed with MEL cells showing that β-globin gene promoter activity is strikingly dependent on the relative levels of FLI-1 and EKLF proteins. Indeed, the repressing effect of FLI-1 on β-globin gene promoter activity is clearly dose dependent and is reversed by cotransfecting increasing amounts of EKLF. Reciprocally, β-globin gene promoter activity is stimulated by the transfection of EKLF, and this stimulation by EKLF is repressed by cotransfecting increasing amounts of FLI-1. In addition, FLI-1 not only represses β-globin gene activity but also abolishes its stimulation in response to HMBA. Taken together, these data strongly suggest that β-globin gene transcription in MEL cells is limited by the quenching of EKLF activity by transfected as well as endogenous FLI-1. An interesting prediction of this finding is that combining HMBA treatment and experimental reduction of the endogenous FLI-1 level should improve terminal erythroid differentiation of MEL cells compared to that obtained by HMBA alone. Further evidence that FLI-1 is indeed a specific antagonist of EKLF is shown by the dose-dependent inhibition of the EKLF-dependent activity of artificial promoter placed under the control of EKLF DNA binding sites. Finally, this functional antagonism is associated with physical interaction between the two proteins. One important question raised by this finding regards the mechanism by which FLI-1 inhibits EKLF activity.
Our data indicate that the ETS domain of FLI-1 is absolutely required to inhibit EKLF activity. Since the FLI-1 ETS domain interacts with the DNA binding domain of EKLF, one possibility could be that FLI-1 inhibits the binding of EKLF to its DNA targets. However, this possibility is inconsistent with our finding that the FLI-1 ETS domain alone not only is unable to repress EKLF activity but in contrast stimulates β-globin gene promoter transcription in MEL cells. Furthermore, we found by gel shift assays that the binding of a GST-EKLF fusion protein to EKLF-specific DNA probe is unaffected by a 10-fold excess of GST-FLI-1 (Fig. 11). We thus consider inhibition of EKLF DNA binding to be a highly improbable possibility. In contrast, our data show that the FLI-1 ETS domain behaves as an autonomous repression domain when it is recruited to the promoter through its fusion to the GAL4 DNA binding domain. Our data also show that repression of EKLF activity requires the association of the ETS domain with, indifferently, the N- or C-terminal domain of FLI-1, which themselves also interact with EKLF. That the N- and the C-terminal domains of FLI-1 are interchangeable suggests that their main contribution to repression is through their own interaction with EKLF. Based on all these data, we strongly favor the possibility that repression is due to the autonomous repression activity of the FLI-1 ETS domain and its indirect tethering to EKLF target promoters by protein-protein interaction involving the terminal domains of FLI-1. This model is further supported by the recent demonstration that the repression of the COL1A2 promoter by FLI-1 also appears to be mediated through the indirect recruitment of FLI-1 by protein-protein interaction with the other Kruppel family transcription factor Sp1 (14).
On the other hand, our data demonstrate that FLI-1 also interacts with GATA-1. However, FLI-1 does not repress but enhances GATA-1 activity. Although this synergism is similarly best explained by the indirect recruitment of FLI-1 by protein-protein interaction with GATA-1, another question is what the difference is in the interaction between FLI-1 and its two different recruitment partners which determines its stimulating or repressing effect on transcription. For example, while FLI-1 interacts with EKLF through both its ETS and terminal domains, the only domain of FLI-1 interacting with GATA-1 is the ETS domain. In other words, the recruitment of FLI-1 by EKLF does not necessarily involve direct interaction with the ETS domain, but this cannot be the case when it is recruited by GATA-1. Since the ETS domain appears to be the active domain responsible for repression, it is therefore very tempting to speculate that repression requires its interaction with other cofactor(s), which cannot occur when the ETS domain directly interacts either with GATA-1 or with DNA.
One additional question raised by our data is the mechanism by which the FLI-1 ETS domain can repress transcription following its indirect recruitment to promoters. One current mechanism used by many repressor domains is to recruit corepressor complexes displaying HDAC activity. For example, the repression activity of the ELK-1 ETS domain is associated with the recruitment of the HDAC-1 and mSin3A proteins and is sensitive to the HDAC inhibitor trichostatin (73). Interestingly, the SAP18 protein, which also participates in the composition of the Sin3/HDAC family repressor complexes, has been identified as a binding partner of the Xenopus FLI-1 protein by yeast two-hybrid assay (18). However, all of our attempts to inhibit FLI-1 repression activity by trichostatin have failed (data not shown), and we have therefore presently no evidence that HDACs are involved in the repression. On the other hand, it has been shown very recently that the repression activity of the other ETS family protein, SPI-1/PU.1, involves protein-protein interaction with the acetyltransferase domain of CBP and inhibition of the CBP-mediated acetylation of many different nuclear factors, including GATA-1 and EKLF (29). According to this last study (29), the SPI-1/PU.1 ETS domain contributes to the inhibition of CBP acetylase activity, thus suggesting that this mechanism might be shared by other ETS family proteins. In agreement with our data indicating that FLI-1 does not inhibit GATA-1 activity, FLI-1 does not inhibit the CBP-mediated acetylation of GATA-1 (29). However, the intriguing possibility remains that FLI-1 might be involved in the specific inhibition of CBP-mediated acetylation of EKLF (79). Whatever the underlying mechanism is, the present study identifies the EKLF protein as a critical target of FLI-1 involved in the inhibition of erythroleukemic cell differentiation.
Implication of the functional cross-antagonism between FLI-1 and EKLF in the control of the erythrocytic versus megakaryocytic differentiation.
The present study also shows that EKLF itself inhibits FLI-1 activity. This inhibition of FLI-1 activity is also dependent on the DNA binding domain of EKLF, which has been shown recently to behave as an autonomous repression domain when it is recruited indirectly to promoters (13). As suggested above for the inhibition of EKLF activity, the inhibition of FLI-1 activity most probably involves the indirect recruitment of EKLF to FLI-1 target promoters by protein-protein interaction. Whatever is the exact underlying mechanism, our study undoubtedly establishes functional cross-antagonism between FLI-1 and EKLF. This functional cross-antagonism between FLI-1 and EKLF has important implications for the understanding of the transcriptional control of erythrocytic versus megakaryocytic differentiation.
Indeed, erythrocytic and megakaryocytic lineages are known to derive from a common bipotential progenitor (16, 58, 64). At least four DNA binding transcription factors have been unambiguously identified to contribute to the positive control of gene-specific transcription between these two lineages. Among them, NF-E2 and GATA-1 are shared by both lineages and activate both erythrocytic and megakaryocytic gene promoters (58, 39). In contrast, EKLF is expressed only in erythroid cells and cooperates with GATA-1 in the activation of erythroid promoters. Symmetrically, FLI-1 is highly expressed in megakaryocytic cells and contributes with GATA-1 to the activation of megakaryocytic promoters (6, 7, 19, 23, 28, 58, 62). Differential expression of FLI-1 and EKLF thus appears as a critical determinant involved in the positive control of erythrocytic versus megakaryocytic transcription. Our finding of functional cross-antagonism between FLI-1 and EKLF strongly suggests that this positive control is doubled by cross-negative control. While the inhibition of erythrocytic differentiation by FLI-1 is well established (2, 3, 36, 51, 63, 66), two concordant results strongly suggest that EKLF itself may actually inhibit megakaryocytic differentiation. First, we show here that EKLF represses the FLI-1-dependent activity of the GPIX gene promoter, which is one of the recently identified megakaryocyte-specific FLI-1 target genes in vivo (28). Furthermore, the decrease in platelet numbers observed in transgenic mice overexpressing EKLF also indicates that EKLF actually inhibits megakaryocytic differentiation in vivo (67). Altogether, these data lead us to propose a working model which summarizes the dual function of FLI-1 and EKLF in the control of the erythroid versus megakaryocytic differentiation of bipotential hematopoietic progenitors (Fig. 12). According to this model, the initial engagement of bipotential progenitors towards either erythroid or megakaryocytic differentiation would be critically determined by subtle (probably stochastic) variation in the ratio between EKLF and FLI-1 protein levels. Once initiated, irreversible engagement towards either lineage would be determined simultaneously by the positive contribution of each factor in the activation of lineage-specific promoters as well as by the cross-negative inhibition of their positive activities. This model is very similar to the model already proposed by others for the transcriptional control of erythroid versus myeloid differentiation based on the functional cross-antagonism between SPI-1/PU.1 and GATA-1 (10, 11, 55). In that respect, the present study strongly supports the emerging view that functional cross-antagonisms between transcription factors play a very important role in hematopoietic cell lineage differentiation and simultaneously provide new insight into mechanisms of maturation arrest seen in leukemic cells overexpressing lineage-specific transcription factors (11, 59, 72).
FIG. 12.
Cross-antagonistic model of EKLF and FLI-1 activities in erythroid versus megakaryocytic differentiation. Schematic representation of the cell fate determination of erythrocytic and megakaryocytic cell types from the common bipotential progenitor. EKLF and FLI-1 cooperate with GATA-1 and contribute in a positive manner to produce erythroid or megakaryocytic cells, respectively. Simultaneously, EKLF and FLI-1 contribute in a negative manner through the cross-inhibition of their positive activities.
Repression activity of the DNA binding domain of dual transcription factors.
This study thus establishes that FLI-1 is a new example of an increasing number of dual DNA binding transcription factors (34) which, like Pit-1 (57), Skin-1a (65), ELK-1 (73), SPI-1/PU.1 (29), EKLF (13), Sp1 (20), YY1 (74), or GATA-2 (49), can alternatively activate or repress transcription depending on their DNA-protein and/or protein-protein interactions. Intriguingly, the DNA binding domain of all these dual transcription factors overlaps with the domain responsible for repression. This observation is particularly striking since their DNA binding domains correspond to very different structures, including zinc fingers (Sp1, EKLF, and YY1), ETS domain (ELK-1, SPI-1/PU.1, and FLI-1), or POU domain (Pit1). Many other transcription factors harboring these three different structures of the DNA binding domain are only known to activate transcription. However, very few studies have addressed directly whether their DNA binding domains can behave as autonomous repressor domains when they are recruited indirectly to promoters. This raises the intriguing possibility that repression activity of unbound DNA binding domains might be an unanticipated common property of many different DNA binding transcription factors, thus extending considerably their regulating potential. If so, an interesting challenge for future studies will be to understand the selection pressure responsible for regrouping DNA binding and transcriptional repression activities in a single domain of transcription factors.
Acknowledgments
We are very grateful to J. Bieker, J. Crispino, M. Crossley, O. Delattre, F. Grosveld, S. Khochbin, T. D. Littlewood, G. J. Roth, and A. D. Sharrocks for reagents. We also thank S. Arnaud and C. Bourgin for their technical advice for GST pull-down and coimmunoprecipitation experiments and B. Guyot for helpful discussion on the manuscript.
This work was supported by grants from ARC (no. 5737 and no. 4484), La Ligue (comité national and comité du Rhône), and Fondation de France.
REFERENCES
- 1.Afrikanova, I., E. Yeh, D. Bartos, S. S. Watowich, and G. D. Longmore. 2002. Oncogene cooperativity in Friend erythroleukemia: erythropoietin receptor activation by the env gene of SFFV leads to transcriptional upregulation of PU.1, independent of SFFV proviral insertion. Oncogene 21:1272-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Athanasiou, M., P. A. Clausen, G. J. Mavrothalassitis, X. K. Zhang, D. K. Watson, and D. G. Blair. 1996. Increased expression of the ETS-related transcription factor FLI-1/ERGB correlates with and can induce the megakaryocytic phenotype. Cell Growth Differ. 7:1525-1534. [PubMed] [Google Scholar]
- 3.Athanasiou, M., G. Mavrothalassitis, L. Sun-Hoffman, and D. G. Blair. 2000. FLI-1 is a suppressor of erythroid differentiation in human hematopoietic cells. Leukemia 14:439-445. [DOI] [PubMed] [Google Scholar]
- 4.Barnache, S., F. Wendling, C. Lacombe, N. Denis, M. Titeux, W. Vainchenker, and F. Moreau-Gachelin. 1998. Spi-1 transgenic mice develop a clonal erythroleukemia which does not depend on p53 mutation. Oncogene 16:2989-2995. [DOI] [PubMed] [Google Scholar]
- 5.Barnache, S., P. Mayeux, B. Payrastre, and F. Moreau-Gachelin. 2001. Alterations of the phosphoinositide 3-kinase and mitogen-activated protein kinase signaling pathways in the erythropoietin-independent Spi-1/PU.1 transgenic proerythroblasts. Blood 98:2372-2381. [DOI] [PubMed] [Google Scholar]
- 6.Bastian, L. S., M. Yagi, C. Chan, and G. J. Roth. 1996. Analysis of the megakaryocyte glycoprotein IX promoter identifies positive and negative regulatory domains and functional GATA and Ets sites. J. Biol. Chem. 271:18554-18560. [DOI] [PubMed] [Google Scholar]
- 7.Bastian, L. S., B. A. Kwiatkowski, J. Breininger, S. Danner, and G. Roth. 1999. Regulation of the megakaryocytic glycoprotein IX promoter by the oncogenic Ets transcription factor Fli-1. Blood 93:2637-2644. [PubMed] [Google Scholar]
- 8.Ben-David, Y., and A. Bernstein. 1991. Friend virus-induced erythroleukemia and the multistage nature of cancer. Cell 66:831-834. [DOI] [PubMed] [Google Scholar]
- 9.Bieker, J. J., and C. M. Southwood. 1995. The erythroid Kruppel-like factor transactivation domain is a critical component for cell-specific inducibility of a beta-globin promoter. Mol. Cell. Biol. 15:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantor, A. B., and S. H. Orkin. 2001. Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 11:513-519. [DOI] [PubMed] [Google Scholar]
- 11.Cantor, A. B., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. [DOI] [PubMed] [Google Scholar]
- 12.Carrere, S., A. Verger, A. Flourens, D. Stehelin, and M. Duterque-Coquillaud. 1998. Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains. Oncogene 16:3261-3268. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X., and J. J. Bieker. 2001. Unanticipated repression function linked to erythroid Kruppel-like factor. Mol. Cell. Biol. 21:3118-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czuwara-Ladykowska, J., F. Shirasaki, P. Jackers, D. K. Watson, and M. Trojanowska. 2001. Fli-1 inhibits collagen type I production in dermal fibroblasts via an Sp1-dependent pathway. J. Biol. Chem. 276:20839-20848. [DOI] [PubMed] [Google Scholar]
- 15.Darby, T. G., J. D. Meissner, A. Ruhlmann, W. H. Mueller, and R. J. Scheibe. 1997. Functional interference between retinoic acid or steroid hormone receptors and the oncoprotein Fli-1. Oncogene 15:3067-3082. [DOI] [PubMed] [Google Scholar]
- 16.Debili, N., L. Coulombel, L. Croisille, A. Katz, J. Guichard, J. Breton-Gorius, and W. Vainchenker. 1996. Characterization of a bipotent erythro-megakaryocytic progenitor in human bone marrow. Blood 88:1284-1296. [PubMed] [Google Scholar]
- 17.Delgado, M. D., P. Gutierrez, C. Richard, M. A. Cuadrado, F. Moreau-Gachelin, and J. Leon. 1998. Spi-1/PU.1 proto-oncogene induces opposite effects on monocytic and erythroid differentiation of K562 cells. Biochem. Biophys. Res. Commun. 252:383-391. [DOI] [PubMed] [Google Scholar]
- 18.Deramaudt, T. B., P. Remy, and P. Stiegler. 2001. Identification of interaction partners for two closely-related members of the ETS protein family, FLI and ERG. Gene 274:169-177. [DOI] [PubMed] [Google Scholar]
- 19.Deveaux, S., A. Filipe, V. Lemarchandel, J. Ghysdael, P. H. Romeo, and V. Mignotte. 1996. Analysis of the thrombopoietin receptor (MPL) promoter implicates GATA and Ets proteins in the coregulation of megakaryocyte-specific genes. Blood 87:4678-4785. [PubMed] [Google Scholar]
- 20.Doetzlhofer, A., H. Rotheneder, G. Lagger, M. Koranda, V. Kurtev, G. Brosch, E. Wintersberger, and C. Seiser. 1999. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19:5504-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donze, D., T. M. Townes, and J. J. Bieker. 1995. Role of erythroid Kruppel-like factor in human gamma- to beta-globin gene switching. J. Biol. Chem. 270:1955-1959. [DOI] [PubMed] [Google Scholar]
- 22.Doubeikovskaia, Z., A. Aries, P. Jeannesson, F. Morle, and A. Doubeikovski. 2001. Purification of human recombinant GATA-1 from bacteria: implication for protein-protein interaction studies. Protein Expr. Purif. 23:426-431. [DOI] [PubMed] [Google Scholar]
- 23.Eisbacher, M., L. M. Khachigian, T. H. Khin, M. L. Holmes, and B. H. Chong. 2001. Inducible expression of the megakaryocyte-specific gene glycoprotein IX is mediated through an Ets binding site and involves upstream activation of extracellular signal-regulated kinase. Cell Growth Differ. 12:435-445. [PubMed] [Google Scholar]
- 24.Gregory, R. C., D. J. Taxman, D. Seshasayee, M. H. Kensinger, J. J. Bieker, and D. M. Wojchowski. 1996. Functional interaction of GATA1 with erythroid Kruppel-like factor and Sp1 at defined erythroid promoters. Blood 87:1793-1801. [PubMed] [Google Scholar]
- 25.Gutman, A., and B. Wasylyk. 1990. The collagenase gene promoter contains a TPA and oncogene-responsive unit encompassing the PEA3 and AP-1 binding sites. EMBO J. 9:2241-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallier, M., A. Tavitian, and F. Moreau-Gachelin. 1996. The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb. J. Biol. Chem. 271:11177-11181. [DOI] [PubMed] [Google Scholar]
- 27.Hallier, M., A. Lerga, S. Barnache, A. Tavitian, and F. Moreau-Gachelin. 1998. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J. Biol. Chem. 273:4838-4842. [DOI] [PubMed] [Google Scholar]
- 28.Hart, A., F. Melet, P. Grossfeld, K. Chien, C. Jones, A. Tunnacliffe, R. Favier, and A. Bernstein. 2000. Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity 13:167-177. [DOI] [PubMed] [Google Scholar]
- 29.Hong, W., A. Y. Kim, S. Ky, C. Rakowski, S. B. Deo, D. Chakravarti, M. Atchison, and G. A. Blobel. 2002. Inhibition of CBP-mediated protein acetylation by the Ets family oncoprotein PU.1. Mol. Cell. Biol. 22:3729-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaishankar, S., J. Zhang, M. F. Roussel, and S. J. Baker. 1999. Transforming activity of EWS/FLI is not strictly dependent upon DNA-binding activity. Oncogene 18:5592-5597. [DOI] [PubMed] [Google Scholar]
- 31.Kawada, H., T. Ito, P. N. Pharr, D. D. Spyropoulos, D. K. Watson, and M. Ogawa. 2001. Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. Int. J. Hematol. 73:463-468. [DOI] [PubMed] [Google Scholar]
- 32.Kihara-Negishi, F., H. Yamamoto, M. Suzuki, T. Yamada, T. Sakurai, T. Tamura, and T. Oikawa. 2001. In vivo complex formation of PU.1 with HDAC1 associated with PU.1-mediated transcriptional repression. Oncogene 20:6039-6047. [DOI] [PubMed] [Google Scholar]
- 33.Knoop, L. L., and S. J. Baker. 2001. EWS/FLI alters 5′-splice site selection. J. Biol. Chem. 276:22317-22322. [DOI] [PubMed] [Google Scholar]
- 34.Latchman, D. S. 2001. Transcription factors: bound to activate or repress. Trends Biochem. Sci. 26:211-213. [DOI] [PubMed] [Google Scholar]
- 35.Lemercier, C., A. Verdel, B. Galloo, S. Curtet, M. P. Brocard, and S. Khochbin. 2000. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275:15594-15599. [DOI] [PubMed] [Google Scholar]
- 36.Lesault, I., C. T. Quang, J. Frampton, and J. Ghysdael. 2002. Direct regulation of BCL-2 by FLI-1 is involved in the survival of FLI-1-transformed erythroblasts. EMBO J. 21:694-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim, S. K., J. J. Bieker, C. S. Lin, and F. Costantini. 1997. A shortened life span of EKLF-/- adult erythrocytes, due to a deficiency of beta-globin chains, is ameliorated by human gamma-globin chains. Blood 90:1291-1299. [PubMed] [Google Scholar]
- 38.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin, D. I., and S. H. Orkin. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 4:1886-1898. [DOI] [PubMed] [Google Scholar]
- 40.Matsumura, I., A. Kawasaki, H. Tanaka, J. Sonoyama, S. Ezoe, N. Minegishi, K. Nakajima, M. Yamamoto, and Y. Kanakura. 2000. Biologic significance of GATA-1 activities in Ras-mediated megakaryocytic differentiation of hematopoietic cell lines. Blood 96:2440-2450. [PubMed] [Google Scholar]
- 41.Melet, F., B. Motro, D. J. Rossi, L. Zhang, and A. Bernstein. 1996. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol. Cell. Biol. 16:2708-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melot, T., N. Gruel, A. Doubeikovski, N. Sevenet, T. L. Teillaud, and O. Delattre. 1997. Production and characterization of mouse monoclonal antibodies to wild-type and oncogenic FLI-1 proteins. Hybridoma 16:457-464. [DOI] [PubMed] [Google Scholar]
- 43.Miller, I. J., and J. J. Bieker. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell. Biol. 13:2776-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreau-Gachelin, F., F. Wendling, T. Molina, N. Denis, M. Titeux, G. Grimber, P. Briand, W. Vainchenker, and A. Tavitian. 1996. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol. Cell. Biol. 16:2453-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreau-Gachelin, F., A. Tavitian, and P. Tambourin. 1988. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature 331:277-280. [DOI] [PubMed] [Google Scholar]
- 46.Nerlov, C., E. Querfurth, H. Kulessa, and T. Graf. 2000. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood 95:2543-2551. [PubMed] [Google Scholar]
- 47.Oikawa, T., T. Yamada, F. Kihara-Negishi, H. Yamamoto, N. Kondoh, Y. Hitomi, and Y. Hashimoto. 1999. The role of Ets family transcription factor PU.1 in hematopoietic cell differentiation, proliferation and apoptosis. Cell Death Differ. 6:599-608. [DOI] [PubMed] [Google Scholar]
- 48.Orkin, S. H. 2000. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1:57-64. [DOI] [PubMed] [Google Scholar]
- 49.Ozawa, Y., M. Towatari, S. Tsuzuki, F. Hayakawa, T. Maeda, Y. Miyata, M. Tanimoto, and H. Saito. 2001. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood 98:2116-2123. [DOI] [PubMed] [Google Scholar]
- 50.Pandya, K., D. Donze, and T. M. Townes. 2001. Novel transactivation domain in erythroid Kruppel-like factor (EKLF). J. Biol. Chem. 276:8239-8243. [DOI] [PubMed] [Google Scholar]
- 51.Pereira, R., C. T. Quang, I. Lesault, H. Dolznig, H. Beug, and J. Ghysdael. 1999. FLI-1 inhibits differentiation and induces proliferation of primary erythroblasts. Oncogene 18:1597-1608. [DOI] [PubMed] [Google Scholar]
- 52.Pereira, R., J. Raingeaud, M. Pironin, J. Ghysdael, and C. T. Quang. 2000. SPI-1 transforming properties depend upon specifically activated forms of the EPOR. Oncogene 19:5106-5110. [DOI] [PubMed] [Google Scholar]
- 53.Quang, C. T., O. Wessely, M. Pironin, H. Beug, and J. Ghysdael. 1997. Cooperation of Spi-1/PU.1 with an activated erythropoietin receptor inhibits apoptosis and Epo-dependent differentiation in primary erythroblasts and induces their Kit ligand-dependent proliferation. EMBO J. 16:5639-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao, G., N. Rekhtman, G. Cheng, T. Krasikov, and A. I. Skoultchi. 1997. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene 14:123-131. [DOI] [PubMed] [Google Scholar]
- 55.Rekhtman, N., F. Radparvar, T. Evans, and A. I. Skoultchi. 1999. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13:1398-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarrazin, S., J. Starck, C. Gonnet, A. Doubeikovski, F. Melet, and F. Morle. 2000. Negative and translation termination-dependent positive control of FLI-1 protein synthesis by conserved overlapping 5′ upstream open reading frames in Fli-1 mRNA. Mol. Cell. Biol. 20:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scully, K. M., E. M. Jacobson, K. Jepsen, V. Lunyak, H. Viadiu, C. Carriere, D. W. Rose, F. Hooshmand, A. K. Aggarwal, and M. G. Rosenfeld. 2000. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science 290:1127-1131. [DOI] [PubMed] [Google Scholar]
- 58.Shivdasani, R. A. 2001. Molecular and transcriptional regulation of megakaryocyte differentiation. Stem Cells 19:397-407. [DOI] [PubMed] [Google Scholar]
- 59.Sieweke, M. H., and T. Graf. 1998. A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev. 8:545-551. [DOI] [PubMed] [Google Scholar]
- 60.Simon, M. C. 1998. PU.1 and hematopoiesis: lessons learned from gene targeting experiments. Semin. Immunol. 10:111-118. [DOI] [PubMed] [Google Scholar]
- 61.Singh, H., R. P. DeKoter, and J. C. Walsh. 1999. PU.1, a shared transcriptional regulator of lymphoid and myeloid cell fates. Cold Spring Harb. Symp. Quant. Biol. 64:13-20. [DOI] [PubMed] [Google Scholar]
- 62.Spyropoulos, D. D., P. N. Pharr, K. R. Lavenburg, P. Jackers, T. S. Papas, M. Ogawa, and D. K. Watson. 2000. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 20:5643-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Starck, J., A. Doubeikovski, S. Sarrazin, C. Gonnet, G. Rao, A. Skoultchi, J. Godet, I. Dusanter-Fourt, and F. Morle. 1999. Spi-1/PU.1 is a positive regulator of the Fli-1 gene involved in inhibition of erythroid differentiation in friend erythroleukemic cell lines. Mol. Cell. Biol. 19:121-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Starck, J., G. Mouchiroud, C. Gonnet, A. Mehlen, D. Aubert, A. Dorier, J. Godet, and F. Morle. 1999. Unexpected and coordinated expression of Spi-1, Fli-1, and megakaryocytic genes in four Epo-dependent cell lines established from transgenic mice displaying erythroid-specific expression of a thermosensitive SV40 T antigen. Exp. Hematol. 27:630-641. [DOI] [PubMed] [Google Scholar]
- 65.Sugihara, T. M., E. I. Kudryavtseva, V. Kumar, J. J. Horridge, and B. Andersen. 2001. The POU domain factor Skin-1a represses the keratin 14 promoter independent of DNA binding. A possible role for interactions between Skn-1a and CREB-binding protein/p300. J. Biol. Chem. 276:33036-33044. [DOI] [PubMed] [Google Scholar]
- 66.Tamir, A., J. Howard, R. R. Higgins, Y. J. Li, L. Berger, E. Zacksenhaus, M. Reis, and Y. Ben-David. 1999. Fli-1, an Ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: evidence for direct transcriptional repression of the Rb gene during differentiation. Mol. Cell. Biol. 19:4452-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tewari, R., N. Gillemans, M. Wijgerde, B. Nuez, M. von Lindern, F. Grosveld, and S. Philipsen. 1998. Erythroid Kruppel-like factor (EKLF) is active in primitive and definitive erythroid cells and is required for the function of 5′HS3 of the beta-globin locus control region. EMBO J. 17:2334-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Truong, A. H., and Y. Ben-David. 2000. The role of Fli-1 in normal cell function and malignant transformation. Oncogene 19:6482-6489. [DOI] [PubMed] [Google Scholar]
- 69.Yamada, T., N. Kondoh., M. Matsumoto, M. Yoshida, A. Maekawa, and T. Oikawa. 1997. Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood 89:1383-1393. [PubMed] [Google Scholar]
- 70.Yamada, T., F. Kihara-Negishi, H. Yamamoto, M. Yamamoto, Y. Hashimoto, and T. Oikawa. 1998. Reduction of DNA binding activity of the GATA-1 transcription factor in the apoptotic process induced by overexpression of PU.1 in murine erythroleukemia cells. Exp. Cell Res. 245:186-194. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto, H., F. Kihara-Negishi, T. Yamada, Y. Hashimoto, and T. Oikawa. 1999. Physical and functional interactions between the transcription factor PU.1 and the coactivator CBP. Oncogene 18:1495-1501. [DOI] [PubMed] [Google Scholar]
- 72.Yamamoto, H., F. Kihara-Negishi, T. Yamada, M. Suzuki, T. Nakano, and T. Oikawa. 2002. Interaction between the hematopoietic Ets transcription factor Spi-B and the coactivator CREB-binding protein associated with negative cross-talk with c-Myb. Cell Growth Differ. 13:69-75. [PubMed] [Google Scholar]
- 73.Yang, S. H., E. Vickers, A. Brehm, T. Kouzarides, and A. D. Sharrocks. 2001. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21:2802-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao, Y. L., W. M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yi, H., Y. Fujimura, M. Ouchida, D. D. Prasad, V. N. Rao, and E. S. Reddy. 1997. Inhibition of apoptosis by normal and aberrant Fli-1 and erg proteins involved in human solid tumors and leukemias. Oncogene 14:1259-1268. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, L., A. Eddy, Y. T. Teng, M. Fritzler, M. Kluppel, F. Melet, and A. Bernstein. 1995. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Mol. Cell. Biol. 15:6961-6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, P., G. Behre, J. Pan, A. Iwama, N. Wara-Aswapati, H. S. Radomska, P. E. Auron, D. G. Tenen, and Z. Sun. 1999. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. USA 96:8705-8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang, P., X. Zhang, A. Iwama, C. Yu, K. A. Smith, B. U. Mueller, S. Narravula, B. E. Torbett, S. H. Orkin, and D. G. Tenen. 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 96:2641-2648. [PubMed] [Google Scholar]
- 79.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zochodne, B., A. H. Truong, K. Stetler, R. R. Higgins, J. Howard, D. Dumont, S. A. Berger, and Y. Ben-David. 2000. Epo regulates erythroid proliferation and differentiation through distinct signaling pathways: implication for erythropoiesis and Friend virus-induced erythroleukemia. Oncogene 19:2296-2304. [DOI] [PubMed] [Google Scholar]