Abstract
Meprins are multidomain zinc metalloproteases that are highly expressed in mammalian kidney and intestinal brush border membranes and in leukocytes and certain cancer cells. Mature meprins are oligomers of evolutionarily related, separately encoded α and/or β subunits. Homooligomers of meprin α are secreted; oligomers containing meprin β are plasma membrane associated. Meprin substrates include bioactive peptides and extracellular matrix proteins. Meprins have been implicated in cancer and intestinal inflammation. Additionally, meprin β is a candidate gene for diabetic nephropathy. To elucidate in vivo functions of these metalloproteases, meprin β null mice were generated by targeted disruption of the meprin β gene on mouse chromosome 18q12. Analyses of meprin β knockout mice indicated that (i) 50% fewer null mice are born than the Mendelian distribution predicts, (ii) null mice that survive develop normally and are viable and fertile, (iii) meprin β knockout mice lack membrane-associated meprin α in kidney and intestine, and (iv) null mice have changes in renal gene expression profiles compared to wild-type mice as assessed by microarray analyses. Thus, disruption of the meprin β allele in mice affects embryonic viability, birth weight, renal gene expression profiles, and the distribution of meprin α in kidney and intestine.
Meprins (meprin A [EC 3.4.24.18] and meprin B [EC 3.4.24.63]), metalloproteases of the astacin family, comprise approximately 5% of kidney brush border membrane protein in mice (2, 3). They are also expressed in intestinal brush border membranes, in leukocytes, and in certain epithelial cancer cells (17, 21). Various growth factors, cytokines, and extracellular matrix proteins are substrates for meprin in vitro; among the best substrates are gastrin, gastrin-releasing peptide, monocyte chemoattractant protein-1, osteopontin, and laminin (1, 15). The nature of these substrates and the expression patterns of the subunits in acute renal failure, intestinal disease, and cancerous cells implicate meprins in the regulation of growth, inflammation, cancer cell metastasis, and matrix remodeling (5, 15, 17, 18, 21, 33).
The meprin subunits, α and β, associate to form homo- or heterooligomers (19). Although the subunits are encoded by distinct genes on different chromosomes (Mep1α on chromosome 6 in humans and 17 in mice and Mep1β on chromosome 18 in both species), under some circumstances there is coordinate regulation of the expression of the genes that results in heterooligomeric protein complexes (4). Meprin α and β subunits are 42% identical at the amino acid level and share the same domain structure, except that meprin α contains an inserted (I) domain which is not present in meprin β (20). The I domain directs the proteolytic cleavage of meprin α during maturation in the endoplasmic reticulum so that the bulk of the subunit is released from the membrane. The consequence of the cleavage is that meprin α is secreted unless it interacts with membrane-associated meprin β. In vivo, meprin α is secreted as a homooligomer and is also found as a heterooligomer at the plasma membrane in association with meprin β, a type I transmembrane protein (10, 20).
Meprin β was chosen for targeted disruption based on the following. (i) Certain inbred strains of mice, such as C3H/He and CBA, do not express meprin α in adult kidney and intestine, while all mouse strains tested express meprin β in the adult and embryonic kidney and intestine; this indicates that meprin β is more likely than meprin α to be essential in adult mice (2, 16). (ii) Linkage analysis has implicated meprin β as a candidate gene for diabetic nephropathy in the Pima Indian population, a population in which 92% of end stage renal disease cases are attributed to diabetic nephropathy (12, 23). (iii) Meprins have been implicated in the progression of pathology in renal ischemia and reperfusion, in ureteral obstruction, and in Alport's syndrome (24, 26, 32, 33). (iv) Lastly, meprin β associates with meprin α to maintain it at the cell surface; thus, in the absence of meprin β the localization of meprin α to the apical plasma membrane could be altered. Due to the consistent expression of meprin β and its implication in disease, it was chosen as a target for disruption. The work reported here details the first characterization of meprin β knockout mice.
MATERIALS AND METHODS
Construction of the targeting vector.
Targeted disruption of the Mep1β gene was accomplished by homologous recombination with a targeting vector that inserted a neomycin resistance cassette into the meprin β catalytic center encoded by exon 7. To accomplish this, primers 5′-ACGTCGACTCAGTGTTCAAGGGCAGTGG-3′ (containing a SalI linker and corresponding to exon 6) and 5′-TTCCCGCGGACAACTCCTGCTTCCCAG-3′ (containing a SacII linker and corresponding to exon 7) were used to generate a 2.1-kb amplicon from 129/Sv R1 genomic DNA. (Underlining indicates the restriction sites for the enzymes specified.) The PCR product was cut with SalI-SacII, gel purified, and ligated into Sa1I-SacII-digested pKO Scramber NTKV-1901 (Lexicon Genetics, Inc.). The resulting plasmid was pKO-Mep1β-2.1. An additional amplicon was derived by using primers 5′-TATCTCGAGGGACAGGATTCAGCCAG-3′ (containing a XhoI linker and corresponding to exon 7) and 5′-CAGAAGCTTCTCCAACATCGCCGTGG-3′ (containing a HindIII linker and corresponding to exon 10). This 4.7-kb PCR fragment was cut with XhoI-HindIII, gel purified, and ligated into XhoI-HindIII-digested pKO-Mep1β-2.1 The resulting vector, pKO-Mep1β, had exon 7 (which encodes the catalytic center) interrupted by a 1.6-kb neomycin resistance gene (Fig. 1A).
FIG. 1.
Strategy for disruption of the mouse Mep1β gene and genotype screening. (A) Schematic diagram of a portion of the exon-intron structures of the wild-type and knockout meprin β alleles. Exons (5 to 10) are represented as black boxes. The neomycin cassette derived from the targeting vector (pKO Scrambler NTKV-1901; Lexicon) is depicted as a gray box in exon 7 of the knockout allele. The 19-amino-acid consensus sequence for the catalytic center of astacin family metalloproteases is also shown. (B) Southern blot analysis of tail-derived genomic DNA. The probe used for Southern blotting detects two XbaI-generated fragments: 8.4 kb from the wild-type allele and 4.0 kb from the knocked-out allele. Lanes: +/+, meprin β wild-type DNA; +/−, meprin β heterozygous DNA; −/−, meprin β null DNA.
Screening of ES cells.
Targeting vector DNA was electroporated into R1 mouse embryonic stem (ES) cells (22), and 480 clones were screened by PCR. A 2.2-kb amplicon represented the wild-type allele, and a 3.7-kb amplicon indicated the targeted allele. Of 480 ES cell clones, 8 carried the targeted allele. Three targeted clones were microinjected into C57BL/6 blastocysts. Of the 43 ES cell-mouse chimeras obtained, 6 transmitted the targeted Mep1β allele to their offspring. Electroporation and selection of targeted ES cells and subsequent blastocyst injections were performed at the University of Michigan Transgenic Animal Facility. Chimeric animals were crossed with C57BL/6J mice and bred to homozygosity at Pennsylvania State University College of Medicine in full compliance with animal use and care regulations.
Genotyping.
Tail biopsies were digested overnight with agitation at 42°C with 6 U of proteinase K in 0.4 ml of buffer (50 mM Tris [pH 7.5], 100 mM EDTA, 125 mM NaCl, and 1% sodium dodecyl sulfate). A saturated NaCl solution (200 μl) was added to the digested samples. Samples were vortexed and then centrifuged for 15 min at 16,000 × g. The DNA was then ethanol precipitated from the resulting supernatant. Southern blot and PCR analyses were used to determine genotype. PCR primers 5′-GACTTCAAGCCTTGGTCCGGAG-3′ and 5′-CCTGGCTGAATCCTGTCCC-3′ generate a 2.2-or 3.7-kb amplicon for wild-type and knocked-out alleles, respectively. XbaI was used to digest 10 μg of DNA for Southern blotting. A 740-bp probe corresponding to exon 8 through exon 9 detects wild-type (8.4-kb) and disrupted (4.0-kb) alleles (Fig. 1B).
Immunohistochemistry.
Tissues were fixed in methyl Carnoy's solution (60% methanol, 30% chloroform, 10% acetic acid), embedded in paraffin wax, and thin sectioned (5 μm). An avidin-biotin blocking kit (Vector Laboratories) was used to block background contributed by endogenous biotin or biotin-binding proteins. Background Buster (Accurate Chemical & Scientific Corp.) was then used as a general blocking agent. Polyclonal rabbit anti-recombinant meprin α or β served as the primary antibody. Preimmune rabbit serum was used in place of primary antibody for negative controls. Biotinylated goat anti-rabbit immunoglobulin G (heavy plus light chains) at a final concentration of 7.5 μg/ml (Vector Laboratories) served as the secondary antibody. Sections were washed and then incubated with Vectastain ABC Elite reagents (Vector Laboratories). 3,3′-Diaminobenzidine (1 mg/ml) in Tris (pH 7.6) with 8.5% hydrogen peroxide was applied to all sections. Sections were counterstained with 1% methyl green, rinsed in 100% ethanol, submerged in xylene, and mounted with Permount (Fisher). Digital photographs were taken with a Nikon Eclipse E600 microscope.
Microarray.
Kidneys from second filial generation (F2) adult male wild-type (n = 3), heterozygous (n = 3), and knockout (n = 4) mice of a mixed C57BL/6J × Sv129 background were individually homogenized in 4 ml of TRIzol (Invitrogen). The preparation of total RNA and the subsequent labeling, fragmentation, and hybridization steps were performed according to the protocols issued by Affymetrix.
In short, total RNA (8 μg) was reverse transcribed with Superscript II (Invitrogen) by using a high-pressure liquid chromatography-purified oligo(dT)24 primer encoding a T7 RNA polymerase promoter site. Double-stranded cDNA was then purified by phenol-chloroform extraction. The DNA was then ethanol precipitated from the aqueous phase, and the precipitate was air dried. All of the obtained cDNA was subjected to in vitro transcription labeling with biotinylated ribonucleotides by using the BioArray High Yield RNA transcript labeling kit (Enzo Diagnostics, Inc.). The resulting labeled RNA was purified by using RNeasy columns (Qiagen) and then quantified. Labeled RNA (20 μg) was fragmented for chip hybridization. Completeness of fragmentation was verified by gel electrophoresis of an aliquot of sample. Fragmented RNA (15 μg) was combined in hybridization buffer with BioB (1.5 pM), BioC (5 pM), BioD (25 pM), cre (100 pM), and B2 oligonucleotide (50 pM) hybridization controls, herring sperm DNA (0.1 mg/ml), and acetylated bovine serum albumin (0.5 mg/ml) (Affymetrix). Test3 chips (Affymetrix) were used to confirm that labeled RNA was properly fragmented and labeled and that 5′, middle, and 3′ portions of transcripts were represented. MG-U74Av2 oligonucleotide probe array chips (Affymetrix) were hybridized with 200 μl of the RNA mixture for 16 h at 45°C with rotation at 60 rpm. Arrays were washed and stained according to protocols utilized by the GeneChip Fluidics Station 400 (Affymetrix). Arrays were scanned and then scaled to an average intensity of 500 and analyzed with Microarray Suite 5.0. Chip files from Microarray Suite 5.0 were entered into a database by using Micro Database 3.0. This database was then analyzed by using Data Mining Tools 3.0 (Affymetrix). The average signal intensity and average fold change were analyzed for statistically significant changes (P < 0.05) by t test analysis.
Western blots of brush border membrane-enriched fractions and urinary meprin.
Renal brush border-enriched fractions were isolated as previously described (29). Total protein of the enriched fractions was determined by using the Bio-Rad Bradford dye standard assay (Bio-Rad). Samples were subjected to 7.5% polyacrylamide gel electrophoresis under reducing and denaturing conditions. Primary antibodies were polyclonal rabbit anti-recombinant meprin α or β. Secondary antibodies were sheep anti-rabbit immunoglobulin G conjugated to horseradish peroxidase.
To determine urinary meprin α protein levels, urine samples from meprin β null and wild-type mice were analyzed by Western blotting. Equal urinary creatinine was loaded onto polyacrylamide gels for each sample being compared. Since urine creatinine secretion is constant for an individual over 24 h, normalizing urine samples based on urine creatinine allows for comparative analyses among different animals. Sample normalization is essential because urine samples collected from different individuals may not be equal in volume and/or concentration (6, 9). Sample normalization using urine creatinine values is based on the assumption that the animals being compared have similar 24-h creatinine secretion. This is a valid assumption for this study because serum creatinine levels of meprin β null and wild-type mice are not statistically different. Meprin α band densities were determined by using NIH Image software. For analyses of urinary meprin α and brush border membrane meprin α and meprin β, samples were obtained from individual age- and sex-matched animals. The blots shown are representative of data observed for at least five different animals per genotype.
Urine and serum analyses.
Urine was collected from animals into sterile microcentrifuge tubes by bladder massage. For analyses of blood urea nitrogen (BUN), total serum protein, serum albumin, serum creatinine, serum glucose, and serology, blood was collected from the vena cava of ether-anesthetized mice. Chemstrip 10 SG dip sticks (Roche) were used for semiquantitative analysis of urinary pH, protein, nitrite, glucose, ketones, urobilinogen, bilirubin, blood, leukocytes, and specific gravity. Urine and serum creatinine levels were measured by using the Infinity Creatinine Assay (Sigma Diagnostics). Total urinary protein was measured by using the Bio-Rad Bradford dye standard assay. Mice can become pathologically proteinuric; under normal circumstances serum proteins are not present in murine urine, while smaller proteins such as the major urinary proteins are typically abundant in mouse urine (6). Serum albumin was determined by using the Sigma Diagnostics albumin reagent. BUN was measured by using the BUN rate reagent (Sigma Diagnostics).
RT-PCR.
Total RNA isolated from mouse kidney by using TRIzol (Invitrogen) was subjected to reverse transcriptase PCR (RT-PCR) with the Superscript One-Step RT-PCR kit (Invitrogen). The amount of amplicon at various cycles was analyzed by using integrated band density data obtained with the Stratagene Eagle-Eye II system equipped with Eagle-Sight software. Glyceraldehyde-3-phosphate dehydrogenase was amplified as a control. For amplification of TRAF-6, the following primers were used: sense primer 5′-GGA AGA TTG GCA AGT TTG GGA TGC-3′ and antisense primer 5′-AGT ACA TGG ACG CTA CAC CCC CG-3′.
Statistical analysis.
For analysis of genotype distributions of the F2 population, a chi-square (χ2) analysis was performed on data obtained from F1 heterozygous crosses. For all other analyses, the unpaired, two-tailed t test was used; P values of <0.05 at a confidence interval of 95% were considered significant.
RESULTS
Meprin β null animals are underrepresented in the F2 population.
Meprin β is encoded by a single gene on mouse chromosome 18q12. The gene contains 15 exons and spans approximately 40 kb (4, 13). Targeted disruption of the mouse meprin β allele was accomplished by introduction of a neomycin resistance gene into exon 7, which encodes the protease domain (Fig. 1A). F1 meprin β+/− mice were crossed to produce F2 mice, which were determined to be meprin β+/+, β+/−, or β−/− by PCR and Southern blot analysis (Fig. 1B). Analysis of the genotype distribution of F2 mice by chi-square (χ2) analysis revealed that the meprin β null mice (β−/−) are underrepresented (P = 0.001) in the F2 population (Table 1). Mendelian genetics predicts that 25% of the animals born should be meprin β null; however, only 12% of the animals born were homozygous null. Postnatal morbidity is unlikely to account for the discrepancy in the number of meprin β null pups born, as all pups were counted on the day of birth as well as every day thereafter until they were genotyped at 21 days.
TABLE 1.
Genotype and sex distributions of offspring (n = 163) from heterozygous matingsa
| Genotype and sex of pups | No. observed | % Predicted | % Observed |
|---|---|---|---|
| Meprin β+/+ | 51 | 25 | 31.3 |
| Meprin β+/− | 92 | 50 | 56.4 |
| Meprin β−/− | 20 | 25 | 12.3 |
| Male | |||
| Total | 87 | 50 | 53.4 |
| Meprin β+/+ | 28 | ||
| Meprin β+/− | 49 | ||
| Meprin β−/− | 10 | ||
| Female | |||
| Total | 76 | 50 | 46.6 |
| Meprin β+/+ | 23 | ||
| Meprin β+/− | 43 | ||
| Meprin β−/− | 10 |
Genotypic frequencies are for progeny of F1 heterozygous crosses. The observed number of meprin β knockout animals does not follow a Mendelian distribution of 1:2:1 (P = 0.001 [confidence interval = 95%; chi-square test]).
General observations for meprin β knockout mice.
The meprin β knockout animals that were born did not exhibit overt physical abnormalities. Necropsies (conducted by Pennsylvania State University Department of Comparative Medicine veterinary pathologist James W. Griffith) which included complete and differential blood counts, gross anatomical and histological examinations, and serum analyses were conducted on a subset of adult null and wild-type mice. Gross anatomical and histological differences were not evident, nor were differences in serology, hematology, serum albumin, serum creatinine, total serum protein, serum glucose, or BUN evident (Table 2). Differences in urinary pH, nitrite, glucose, ketones, urobilinogen, bilirubin, leukocytes, and specific gravity were not observed. Urinary protein secretion was also analyzed; wild-type (n = 21) and knockout (n = 28) mice did not display statistically different urinary protein levels based on urine protein/urine creatinine ratios (average = 0.14 for both groups) (6, 9). Additionally, both sexes were fertile and equally represented in both the total F2 population and in the knockout subset of the F2 population (Table 1).
TABLE 2.
Serum chemistry of meprin β null and wild-type micea
| Genotype | BUN (mg/dl) | Serum creatinine (mg/dl) | Total serum protein (g/dl) | Serum albumin (mg/dl) | Serum glucose (mg/dl) |
|---|---|---|---|---|---|
| Meprin β−/− | 29.67 ± 2.24 (n = 15) | 0.45 ± 0.03 (n = 15) | 5.17 ± 0.16 (n = 15) | 3.13 ± 0.05 (n = 15) | 275.5 ± 7.6 (n = 12) |
| Meprin β+/+ | 25.83 ± 2.24 (n = 12) | 0.38 ± 0.02 (n = 12) | 4.99 ± 0.21 (n = 12) | 3.06 ± 0.05 (n = 12) | 259.4 ± 7.4 (n = 12) |
Values are expressed as means ± standard errors.
Meprin β knockout pups weighed more at birth than wild-type pups (P = 0.0001) and null females weighed more than wild-type females at time of weaning (age 21 days) (P = 0.046) (Table 3). By 150 days of age, the body weights of the different genotypes were not statistically different. Male weights were not statistically different among the genotypes at weaning or age 150 days (Table 3). For the data in Table 3, n decreases between 21 and 150 days because animals were either breeding or being used in experiments; thus, the decrease is not a reflection of increased mortality. Pregnant knockout females exhibited a standard gestation of 19 to 21 days, and they had litters of comparable size to those of wild-type mice. While the average litter sizes of knockout mice (6.25 ± 0.51 [mean ± standard error; n = 16]) and wild-type mice (7.45 ± 0.85 [n = 11]) were not statistically different (P = 0.2), the distribution of the litter sizes indicated a tendency for null litters to be smaller than wild-type litters (Fig. 2). For knockout mice, 75% of the litters were smaller than the average wild-type litter. This trend may become statistically significant as more litters are observed.
TABLE 3.
Body weights of wild-type, heterozygous, and meprin β null mice at birth, 21 days (weaning), and 150 days
| Time | Genotype and sex | n | Body wt (g)
|
|||
|---|---|---|---|---|---|---|
| Mean ± SE | Minimum | Maximum | Median | |||
| Birth | Wild type | 24 | 1.43 ± 0.02 | 1.25 | 1.65 | 1.40 |
| Meprin β null | 21 | 1.59 ± 0.03a | 1.35 | 1.80 | 1.60 | |
| Day 21 | Female | |||||
| Wild type | 19 | 9.31 ± 0.27 | 7.60 | 11.45 | 9.20 | |
| Heterozygous | 36 | 9.62 ± 0.25 | 6.75 | 12.75 | 9.25 | |
| Meprin β null | 12 | 10.11 ± 0.37a | 8.15 | 11.50 | 10.15 | |
| Male | ||||||
| Wild type | 22 | 9.77 ± 0.1 | 8.10 | 11.55 | 9.67 | |
| Heterozygous | 40 | 10.20 ± 0.21 | 7.65 | 12.60 | 10.20 | |
| Meprin β null | 11 | 9.65 ± 0.39 | 7.65 | 11.65 | 9.25 | |
| Day 150 | Female | |||||
| Wild type | 12 | 24.29 ± 0.84 | 17.85 | 27.45 | 24.52 | |
| Heterozygous | 27 | 24.77 ± 0.51 | 19.55 | 32.25 | 24.60 | |
| Meprin β null | 6 | 25.55 ± 0.91 | 21.65 | 28.00 | 25.50 | |
| Male | ||||||
| Wild type | 18 | 31.54 ± 0.48 | 27.50 | 35.25 | 31.77 | |
| Heterozygous | 29 | 32.70 ± 0.47 | 27.50 | 39.30 | 32.90 | |
| Meprin β null | 8 | 31.18 ± 1.23 | 27.15 | 37.20 | 30.62 | |
P < 0.05 (confidence interval = 95%; t test for wild-type versus meprin β null mice).
FIG. 2.
Distribution of meprin β null and wild-type litter sizes. Data were obtained from crosses of homozygous meprin β null mice or homozygous meprin β wild-type mice. Horizontal lines in each data set represent the mean, the 75th percentile, or the 25th percentile of the given population.
Effects of the disruption of the meprin β gene on meprin α and β mRNA expression and protein localization.
RT-PCR analyses of kidney RNA revealed that meprin β mRNA was absent in knockout mice, as expected, and present at approximately half the wild-type level in β+/− mice; meprin α mRNA levels in the kidney and intestine were found to be similar among the three genotypes (data not shown). Microarray analyses confirmed that kidney meprin α mRNA levels were not statistically different among the three genotypes and that meprin β was absent in the knockout animals and present in heterozygous samples at half the level seen in wild-type mouse kidney (Table 4). Based on signal intensity values from microarray analyses, both meprin α and meprin β exhibit signal intensity levels indicative of abundant expression in wild-type kidney, as predicted by previous studies of kidney meprin protein.
TABLE 4.
Microarray analyses of kidney RNAs from meprin β knockout, heterozygous, and wild-type mice
| Probe set | Avg signal Intensity ± SE
|
||
|---|---|---|---|
| β−/− (n = 4) | β+/− (n = 3) | β+/+ (n = 3) | |
| Meprin α | 917.0 ± 23.7 | 1,073.9 ± 110.8 | 1,034.3 ± 150.5 |
| Meprin βa | 19.5 ± 6.9 | 641.6 ± 85.1 | 1,431.4 ± 39.4 |
Ratio of signal intensity of β+/− to β+/+ = 0.45.
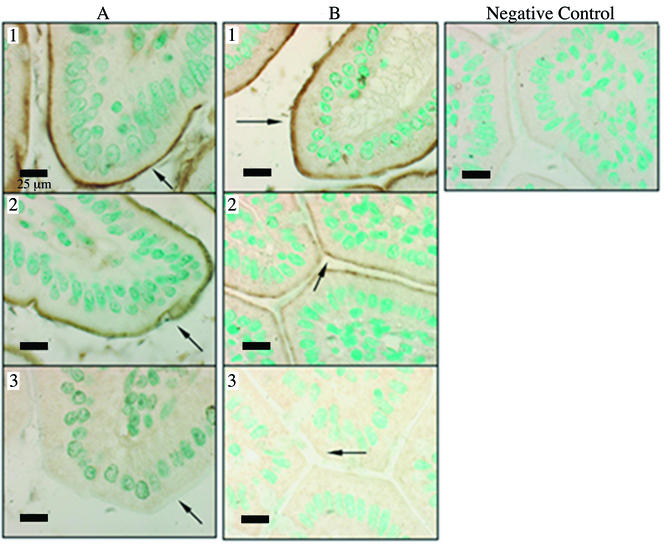
Immunohistochemical analyses of kidney and intestine from meprin β knockout animals showed that neither meprin α nor meprin β was membrane associated in these tissues in the adult animal (Fig. 3). In meprin β heterozygous and meprin β wild-type animals there was brown staining evident on the apical membranes of intestinal villi, indicating the presence of meprin α and β (Fig. 3). Staining for meprin subunits in kidney sections from the three genotypes was similar to that seen for intestinal sections; the apical membranes of kidney proximal tubule cells from heterozygous and wild-type mice stained positively for meprin α, while kidney sections from meprin β null mice did not exhibit membrane-associated meprin α or meprin β (Fig. 4). Quantitative Western blotting for meprin α and β protein in kidney brush border-enriched fractions from wild-type and heterozygous mice demonstrated that when a single meprin β allele is disrupted, the commensurate halving of the β protein at the membrane is concomitant with the presentation of half as much meprin α on the brush border membrane (Fig. 5). As expected, neither meprin α nor meprin β was detectable in renal brush border fractions from meprin β knockout mice.
FIG. 3.
Immunohistochemistry to detect meprin α and β in intestinal sections from wild-type, heterozygous, and meprin β knockout mice. (A) Immunohistochemistry for meprin α. (B) Immunohistochemistry for meprin β. Panels 1, wild-type (β+/+) intestine; panels 2, meprin β heterozygous (β+/−) intestine; panels 3, meprin β knockout (β−/−) intestine. Preimmune serum was added in place of antimeprin primary antibody as a negative control. Arrows point to apical membrane staining in wild-type and heterozygous mice and lack of staining in meprin β knockout intestine. Sections were counterstained for nuclei with methyl green and digitally photographed. Magnification, ×100.
FIG. 4.
Immunohistochemistry to detect meprin α and β in kidney sections from wild-type, heterozygous, and meprin β knockout mice. (A) Immunohistochemistry for meprin α. (B) Immunohistochemistry for meprin β. Panels 1, wild-type (β+/+) kidney; panels 2, meprin β heterozygous (β+/−) kidney; panels 3, meprin β knockout (β−/−) kidney. Preimmune serum was added in place of antimeprin primary antibody as a negative control. Arrows point to apical membrane staining in wild-type and heterozygous mice and lack of staining in meprin β knockout kidney. Sections were stained for nuclei with methyl green and digitally photographed. Magnification, ×40.
FIG. 5.
Semiquantitative Western blotting of kidney brush border-enriched fractions from wild-type and meprin β heterozygous mice. (A) Western blot to detect meprin α. (B) Western blot to detect meprin β. Graphs show band densities versus the amount of kidney brush border protein analyzed for wild-type (β+/+) and heterozygous (β+/−) mice. O.D., optical density.
Based on immunohistochemistry, RT-PCR, and microarray data, there was a difference between the membrane meprin α protein levels and the meprin α mRNA levels. Membrane-associated meprin α is not detected in meprin β knockout animals, while the meprin α mRNA levels are not statistically decreased compared to those in wild-type or heterozygous mice. This difference is explained by the observation that meprin α is secreted into the urine at greater levels in meprin β knockout mice than in wild-type animals. Semiquantitative Western blots showed that on average meprin β knockout animals have approximately twice the amount of urinary meprin α as wild-type mice (Fig. 6). Urinary meprin α subunits from knockout and wild-type animals form oligomeric complexes (from dimers to dodecamers) as evidenced by size exclusion chromatography (data not shown). Additionally, urinary meprin α is secreted primarily as an activatable proprotein in both knockout and wild-type animals.
FIG. 6.
Semiquantitative Western blotting of urinary meprin α. (A) Urine samples from wild-type and meprin β knockout mice were equivalently loaded for creatinine (1, 2, and 3 μg). (B) Western blotting for meprin α followed by analysis of band densities revealed that meprin β knockout mice have approximately twice the amount of meprin α in their urine as do wild-type mice.
Effects of the disruption of the meprin β gene on renal gene expression profiles.
Kidney mRNA from three wild-type mice and four null mice was assessed by using Affymetrix microarray chips. Of the 12,473 genes represented on the chips, 45 to 48% were found to be expressed in the tested samples. A total of 39 genes changed threefold or more (P < 0.05) in expression for meprin β null kidney samples compared to wild-type samples (Table 5). Of the 39 genes, 11 were expressed sequence tags (ESTs) and 1 was a pseudogene (data not shown). Genes that have been characterized were assigned to five categories based on gene product functions that are annotated in PubMed and GenBank (Table 5). Most of the genes that changed 3.0-fold or more were increased in expression in the meprin β knockout mice. Meprin β represented the largest fold change (73.4-fold decrease).
TABLE 5.
Microarray analysis of kidney RNA: functional classification of genes which changed in expression at least threefold in comparisions of meprin β knockout and wild-type mice
| Genea | Fold increase | Fold decrease | P value |
|---|---|---|---|
| Meprin β (Mep1b) | 73.4 | 0.001 | |
| Immune response related | |||
| CD1d2 antigen | 3.7 | 0 | |
| Tumor necrosis factor receptor-associated factor (TRAF-6) | 3.3 | 0.022 | |
| Killer cell lectin-like receptor, subfamily A member 8 (Klra 8) | 3.2 | 0.004 | |
| Casein alpha (Csna) | 3.2 | 0.029 | |
| Tumor necrosis factor (ligand) superfamily, member 7 (Tnfsf 7) | 3.1 | 0.035 | |
| Toll-like receptor 6 (Tlr 6) | 3.1 | 0.006 | |
| Cell growth or cell cycle related | |||
| 18S rRNA (translation) | 7.0 | 0.003 | |
| Cyclin-dependent kinase (CDK 2) | 3.7 | 0.019 | |
| Growth factor receptor-bound protein 10 (Grb 10) | 3.5 | 0.025 | |
| Cell cycle regulatory transcription factor DP1 | 3.3 | 0.011 | |
| Paired box gene 4 (Pax 4) (transcription factor) | 3.3 | 0.030 | |
| Paired-like homeodomain transcription factor 2 (Pitx 2) | 3.1 | 0.016 | |
| Cell division cycle control protein 25C (Cdc25c) | 3.0 | 0.030 | |
| Tumor susceptibility gene 101 (Tsg 101) | 3.0 | 0.036 | |
| Lung carcinoma myc-related oncogene 1 (L-myc 1) | 4.5 | 0 | |
| Nervous system related | |||
| Nicotinic acetylcholine receptor gamma (Chrng) | 5.4 | 0.034 | |
| Potassium voltage-gated channel, shaker-related subfamily 7 (Kcna 7) | 5.1 | 0.008 | |
| Potassium chloride cotransporter 2 (KCC 2) | 3.0 | 0.003 | |
| Chloride channel 1 (Clc1) | 3.2 | 0.024 | |
| Posttranslational modification related | |||
| Heparan sulfate N-deacetylase/N-sulfotransferase 1 (Ndst 1) | 5.2 | 0.034 | |
| Mannoside acetyl glucosaminyl transferase 3 (Mgat 3) | 3.3 | 0.002 | |
| Cytoskeleton-related kinesin-like protein 9 (Kif9) | 5.2 | 0.030 | |
| Not assigned | |||
| Recombining binding protein supressor of hairless-like (Rbpsuh 1) | 3.6 | 0.024 | |
| Transferrin receptor (Trfr) | 3.3 | 0.033 | |
| Testis-specfic gene 1 (Tpx 1) | 3.2 | 0.042 | |
| Sperm tail-associated protein (Stap) | 3.1 | 0.037 |
Genes which increased or decreased in expression at least threefold with a P value of <0.05 are reported.
The microarray chips utilized have approximately 6,000 characterized genes represented; these include genes for 784 characterized proteases. None of the functionally characterized genes in Table 5 encode a protease or protease inhibitor. Genes related to cell growth and the cell cycle constituted 23% of the changes, while genes related to the immune system comprised 15% of the changes. Among the observed changes are genes that encode membrane-associated receptors, adapter proteins for signaling cascades, and transcription factors. Of particular interest was the increased expression of the signaling protein tumor necrosis factor receptor-associated factor-6 (TRAF-6). The MATH (for meprin and TRAF homology) and AM (for after MATH) domains of meprins share 30% identity with the TRAFC domains of TRAFs (34, 36). Semiquantitative RT-PCR was performed to test the validity of the microarray data; the RT-PCR data confirmed that TRAF-6 is increased in meprin β null kidney compared to wild-type kidney (data not shown).
DISCUSSION
This work establishes that targeted disruption of the meprin β gene in mice does not generate an overt pathological phenotype in animals that survive to birth. However, the underrepresentation of meprin β null mice in the F2 population indicates that meprin β is important for either embryonic development, implantation, or fertilization. Previous work demonstrated that meprin α and β are expressed in the kidney and intestine during embryogenesis (16). Furthermore, EST data indicate that meprin β is expressed as early as the first blastocyst stage as well as in adult testis; this raises the possibility that meprin β could be important for sperm maturation or function. (WashU-HHMI Mouse EST Project, db EST Id 1337387 and EST Id 3378734, at www.ncbi.nlm.nih.gov). The microarray data here revealed that two genes, which are expressed in testis (sperm tail-associated protein and testis-specific gene-1), are increased in expression in the kidney in the absence of meprin β. It is possible that the expression of these genes is also altered in testis in the absence of meprin β and could thereby affect sperm function. The EST information is consistent with observations that several astacin family members are important in the early development of mammals and in hardening of the chorion in oviparous species (3, 27).
In mice, implantation occurs 4 to 5 days following fertilization; for successful implantation to occur, the zona pellucida must be shed (28). Implantation is coupled with a surge in progesterone levels; this surge could induce meprin β expression, as it has been shown that meprin β expression is induced by glucocorticoids (11). If meprin β is important for implantation, then one may predict that litter sizes of meprin β null animals would be small compared to litter sizes of wild-type mice. While the average litter sizes of knockout and wild-type mice were not statistically different, 75% of meprin β knockout litters were smaller than the average wild-type litter.
The litter sizes of the heterozygous F1 crosses were not statistically different from those of the wild-type crosses. One may expect that if approximately 10% of the F2 population is dying in utero, a concomitant decrease in the litter sizes of heterozygous F1 crosses compared to wild-type crosses would be observed. There are two possible explanations as to why this was not observed. The first is that for a 10% difference in litter sizes to be statistically significant, a very large number of litters must be observed. The second is that if the in utero discrepancy arises preimplantation, then wild-type or heterozygous blastocysts could implant and a smaller litter size may not be fully realized. This compensatory implantation would not occur in homozygous null crosses, since there would not be wild-type and heterozygous blastocysts present to implant.
With respect to postnatal growth, it was observed that meprin β knockout pups weighed more than wild-type pups at birth. Meprins are known to cleave various peptide hormones, such as parathyroid hormone, α-melanocyte-stimulating hormone, insulin B chain, gastrin, and gastrin-releasing peptide, in vitro; thus, alterations in peptide hormone degradation in meprin β null mice may have an impact on fetal growth (1). Furthermore, maternal circulating glucose levels during pregnancy can influence birth weight; high maternal glucose levels have been correlated with increased birth weights (14, 26). While circulating glucose levels in adult, nonpregnant meprin β knockout and wild-type mice did not appear to be statistically different, it is possible that during pregnancy circulating glucose levels differ between the genotypes and consequently have an impact on fetal growth. Additionally, meprin β knockout female pups weighed more at weaning (21 days) than wild-type female pups, while differences among male pups were not observed. This observation implies that meprin β differentially affects growth of males and females in the early postnatal stage.
The data here also demonstrate that the presence of meprin β is obligatory for meprin α to be membrane associated in the kidney and intestine. The most obvious implication of meprin α localization being altered upon the disruption of the meprin β gene is that under normal conditions meprin α is not likely to appreciably interact with other membrane proteins in the kidney or intestine. Additionally, halving of the amount of meprin β on the kidney brush border is accompanied by halving of the amount of meprin α on the brush border, indicating that the interaction of meprin α with meprin β appears to have a regulated stoichiometry of oligomerization in vivo. These observations are consistent with previous data on the oligomerization of meprins (19). Meprin α mRNA expression is not altered when the meprin β gene is disrupted. Allelic dose compensation for meprin β was not observed; in meprin β heterozygous mice, a single functional copy of the meprin β gene is insufficient to restore kidney meprin β expression to the wild-type level.
While meprin α is not detected on the apical membranes of renal proximal tubule cells in meprin β null mice, it is clear that meprin α is still apically targeted in these cells, since it is secreted into the urine. The observation that urinary meprin α from both wild-type and meprin β null mice is predominantly a zymogen is possibly a protective mechanism, since active meprin α has been shown to be cytotoxic in vivo and in vitro (5). Characterizing the localization and expression of meprin α in the meprin β null mice is important, since in certain disease states the expression and localization of meprin α is altered. For example, in colon cancer and renal ischemia-reperfusion injury, the localization of meprins is altered; a basolateral versus apical distribution is observed (18, 35). It has been proposed that the redistribution of meprins to the basolateral surface exacerbates injury.
Linkage analysis of the Pima Indian population by Imperatore et al. identified meprin β as a candidate gene for diabetic nephropathy. For the Pima Indian study, the level of proteinuria of study participants was used as an assessment of renal function (12). Analyses of urine protein/urine creatinine level ratios of wild-type and meprin β knockout mice did not reveal any statistical differences. Likewise, additional markers of kidney function, such as BUN and serum creatinine levels, were not statistically changed between the genotypes. It is possible, however, that a difference in renal function may be observed if diabetes is induced in these mice or in response to a renal stress. In several models of kidney disease, such as experimental hydronephrosis, toxin-induced nephrosis, and collagen IV α 3 knockout mice (which develop Alport's syndrome), meprin β expression is decreased (24, 25, 32, 33). Additionally, it is known that interstitial fibrosis, which characterizes the aforementioned renal diseases, is accompanied by alterations in various metalloproteinase activities as well as changes in metalloprotease inhibitors (8, 31). It has been suggested that decreases in metalloproteinases such as meprin can lead to tissue damage due to a reduced capacity to turn over or remodel basement membranes and extracellular matrix components (24, 33). Furthermore, the capacity of meprins to degrade certain cytokines implicates these metalloproteases in inflammation.
Assessing the response of meprin β null mice to the various renal challenges will be important, especially in the context of evidence presented here, in that renal gene expression profiles are changed upon disruption of the meprin β gene. Based on the types of renal gene expression changes that were observed, it is possible that meprins affect or interact with signaling proteins or other proteins at the cell surface that are involved in cell growth and the immune response. Such an interaction could be direct or indirect. The most dramatic changes in gene expression in meprin β null kidneys arise for genes related to the cell growth and cell cycle processes and the immune system. These observations may relate to the ability of meprins to degrade various growth factors and to the point that meprins are expressed by certain cancer cells and leukocytes (17, 18, 21). Additionally, changes in the expression of TRAF-6 and Toll-like receptor-6 have implications with respect to the activation of c-Jun N-terminal kinase and NF-κB (7, 30).
This work has demonstrated that meprins are likely to be involved in development and growth. Furthermore, it has provided a baseline for future studies of the meprin β null mice. It has also called into question the necessity for meprin β in normal adult mice. Lastly, several genes related to the immune response and to cell growth have been identified as possible genetic compensation factors for disruption of the meprin β gene. Further studies, including physiological challenges to the meprin β knockout mice, are necessary to elucidate the in vivo function of meprins and their roles in disease states such as end stage renal disease and cancer.
Acknowledgments
We thank Sarah Bronson for advice on generating and breeding the knockout mice; Gary Clawson for use of equipment and software for microarray and immunohistochemistry analyses; Andras Nagy, Reka Nagy, and Wanda Abramow-Newerly for the R1 ES cells; James W. Griffith for performing necropsies; Deborah Kees-Folts for assistance in analyzing tissue sections; Anita K. Hopper for reviewing the manuscript; and Xiaojing Wang for technical assistance.
This work was supported by NIH grants DK54625 and DK19691 (to J. S. Bond). The equipment and software for the microarray and immunohistochemistry analyses are supported by NIH grant CA40145 (to G. Clawson).
REFERENCES
- 1.Bertenshaw, G. P., B. E. Turk, S. J. Hubbard, G. L. Matters, J. E. Bylander, J. M. Crisman, L. C. Cantley, and J. S. Bond. 2001. Marked differences between metalloproteases meprin A and B in substrate and peptide bond specificity. J. Biol. Chem. 276:13248-13255. [DOI] [PubMed] [Google Scholar]
- 2.Bond, J. S., and R. J. Beynon. 1986. Meprin: a membrane-bound metallo-endopeptidase Curr. Top. Cell Reg. 28:263-290. [DOI] [PubMed] [Google Scholar]
- 3.Bond, J. S., and R. J. Beynon. 1995. The astacin family of metalloendopeptidases. Protein Sci. 4:1247-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond, J. S., K. Rojas, J. Overhauser, H. Y. Zoghbi, and W. Jiang. 1995. The structural genes, MEP1A and MEP1B, for the alpha and beta subunits of the metalloendopeptidase meprin map to human chromosomes 6p and 18q, respectively. Genomics 25:300-303. [DOI] [PubMed] [Google Scholar]
- 5.Carmago, S., S. V. Shah, and P. D. Walker. 2002. Meprin, a brush-border enzyme, plays an important role in hypoxic/ischemic acute renal tubular injury in rats. Kidney Int. 61:959-966. [DOI] [PubMed] [Google Scholar]
- 6.Chen, A., C. H. Wei, L. F. Sheu, S. L. Ding, and W. H. Lee. 1995. Induction of proteinuria by adriamycin or bovine serum albumin in the mouse. Nephron 69:293-300. [DOI] [PubMed] [Google Scholar]
- 7.Chung, J. Y., Y. C. Park, H. Ye, and H. Wu. 2002. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115:679-688. [DOI] [PubMed] [Google Scholar]
- 8.Eddy, A. A., H. Kim, J. Lopez-Guisa, T. Oda, and P. D. Soloway. 2000. Interstitial fibrosis in mice with overload proteinuria: deficiency of TIMP-1 is not protective. Kidney Int. 58:618-628. [DOI] [PubMed] [Google Scholar]
- 9.Ginsberg, J. M., B. S. Chang, R. A. Matarese, and S. Garella. 1983. Use of single voided urine samples to estimate quantitative proteinuria. N. Engl. J. Med. 309:1543-1546. [DOI] [PubMed] [Google Scholar]
- 10.Grunberg, J., E. Dumermuth, J. A. Eldering, and E. E. Sterchi. 1993. Expression of the alpha subunit of PABA peptide hydrolase (EC 3.4.24.18) in MDCK cells. Synthesis and secretion of an enzymatically inactive homodimer. FEBS Lett. 335:376-379. [DOI] [PubMed] [Google Scholar]
- 11.Henning, S. J., T. J. Oesterreicher, D. E. Osterholm, D. Lottaz, D. Hahn, and E. E. Sterchi. 1999. Meprin mRNA in rat intestine during normal and glucocorticoid-induced maturation: divergent patterns of expression of alpha and beta subunits. FEBS Lett. 462:368-372. [DOI] [PubMed] [Google Scholar]
- 12.Imperatore, G., W. C. Knowler, R. G. Nelson, and R. L. Hanson. 2001. Genetics of diabetic nephropathy in the Pima Indians. Curr. Diabetes Rep. 1:275-281. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, W., J. M. Kumar, G. L. Matters, and J. S. Bond. 2000. Structure of the mouse metalloprotease meprin beta gene (Mep1b): alternative splicing in cancer cells. Gene 248:77-87. [DOI] [PubMed] [Google Scholar]
- 14.Jovanovic-Peterson, L., C. M. Peterson, G. F. Reed, B. E. Metzger, J. L. Mills, R. H. Knopp, and J. H. Aarons. 1991. Maternal postprandial glucose levels and infant birth weight: the Diabetes in Early Pregnancy Study. Am. J. Obstet. Gynecol. 164:103-111. [DOI] [PubMed] [Google Scholar]
- 15.Kaushal, G. P., P. D. Walker, and S. V. Shah. 1994. An old enzyme with a new function: purification and characterization of a distinct matrix-degrading metalloproteinase in rat kidney cortex and its identification as meprin. J. Cell Biol. 126:1319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, J. M., and J. S. Bond. 2001. Developmental expression of meprin metalloprotease subunits in ICR and C3H/He mouse kidney and intestine in the embryo, postnatally and after weaning. Biochim. Biophys. Acta 1518:106-114. [DOI] [PubMed] [Google Scholar]
- 17.Lottaz, D., D. Hahn, S. Müller, C. Müller, and E. E. Sterchi. 1999. Secretion of human meprin from intestinal epithelial cells depends on differential expression of the alpha and beta subunits. Eur. J. Biochem. 259:496-504. [DOI] [PubMed] [Google Scholar]
- 18.Lottaz, D., C. A. Maurer, D. Hahn, M. W. Buchler, and E. E. Sterchi. 1999. Nonpolarized secretion of human meprin alpha in colorectal cancer generates an increased proteolytic potential in the stroma. Cancer Res. 59:1127-1133. [PubMed] [Google Scholar]
- 19.Marchand, P., J. Tang, and J. S. Bond. 1994. Membrane association and oligomeric organization of the alpha and beta subunits of mouse meprin A. J. Biol. Chem. 269:15388-15393. [PubMed] [Google Scholar]
- 20.Marchand, P., J. Tang, G. D. Johnson, and J. S. Bond. 1995. COOH-terminal proteolytic processing of secreted and membrane forms of the alpha subunit of the metalloprotease meprin A. Requirement of the I domain for processing in the endoplasmic reticulum. J. Biol. Chem. 270:5449-5456. [DOI] [PubMed] [Google Scholar]
- 21.Matters, G. L., and J. S. Bond. 1999. Expression and regulation of the meprin beta gene in human cancer cells. Mol. Carcinog. 25:169-178. [PubMed] [Google Scholar]
- 22.Nagy, A., J. Rossant, R. Nagy, W. Abramow-Newerly, and J. C. Roder. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90:8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson, R. G., J. M. Newman, W. C. Knowler, M. L. Sievers, C. L. Kunzelman, D. J. Pettitt, C. D. Moffett, S. M. Teutsch, and P. H. Bennett. 1988. Incidence of end-stage renal disease in type 2 (non-insulin-dependent) diabetes mellitus in Pima Indians. Diabetologia 31:730-736. [DOI] [PubMed] [Google Scholar]
- 24.Ricardo, S. D., J. S. Bond, G. D. Johnson, J. Kaspar, and J. R. Diamond. 1996. Expression of subunits of the metalloendopeptidase meprin in renal cortex in experimental hydronephrosis. Am. J. Physiol. 270:F669-F676. [DOI] [PubMed] [Google Scholar]
- 25.Sampson, N. S., S. T. Ryan, D. A. Enke, D. Cosgrove, V. Koteliansky, and P. Gotwals. 2001. Global gene expression analysis reveals a role for the alpha 1 integrin in renal pathogenesis. J. Biol. Chem. 276:34182-34188. [DOI] [PubMed] [Google Scholar]
- 26.Scholl, T. O., M. Sowers, X. Chen, and C. Lenders. 2001. Maternal glucose concentration influences fetal growth, gestation, and pregnancy complications. Am. J. Epidemiol. 154:514-520. [DOI] [PubMed] [Google Scholar]
- 27.Shibata, Y., T. Iwamatsu, Y. Oba, D. Kobayashi, M. Tanaka, Y. Nagahama, N. Suzuki, and M. Yoshikuni. 2000. Identification and cDNA cloning of alveolin, an extracellular metalloproteinase, which induces chorion hardening of medaka (Oryzias latipes) eggs upon fertilization. J. Biol. Chem. 275:8349-8354. [DOI] [PubMed] [Google Scholar]
- 28.Snell, G. D., and L. C. Stevens. 1966. Early Embryology, p. 205-246. In E. L. Green (ed.), Biology of the laboratory mouse, 2nd ed. McGraw-Hill Book Company, New York, N.Y.
- 29.Stroupe, S. T., S. S. Craig, C. M. Gorbea, and J. S. Bond. 1991. Sex-related differences in meprin-A, a membrane-bound mouse kidney proteinase. Am. J. Physiol. 261:E354-E361. [DOI] [PubMed] [Google Scholar]
- 30.Takeuchi, O., T. Kawai, H. Sanjo, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, K. Takeda, and S. Akira. 1999. TLR6: a novel member of an expanding toll-like receptor family. Gene 231:59-65. [DOI] [PubMed] [Google Scholar]
- 31.Tang, W. W., L. Feng, Y. Xia, and C. B. Wilson. 1994. Extracellular matrix accumulation in immune-mediated tubulointerstitial injury. Kidney Int. 45:1077-1084. [DOI] [PubMed] [Google Scholar]
- 32.Trachtman, H., R. Greenwald, S. Moak, J. Tang, and J. S. Bond. 1993. Meprin activity in rats with experimental renal disease. Life Sci. 53:1339-1344. [DOI] [PubMed] [Google Scholar]
- 33.Trachtman, H., E. Valderrama, J. M. Dietrich, and J. S. Bond. 1995. The role of meprin A in the pathogenesis of acute renal failure. Biochem. Biophys. Res. Commun. 208:498-505. [DOI] [PubMed] [Google Scholar]
- 34.Uren, A. G., and D. L. Vaux. 1996. TRAF proteins and meprins share a conserved domain. Trends Biochem. Sci. 21:244-245. [PubMed] [Google Scholar]
- 35.Walker, P. D., G. P. Kaushal, and S. V. Shah. 1998. Meprin A, the major matrix degrading enzyme in renal tubules, produces a novel nidogen fragment in vitro and in vivo. Kidney Int. 53:1673-1680. [DOI] [PubMed] [Google Scholar]
- 36.Zapata, J. M., K. Pawlowski, E. Haas, C. F. Ware, A. Godzik, and J. C. Reed. 2001. A diverse family of proteins containing tumor necrosis factor receptor-associated factor domains. J. Biol. Chem. 276:24242-24252. [DOI] [PubMed] [Google Scholar]