Abstract
During the shutdown of proliferation and onset of differentiation of HL-60 promyelocytic leukemia cells, expression of the cell cycle-dependent histone genes is downregulated at the level of transcription. To address the mechanism by which this regulation occurs, we examined the chromatin structure of the histone H4/n (FO108, H4FN) gene locus. Micrococcal nuclease, DNase I, and restriction enzymes show similar cleavage sites and levels of sensitivity at the H4/n locus in both proliferating and differentiated HL-60 cells. In contrast, differentiation-related activation of the cyclin-dependent kinase inhibitor p21cip1/WAF1 gene is accompanied by increased nuclease hypersensitivity. Chromatin immunoprecipitation assays of the H4/n gene reveal that acetylated histones H3 and H4 are maintained at the same levels in proliferating and postproliferative cells. Thus, the chromatin of the H4/n locus remains in an open state even after transcription ceases. Using ligation-mediated PCR to visualize genomic DNase I footprints at single-nucleotide resolution, we find that protein occupancy at the site II cell cycle element is selectively diminished in differentiated cells while the site I element remains occupied. Decreased occupancy of site II is reflected by loss of the site II binding protein HiNF-P. We conclude that H4 gene transcription during differentiation is downregulated by modulating protein interaction at the site II cell cycle element and that retention of an open chromatin conformation may be associated with site I occupancy.
Cell proliferation and the onset of differentiation are associated with complex and interdependent biochemical events involving cell cycle stage-specific modifications in gene expression. Histone genes, which encode proteins that form the nucleosome, represent a key paradigm for understanding cell cycle-dependent regulatory mechanisms. Histone gene transcription is upregulated at the G1/S phase transition and downregulated at the onset of differentiation (3, 33, 35). There is limited insight into the molecular mechanisms by which cells transcriptionally inactivate G1/S phase-related genes during cessation of cell proliferation.
Transcription of genes required for cell cycle progression may be silenced in differentiated cells by modifying the competency of transcription factors to interact with promoters and/or by altering the higher-order organization of chromatin. In general, chromatin of transcriptionally active genes is nuclease hypersensitive and in a relatively open conformation, whereas many silent genes are nuclease resistant, reflecting closed chromatin (10, 15). Transcriptional activity is influenced by dynamic alterations in chromatin structure (5, 9, 10, 25, 27, 32). Chromatin opening and gene activation involve posttranslational modifications of histones (e.g., acetylation or phosphorylation), interactions of specific transcription factors, activities of chromatin remodeling complexes, and localization of genes and cognate regulatory factors in nuclear compartments permissive for transcription (17, 25, 31, 32, 34, 36).
Human HL-60 promyelocytic leukemia cells can be induced to differentiate into the macrophage lineage by treatment with phorbol-12-myristate-13-acetate (PMA), and differentiation is accompanied by adherence, growth arrest, and the expression of a number of phenotypic markers (12, 29). During the shutdown of proliferation and onset of differentiation, expression of the cell cycle-dependent histone genes is downregulated at the level of transcription (7, 33). Among the multiple genes encoding histone H4 (H4a to -o), transcriptional mechanisms that regulate expression of the H4/n (FO108, H4FN) gene have been studied extensively (e.g., reference 18) (4, 11, 24, 26, 38-40). Two proximal promoter regions, designated site I and site II, are principal components that mediate H4 gene activation (24, 35). The site II domain is responsible for cell cycle regulation of transcription during S phase, while the enhancer element site I is required for maximal activation of transcription (18, 26). Studies using HeLa cells revealed changes in the chromatin structure of the H4/n gene that are functionally related to modulations in transcription during the cell cycle (6, 21). However, it remains to be determined whether there is an obligatory reorganization of chromatin structure to silence histone gene transcription in differentiating cells.
In this study, we have assessed the functional coupling between chromatin organization and regulation of histone H4/n gene expression during HL-60 differentiation into the monocyte/macrophage lineage. The higher-order chromatin architecture of the H4/n gene was analyzed by monitoring sensitivity to micrococcal nuclease, DNase I, and restriction enzymes in the transcriptionally active and inactive states in proliferating and differentiated HL-60 cells, respectively. In vivo protein-DNA interactions were detected by genomic DNase I footprinting by using ligation-mediated PCR, and histone acetylation status at the H4/n locus was assessed using chromatin immunoprecipitation assays. We find that the chromatin structure of the histone H4/n gene remains in an open state during the proliferation-differentiation transition and that the H4 gene is regulated primarily by selective modulation of protein occupancy at the site II cell cycle regulatory element.
MATERIALS AND METHODS
Preparation of nuclei.
HL-60 human promyelocytic leukemia cells were grown in RPMI medium supplemented with 20% fetal bovine serum, penicillin (50 U/ml), streptomycin (50 μg/ml), and l-glutamine (2 mM). Cells were induced to differentiate by treatment with 16 nM PMA (Sigma, St. Louis, Mo.). Nuclei were isolated from 108 treated or control cells by Dounce homogenization (loose pestle) in 40 ml of RSB buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2) with 0.5% (vol/vol) NP-40 and protease inhibitors (Complete; Roche Molecular Biochemicals, Indianapolis, Ind.). Nuclei were washed three times with RSB buffer, and cell lysis was evaluated by staining with 0.4% trypan blue.
Nuclease hypersensitivity assays.
DNase I hypersensitivity at the histone H4/n gene locus was analyzed using modification of a previously described procedure (20). In brief, nuclear pellets were resuspended in 3 ml of RSB buffer and the DNA concentration was estimated by absorption at 260 nm. Aliquots of 20 A260 units were digested with increasing concentrations of DNase I (0 to 10 U) (Worthington Biochemicals, Freehold, N.J.) in a 400-μl final volume of RSB buffer with 1 mM CaCl2 for 5 min at room temperature. The reaction was stopped by adding 400 μl of Stop buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 50 mM EDTA, 0.3% sodium dodecyl sulfate [SDS]) and incubated with 10 U of RNase ONE RNase (Promega, Madison, Wis.) at 37°C for 1 h. The samples were extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1). Nucleic acids were precipitated with 0.7 volumes of isopropanol, washed with 70% ethanol, and resuspended in 1× TE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA).
Sensitivity to micrococcal nuclease (MNase) was determined using a freshly prepared suspension of nuclei in MNase digestion buffer (15 mM Tris-HCl [pH 7.5], 60 mM KCl, 15 mM NaCl, 1 mM CaCl2, 3 mM MgCl2, 20% glycerol, 15 mM 2-mercaptoethanol). DNA concentrations were estimated by absorption at 260 nm. Equal amounts of nuclei were digested with increasing concentrations of MNase I (0 to 150 U) (Worthington, Lakewood, N.J.) in a 400-μl final volume for 5 min at 20°C. Reactions were stopped and DNA was extracted as described above.
For restriction enzyme accessibility assays, nuclei were isolated as described above and digested with 25 to 150 U of restriction enzyme in 300 μl of the corresponding reaction buffer (New England Biolabs, Beverly, Mass.) for 30 min at 37°C. Reactions were terminated and DNA was extracted as described above. After purification, the DNA was digested to completion with PstI and BamHI. The products were resolved on agarose gels and detected by Southern blotting using the indirect end-labeling method. Data are expressed as percent DNA digested (the ratio of the amount of the digested fragment to total DNA). Two-tailed paired t tests were used for comparison of mean percent digestions between proliferating and differentiated samples. Differences were considered statistically significant when P was ≤0.015.
Analysis of chromatin by indirect end labeling.
Nucleosomal mapping by indirect end labeling was performed using specific probes spanning the 5′ and 3′ flanking regions of the H4/n gene. The purified DNA (15 μg) was subjected to a secondary digestion by HindIII, PstI, or HpaI, then electrophoresed in 1.3% agarose gels in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA; pH 8.0), and transferred to Hybond N+ membrane (Amersham Pharmacia Biotech, Arlington Heights, Ill.). Histone H4 gene fragments were detected by hybridization with radioactive probes obtained by random priming or PCR labeling. The following primer sets were used to amplify probes anchored at HindIII or HpaI sites: HindIII-F, 5′-AAGCACGGCTCTGAATCC; HindIII-R, 5′-GAGAATTCAGATCCCAACC; HpaI-F, 5′-CTAGGCGCCGCTCCAG; HpaI-R, 5′-CCGTTAACTTCAGATCC. The PstI probe was extracted from agarose gels after restriction enzyme digestion of plasmid DNA by XbaI and PstI. Prehybridization and hybridization were performed at 65°C, using 5× SSC (10× SSC is 1.5 M NaCl, 0.15 M sodium citrate; pH 7.4), 0.7% SDS, and 5× Denhardt solution (5× is 0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin), followed by three sequential washes under stringent conditions. The blots were analyzed by autoradiography or by using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Genomic DNase I footprinting using LM-PCR.
Ligation-mediated PCR (LM-PCR) was carried out essentially by the method of Mueller et al. (22) with minor modifications. Initial extension, amplification, and labeling were performed with Vent DNA polymerase (New England Biolabs). The reaction mixture was diluted and linker ligation was performed with 3 U of T4 DNA ligase (Promega) in the buffer recommended by the manufacturer. Primer labeling was done using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). The primers used were the following: LM7 (+10 to +35), 5′-GACATGACCGCTGGAGCCCGATA-3′; LM8 (−2 to +22), 5′-GCTGGAGCCCGATAGACAGCTTTCTG-3′; LM9 (−6 to +22), 5′-GCTGGAGCCCGATAGACAGCTTCTGTCA-3′.
The initial extension reaction was performed using 4 μg of DNA. After linker ligation and precipitation, half of the sample was used for the amplification step. The annealing temperatures used for the primers were as follows: LM7, 64°C; LM8, 66°C; and LM9, 68°C. Samples were amplified for 20 cycles, and PCR products were labeled for two cycles using 32P-labeled primers. DNA was extracted with phenol-chloroform, precipitated by ethanol, and analyzed using 6% denaturing polyacrylamide gels.
EMSAs.
Nuclear proteins were isolated from HL-60 cells using standard protocols (1, 39). Electrophoretic mobility shift assays (EMSAs) for detection of site II binding proteins were performed as described previously (2, 39). The histone H4 site II probe for detecting the HiNF-D complex was the EcoRI-HindIII insert from pFP202 (37). The oligonucleotides used as double-stranded probes were the following: Sp1 consensus, 5′-ATTCGATCGGGGCGGGGCGAGC-3′; AP-1 consensus, 5′-CGTGACTCAGCGCGCG-3′; HiNF-P wild type, 5′-GATCTTCGGTTTTCAATCTGGTCCGAT-3′; HiNF-P mutant, 5′-GATCTTCGGTTTTCAATCTtctaCGAT-3′; HiNF-M wild type, 5′-GATCCGGGCGCGCTTTCGGTTTTCA-3′; HiNF-M mutant, 5′-GATCCCGGCGCGCTTcaGGTTTCA-3′. Portions of sequences shown in lowercase and boldface represent mutated nucleotides.
RT-PCR.
Total RNA was prepared using Trizol (Life Technologies) according to the manufacturer's instructions. Forward and reverse primers used for PCR analysis were as follows: H4/n Fw (5′-TATCGGGCTCCAGCGGTCATGTC) and Rev (5′-GGATCGAAACGTAAGCTGGAG); human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Fw (5′-TCACCATCTTCCAGGAGCG) and Rev (5′-CTGCTTCACCACCTTCTTGA). PCRs were performed in a total volume of 50 μl using the Titan One-Tube reverse transcription-PCR (RT-PCR) system (Roche) according to the manufacturer's instructions.
Northern blot analysis.
Total RNA (10 μg) was electrophoresed in 1% denaturing formaldehyde gels, transferred to Hybond N membrane (Amersham Pharmacia Biotech) with 10× SSC, subjected to UV cross-linking, and hybridized with 32P-labeled p21WAF1 cDNA or a DNA probe spanning the H4/n coding region. The hybridization conditions were similar to those used for Southern blot hybridization.
Chromatin immunoprecipitation (ChIP) assays.
HL-60 cells were incubated in cross-linking buffer (1% formaldehyde in RPMI medium) at room temperature for 10 min. Cells were harvested, washed once with ice-cold 1× phosphate-buffered saline (PBS), and resuspended in 500 μl of lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1% NP-40, 25 μM MG132, 1× Complete protease inhibitor). Cells were incubated on ice for 20 min and sonicated for 10 s five to six times (setting of 3.5 for Fisher Scienific sonicator with a [1/4]-in. tip). The resulting lysate was precleared by centrifugation for 10 min at 4°C and incubated with 40 μl of protein A/G Plus-Sepharose beads (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.), 2 μg of sheared salmon sperm DNA, and 2 μg of normal rabbit immunoglobulin G for 1 h. After centrifugation at 4,000 rpm in an Eppendorf microcentrifuge for 3 min, the supernatant was transferred to a new tube and incubated with 2 μg of antibody for 16 h at 4°C at 30 rpm. Immune complexes were mixed with 40 μl of a protein A/G-agarose suspension followed by incubation for 2 h at 4°C while rotating. Immunoprecipitates were collected by microcentrifugation at 3,000 rpm for 2 min at 4°C. The bead pellets were sequentially washed with 500 μl each of the following buffers: low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl [pH 8.1], 150 mM NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-Cl [pH 8.1], 500 mM NaCl), and LiCl wash buffer (0.25 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-Cl [pH 8.1]). The beads were then washed three times using 500 μl of TE buffer (Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]). The immune complexes were eluted twice with 100 μl of freshly prepared elution buffer (1% SDS, 100 mM NaHCO3), each time rotating the sample for 10 min at 80 rpm, with 2 min of microcentrifugation between the two elutions at 3,000 rpm. The cross-linking reaction was reversed by overnight incubation of the elutants at 65°C, and the DNA was recovered by phenol-chloroform extraction and ethanol precipitation. Each DNA sample was dissolved in 50 μl of TE buffer. Approximately 5% of the bound DNA fraction was used for quantitative PCR to detect the presence of specific segments of the H4/n locus. The following primer pairs were used: −1582/−1271 F-, CGGGGAGGGAGAATTGCTCC; R-, GAGAGTCTATGAGAAAATCTTCTGG; −999/−797 F-, GAAACGGATGCACAGAATATCC; R-, CTATGCACATCCTCCCGTGTAC; −404/−247 F-, CACGGCTCTGAATCCGCTCGTC; R-, TGGCCTGTGCTCTGGTTCTGAGGA; −221/+29 F-, GATCTGAATTCTCCCGGGGACTGT; R-, GACATGACCGCTGGAGCCCGATA; +664/+932 F-, CACACCTAGAGGGCCACGTCAG; R-, AGGCATCTAGTCCACCCGCTTG.
The antibodies used in our study recognize acetylated histones and are available commercially (Upstate Biotechnology, Lake Placid, N.Y.). Antibodies referred to as H4 (catalog no. 06-598) recognize acetylated forms of histone H4, whereas the H3 (catalog no. 06-599) recognizes acetylated histone H3. Normal rabbit serum (Santa Cruz Biotechnology Inc.) was used as a negative control.
Western blot analysis.
Nuclear extracts were prepared from proliferating and differentiated HL-60 cells, and 10 μg of protein was subjected to SDS-polyacrylamide gel electrophoresis in 8% polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes (Millipore) using a semidry transfer apparatus. Membranes were blocked with 5% nonfat milk powder in PBS buffer (sodium phosphate, 10 mM; potassium phosphate, 2 mM; KCl, 2.7 mM; NaCl,137 mM; pH 7.4) for 1 h at 4°C and incubated with primary antibody (peptide-affinity-purified HiNF-P antibody) and lamin B (catalog no. SC-6217; Santa Cruz Biotechnology) at a dilution of 1:2,000 in PBS for 1 h at room temperature, followed by appropriate horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology; 1:5,000 in PBS; 1 h at room temperature). Blots were developed using Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences, Boston, Mass.).
RESULTS
MNase mapping reveals distinct and constitutive nucleosomal distribution at the histone H4/n locus.
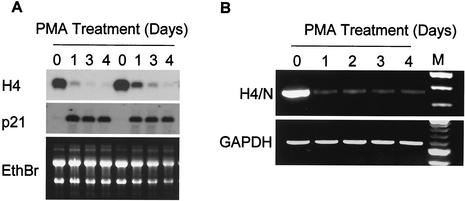
Treatment of HL-60 cells with PMA results in growth arrest and differentiation towards the macrophage lineage concomitant with a dramatic downregulation of histone H4 gene transcription (13, 33). Because histone H4 mRNAs are transcribed from multiple genes, we assessed total H4 mRNA levels by Northern blot analysis and used RT-PCR to detect specifically H4/n gene transcripts (Fig. 1). These data show that histone H4 genes (Fig. 1A), and specifically the H4/n gene (Fig. 1B), are downregulated during differentiation and cessation of cell growth of HL-60 cells in response to PMA.
FIG. 1.
Expression of the histone H4/n gene during PMA-induced differentiation of HL-60 cells. (A) Northern blot analysis was performed with total RNA prepared from PMA-treated (differentiated) and untreated (proliferating) HL-60 cells at the indicated times of treatment from two separate experiments. RNAs were hybridized with probes spanning the coding regions of the genes for H4/n and p21. The H4/n probe detects histone H4 mRNAs derived from multiple members of the histone H4 gene family. The ethidium bromide (EthBr)-stained gel is shown as a control for RNA integrity and quantitation. (B) Semiquantitative RT-PCR was performed with the same RNA samples and primers that detect transcripts specific for the H4/n gene. GAPDH was used as an internal control. The right lane (M) shows a 100-bp repeat marker.
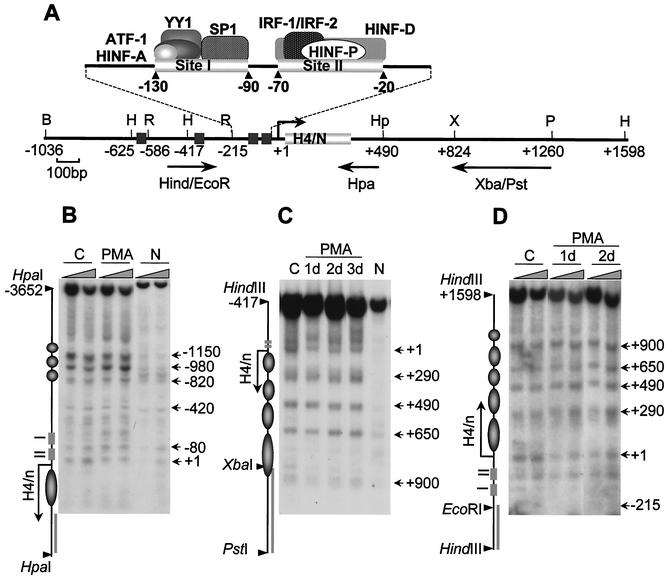
To understand the contribution of chromatin structure to transcriptional regulation of histone H4 genes, we analyzed the nucleosomal organization of the H4/n locus during HL-60 differentiation. We employed MNase to monitor cleavage sensitivity at the 5′ and 3′ regions of the H4/n gene and detected the digestion products by indirect end labeling (Fig. 2A). MNase digestion of chromatin revealed that the H4 gene exhibits nuclease-hypersensitive sites at variable intervals (Fig. 2B to D). Although a subset of these sites is also present in naked DNA, the overall digestion pattern indicates a series of positioned nucleosomes in the distal 5′ promoter region (Fig. 2B) (−1150 to −820) and in the coding and 3′ region of the H4/n locus (Fig. 2C and D) (+1 to +900). No specific sites of digestion were observed in the region spanning nucleotides −820 to −420. In contrast, the region between −420 and +1, encompassing the proximal promoter and the transcription start site, shows multiple, closely spaced sites of MNase sensitivity. Our results suggest that the −800 to +300 segment of the H4 gene has a noncanonical nucleosome organization. Differentiation of HL-60 cells with PMA does not alter the pattern of MNase sensitivity at the H4/n locus. Thus, the nucleosomal organization of this gene is not changed when H4 expression is silenced.
FIG. 2.
Maintenance of nucleosomal organization at the H4/n gene locus during HL-60 differentiation. (A) Restriction map of the human histone H4/n gene locus and functional organization of the proximal promoter. Arrows show DNA probes used for indirect end labeling to map nuclease accessibility of the H4/n locus. (B to D) Nuclei isolated from control (C) and PMA-treated (PMA) HL-60 cells, as well as naked DNA (N), were incubated with increasing concentrations of MNase for 5 min at room temperature. Purified DNA was digested with HpaI (B), PstI/HindIII (C), and HindIII (D). Southern blots were hybridized to radioactively labeled DNA fragments as follows: Hpa PCR probe (B), XbaI/PstI (C), and HindIII/EcoRI (D). The diagrams at the left of panels B, C, and D indicate the locations of transcription regulatory elements (rectangles), the positions of putative nucleosomes (circles and ovals), and the probes used for hybridization (solid bars). The arrows indicate the transcription start site and length of the protein-coding region. Numbers at the right indicate positions of MNase-sensitive sites.
Differentiated HL-60 cells retain DNase I-hypersensitive sites at the H4/n locus.
We next addressed whether there are modifications in DNase I hypersensitivity during the shutdown of histone H4/n gene transcription as HL-60 cells differentiate. Nuclei from PMA-treated and untreated HL-60 cells were isolated and subjected to digestion with increasing concentrations of DNase I. Genomic DNA was purified, cleaved with appropriate restriction enzymes, and analyzed by indirect end labeling (Fig. 3). We detected at least five DNase I-hypersensitive sites in both untreated and PMA-treated HL-60 cells (Fig. 3A). Three of these hypersensitive sites are located in the proximal H4 promoter region surrounding regulatory elements sites I and II (Fig. 3B) (−130, −90, and +1); the other two are located in the coding and 3′ regions (Fig. 3A) (+290 and +490). Although the overall pattern of DNase I digestion was very similar, we reproducibly observed subtle changes in the specific locations of DNase I hypersensitivity at the site II region following PMA-dependent HL-60 differentiation (Fig. 3A, compare lanes 1 and 3, or B, compare lanes 2 and 5). Therefore, these data indicate that although there are local alterations in the proximal promoter, the chromatin architecture of the histone H4/n gene remains open when expression of the gene has been downregulated.
FIG. 3.
Multiple DNase I-hypersensitive sites at the H4/n locus. Nuclei were digested with increasing concentrations of DNase I. DNA was purified and cleaved with PstI/BamHI (A) or HpaI (B). After Southern blotting the filter was hybridized with the XbaI-PstI fragment (A) or the HpaI PCR probe fragment (+336/+500) (B) (see Fig. 2A). The diagram at the left indicates the positions of transcription regulatory elements and the transcription start site. The + sign designates the area of altered nuclease sensitivity. Other symbols and abbreviations are described in the legend to Fig. 2.
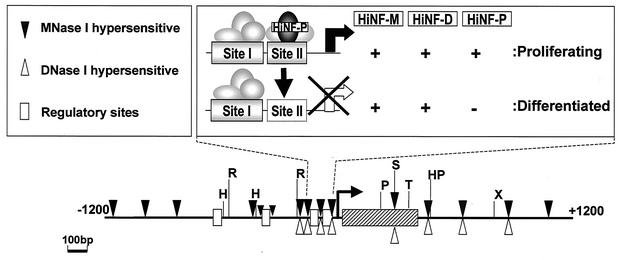
There is a remarkable congruence in the positions of the DNase I- and MNase-hypersensitive sites in the proximal promoter, coding region, and 3′ end of the H4/n gene (see summary in Fig. 9, below). Interestingly, the locations of these sites correspond with the presence of gene regulatory elements involved in transcriptional control and 3′-end processing of RNA transcripts. Hypersensitive sites are found at the same locations in proliferating HeLa cervical carcinoma cells, T98G glioblastoma cells, and IMR90 fibroblasts (data not shown). Thus, nuclease-hypersensitive sites are present in the H4/n gene regardless of the phenotype or proliferative status of the cell.
FIG. 9.
Chromatin organization of the H4/n locus. The diagram depicts MNase-sensitive sites (filled triangles), DNase I-hypersensitive sites (open triangles), and restriction enzyme cleavage sites (black vertical bars). Also indicated are the positions of cis regulatory elements (open boxes), the H4/n coding region (hatched box), and the mRNA start site (hooked arrow). The proximal promoter is expanded at the top right to show occupancy of sites I and II (solid box is occupied; dashed box is vacant) in proliferating and differentiated HL-60 cells. Factors known to interact with the H4/n promoter are depicted as ovals. Selective loss of protein-DNA interactions at site II is reflected by downregulation of HiNF-P binding activity and altered sensitivity to cleavage by AvaII (vertical arrow).
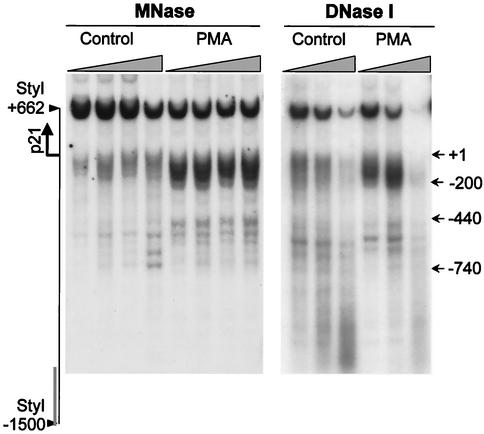
As a control, we analyzed the chromatin structure of the gene for the cyclin-dependent kinase inhibitor p21cip1/WAF1, which is transcriptionally upregulated during PMA-induced HL-60 cell differentiation (14, 41). When we probed the chromatin structure of the p21 promoter with MNase and DNase I, we observed that PMA treatment results in a dramatic increase in nuclease sensitivity (Fig. 4). This alteration in chromatin structure contrasts with the persistent nuclease hypersensitivity of the histone H4/n locus (Fig. 2 and 3). Hence, our findings show that induction of HL-60 differentiation is accompanied by chromatin remodeling of the cell cycle-related p21 gene, but not the histone H4/n gene.
FIG. 4.
Altered chromatin organization of the p21cip1/WAF1 gene locus during HL-60 cell differentiation. Nuclei isolated from control and PMA-treated (2 days) HL-60 cells were incubated with increasing concentrations of MNase or DNase I for 5 min at room temperature. Purified DNA was digested with StyI, and hypersensitive sites were detected by hybridization with a radiolabeled PCR fragment spanning the p21 promoter region (−1500/−1250). The diagram at the left indicates the position of the transcription start site (arrow) and the probe (bar). Numbers at the right indicate positions of nuclease-sensitive sites relative to the transcription start site of the p21 gene.
Restriction enzyme accessibility at the proximal promoter of the histone H4/n gene persists during HL-60 differentiation.
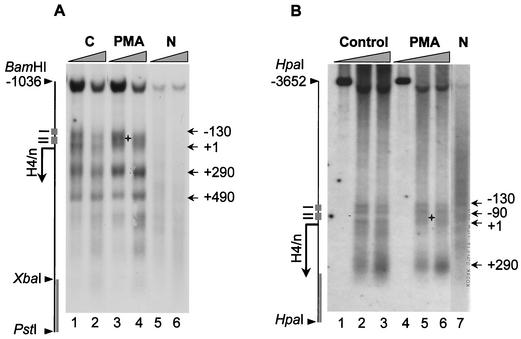
The sensitivity of the histone H4/n locus to cleavage by restriction endonucleases was assessed to quantitate changes in chromatin structure during HL-60 differentiation. Nuclei isolated from proliferating or PMA-treated HL-60 cells were subjected to limited digestion with a panel of restriction enzymes with recognition sites at different locations in the 5′, coding, and 3′ regions of the histone H4/n gene (Fig. 5A). We found that restriction sites at a distance upstream or downstream from the H4/n gene (e.g., HindIII/−625 and XbaI/+824) are cleaved less efficiently than those located closer to the H4 coding and regulatory sequences (e.g., SgrAI/−74 and BanII/+14) (Fig. 5B). We also observed that sites within the distal H4/n promoter (HindIII/−625 and EcoRI/−586) exhibit less sensitivity than those sites located more proximally in the promoter (HindIII/−417 and EcoRI/−215). However, overall restriction enzyme accessibility is similar in actively growing and postproliferative HL-60 cells.
FIG. 5.
Differences in the restriction enzyme accessibility at the H4/n gene in proliferating and differentiated HL-60 cells. (A) Nuclei were isolated from proliferating (P) and differentiated (D; 2 days of PMA treatment) HL-60 cells and digested with 75 to 500 U of restriction enzyme/ml. Purified DNA was digested to completion with PstI and BamHI. Products were detected by Southern blotting using the XbaI/PstI probe. (B) The relative nuclease sensitivity of restriction enzyme sites was quantitated, and the intensities of radioactive bands were used to calculate the percentage of DNA digested. The relative positions of the restriction enzyme sites are shown in the diagram. The asterisk indicates statistical significance at the P ≤ 0.015 level using the two-tailed paired t test.
We noted a modest but significant change between proliferating and differentiated cells in the relative efficiency with which AvaII cleaves its cognate sequences (−45 and −160) located, respectively, within and outside the site II cell cycle domain of the H4 promoter. The AvaII recognition sequence within site II is cleaved preferentially in nuclei from differentiated cells (P ≤ 0.015). In contrast, there is no apparent difference during HL-60 differentiation in MunI cleavage at the 3′ edge of site I. The results are consistent with the subtle changes in DNase I sensitivity of the H4/n proximal promoter during HL-60 differentiation. These localized changes in nuclease sensitivity may directly reflect modifications in protein-DNA interactions at H4/n gene regulatory elements when transcription ceases (see below).
Patterns of histone H3 and H4 acetylation at the histone H4/n locus are sustained during HL-60 differentiation.
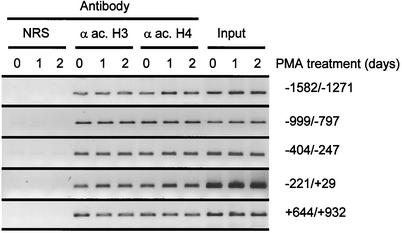
To address whether histone acetylation at the H4/n locus is modified when histone gene expression is silenced during HL-60 differentiation, we performed ChIP assays, using a panel of PCR primers that amplify segments of the histone H4/n gene. In proliferating HL-60 cells, acetylated histones H3 and H4 are associated with DNA segments spanning sequences between −1600 and +900 bp (Fig. 6). Following induction of HL-60 differentiation by PMA treatment, we found that levels of acetylated histones H3 and H4 at the histone H4/n gene are similar to those in proliferating cells (Fig. 6 and data not shown). Thus, the open chromatin structure of the H4/n locus is associated with comparable levels of acetylated histones whether or not the gene is transcriptionally active.
FIG. 6.
Association of acetylated histones at the histone H4/n locus during HL-60 differentiation. ChIP assays were performed with proliferating HL-60 cells (0) or cells differentiated by treatment with PMA for 1 or 2 days. The antibodies used recognize acetylated histone H4 (α ac. H4) or acetylated histone H3 (α ac. H3). Normal rabbit serum (NRS) was used as a negative control. Precipitated DNA was amplified by PCR using primers detecting specific regions of the H4/n gene, as indicated. A representative set of PCR data from one of two experiments is shown.
Protein occupancy of the H4/n site II regulatory region is modified during HL-60 differentiation.
Our laboratory has previously shown that histone H4 transcription is silenced upon HL-60 differentiation (reference 33 and data not shown). The data presented above indicate that this downregulation of histone H4/n gene transcription does not involve changes in overall chromatin organization. Therefore, we used LM-PCR-assisted genomic DNase I footprinting analysis to address whether alterations in the in vivo occupancy of sites I and II of the histone H4/n promoter are associated with transcriptional inactivation. The site I proximal promoter region of the H4/n gene interacts with ubiquitous transcription factors (i.e., Sp1, ATF-1, and YY1), while site II interacts with cell growth-related proteins (i.e., HiNF-D/CDP, HiNF-M/IRF, and HiNF-P/H4TF-2) (35).
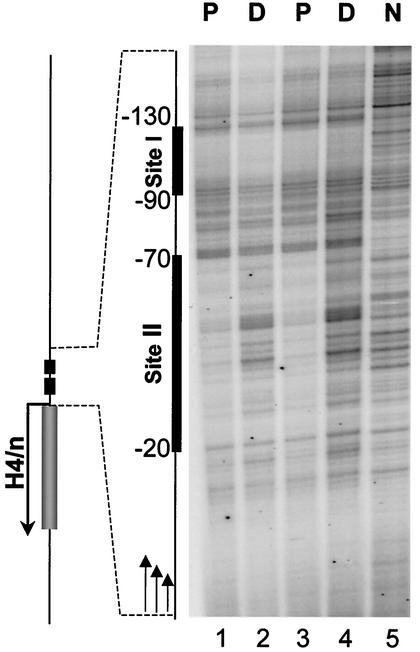
Our results indicate that proliferating HL-60 cells exhibit two genomic DNase I footprints, which are located at nucleotides −130 to −90 and −70 to −20 relative to the transcriptional start site (Fig. 7, lanes 1 and 3). These footprints are in agreement with sites of in vivo protein-DNA interactions (site I and site II) previously observed for the same gene in HeLa S3 cells (24). Following HL-60 differentiation, we found that the site I domain remains protected from DNase I digestion, showing occupancy by the cognate factors (Fig. 7, lanes 2 and 4). In contrast, the site II region becomes sensitive to DNase I digestion in differentiated HL-60 cells, indicating that occupancy of site II by the corresponding sequence-specific transcription factors is significantly reduced. Thus, there is a selective loss of in vivo protein-DNA interactions at the site II element of the H4/n gene upon transcriptional downregulation during HL-60 differentiation.
FIG. 7.
In vivo protein occupancy at the site II cell cycle element of the H4/n gene is selectively downregulated during HL-60 differentiation. Genomic occupancy of the site I and site II regulatory elements of the H4/n gene was analyzed by in vivo DNase I footprinting using LM-PCR. Nuclei from proliferating (P) or differentiated (D) HL-60 cells were digested with DNase I and DNA was purified before LM-PCR analysis. Deproteinized and digested DNA is shown in the right lane (N).
HiNF-P binding activity is selectively downregulated during HL-60 differentiation.
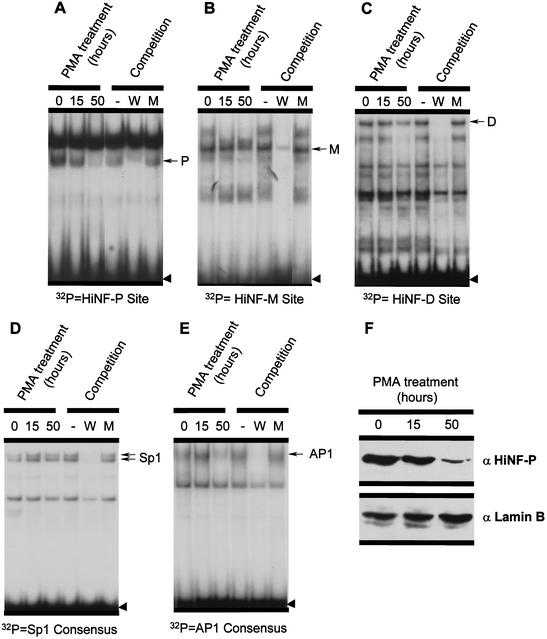
We addressed the mechanism by which changes in genomic occupancy at the site II cell cycle domain are mediated by monitoring the activity of site II binding proteins in proliferating and differentiated HL-60 cells. Using gel mobility shift assays, we found that DNA binding activity of the site II transcription factor HiNF-P is downregulated to undetectable levels in differentiated cells (Fig. 8A). In contrast, factors HiNF-M, HiNF-D, and the site I binding protein Sp1 are found in both proliferating and differentiated cells, although at late stages of differentiation HiNF-D activity also begins to decrease (Fig. 8). For comparison, AP1 activity is low or undetectable in proliferating HL-60 cells but is induced following PMA treatment (Fig. 8E and data not shown). We then performed Western blot analysis to assess HiNF-P protein levels. Our results show that the loss of HiNF-P DNA binding activity during HL-60 differentiation is paralleled by decreasing levels of HiNF-P nuclear proteins (Fig. 8F; compare with Fig. 8A). We conclude that HiNF-P is selectively downregulated during HL-60 differentiation, and this event may account for the cessation of histone gene transcription.
FIG. 8.
EMSA of protein-DNA interactions at site I and site II of the H4/n promoter. Radiolabeled oligonucleotides detecting the site II binding proteins HiNF-P (A), HiNF-M (B), and HiNF-D (C), the site I binding protein Sp1 (D), and the ubiquitous AP1 factor (E) were each incubated with 4 μg of nuclear protein. Nuclear extracts were prepared from proliferating HL-60 cells (0) or HL-60 cells that were treated with PMA for 15 or 50 h. The same nuclear extracts were used for Western immunoblot analysis (F) to assess HiNF-P protein levels. Lamin B was used as loading control. W, wild-type oligo; M, mutant oligo. Arrows designate specific DNA-protein complexes, and the arrowhead shows free probe.
DISCUSSION
Downregulation of proliferation-specific gene transcription in differentiating cells involves the removal of gene activators, recruitment of repressors, and/or alterations in chromatin structure. Here, we have assessed the mechanisms by which the cell growth-and cell cycle-regulated histone H4/n gene is transcriptionally silenced during PMA-induced HL-60 differentiation. Our findings establish that transcriptional inactivation of this gene is mediated by modifications in protein-DNA interactions without substantially altering overall chromatin conformation.
Classical studies of the globin, lysozyme, and ovalbumin loci, as well as more recent studies of many other genes (e.g., references 10, 16, 19, 28, and 30) support the concept that gene activation involves increased sensitivity to nuclease digestion and association with acetylated histones (8, 17, 32). Our examination of the histone H4/n gene using MNase, DNase I, and restriction enzymes, as well as ChIP assays, indicates that this promoter is organized in an open chromatin structure independent of its transcriptional status (Fig. 9). Thus, chromatin of the H4/n gene remains poised for transcription in differentiated cells, perhaps to support rapid induction of histone mRNA synthesis in response to postproliferative demands for histone protein. Furthermore, an open chromatin organization of the H4/n locus is also evidenced by association with acetylated histones H3 and H4. This finding parallels previous observations that acetylation of the histone H4 protein is sustained at the c-myc and c-fos loci during HL-60 differentiation (23). Thus, elevated levels of histone acetylation remain constitutively present at multiple proliferation-associated genes following differentiation of HL-60 cells.
This report provides additional evidence for the concept that nuclease hypersensitivity and histone acetylation are not necessarily indicative of gene activity. While nuclease sensitivity of the H4/n locus persists in differentiated HL-60 cells, the gene is not transcribed (reference 33 and unpublished observations). Genomic DNase I footprinting shows that there is a selective loss of key protein-DNA interactions in the proximal promoter at the site II cell cycle regulatory element. Work from our laboratory has identified three distinct factors that interact at site II: HiNF-M(IRF-2), the CDP-cut-containing HiNF-D complex, and the H4 subtype-specific factor HiNF-P (35). Mutagenesis studies indicate that each of these factors contributes to H4 gene transcription through partially overlapping recognition sequences. However, the loss of site II occupancy in vivo, observed here for differentiated HL-60 cells, correlates with downregulation of DNA binding activity and protein levels of HiNF-P, a principal regulator of histone H4 gene transcription via the cyclin E/CDK2/NPAT pathway (39; P. Mitra, R.-L. Xie, R. Medina, H. Hovhannisyan, S. K. Zaidi, Y. Wei, J. W. Harper, J. L. Stein, A. J. van Wijnen, and G. S. Stein, submitted for publication).
In summary, we have analyzed the regulatory mechanism(s) by which histone H4 gene transcription is silenced during myeloid differentiation of HL-60 cells. Changes in transcription are mediated by modifications in genomic occupancy of the site II cell cycle element and modulations in the activities and protein levels of the cognate factors.
Acknowledgments
We thank S. K. Zaidi for help in preparing the figures, D. Young for assistance with statistical analysis, and J. Rask for manuscript preparation.
This work was supported by NIH grant GM32010.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1997. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Aziz, F., A. J. van Wijnen, P. S. Vaughan, S. Wu, A. R. Shakoori, J. B. Lian, K. J. Soprano, J. L. Stein, and G. S. Stein. 1998. The integrated activities of IRF-2 (HiNF-M) CDP/cut (HiNF-D) and H4TF-2 (HiNF-P) regulate transcription of a cell cycle controlled human histone H4 gene: mechanistic differences between distinct H4 genes. Mol. Biol. Rep. 25:1-12. [DOI] [PubMed] [Google Scholar]
- 3.Baumbach, L. L., G. S. Stein, and J. L. Stein. 1987. Regulation of human histone gene expression: transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry 26:6178-6187. [DOI] [PubMed] [Google Scholar]
- 4.Birnbaum, M. J., K. L. Wright, A. J. van Wijnen, A. L. Ramsey-Ewing, M. T. Bourke, T. J. Last, F. Aziz, B. Frenkel, B. R. Rao, N. Aronin, G. S. Stein, and J. L. Stein. 1995. Functional role for Sp1 in the transcriptional amplification of a cell cycle regulated histone H4 gene. Biochemistry 34:7648-7658. [DOI] [PubMed] [Google Scholar]
- 5.Bulger, M., and M. Groudine. 1999. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 13:2465-2477. [DOI] [PubMed] [Google Scholar]
- 6.Chrysogelos, S., D. E. Riley, G. Stein, and J. Stein. 1985. A human histone H4 gene exhibits cell cycle-dependent changes in chromatin structure that correlate with its expression. Proc. Natl. Acad. Sci. USA 82:7535-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collart, D. G., K. L. Wright, A. J. van Wijnen, A. L. Ramsey, J. Lian, J. L. Stein, and G. S. Stein. 1988. The human H1 histone gene FNC16 is functionally expressed in proliferating HeLa S3 cells and is down-regulated during terminal differentiation in HL60 cells. J. Biol. Chem. 263:15860-15863. [PubMed] [Google Scholar]
- 8.Forsberg, E. C., and E. H. Bresnick. 2001. Histone acetylation beyond promoters: long-range acetylation patterns in the chromatin world. Bioessays 23:820-830. [DOI] [PubMed] [Google Scholar]
- 9.Francis, N. J., and R. E. Kingston. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2:409-421. [DOI] [PubMed] [Google Scholar]
- 10.Gross, D. S., and W. T. Garrard. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57:159-197. [DOI] [PubMed] [Google Scholar]
- 11.Guo, B., J. L. Stein, A. J. van Wijnen, and G. S. Stein. 1997. ATF1 and CREB trans-activate a cell cycle regulated histone H4 gene at a distal nuclear matrix associated promoter element. Biochemistry 36:14447-14455. [DOI] [PubMed] [Google Scholar]
- 12.Harris, P. E., P. Ralph, J. Gabrilove, K. Welte, R. Karmali, and M. A. Moore. 1985. Distinct differentiation-inducing activities of gamma-interferon and cytokine factors acting on the human promyelocytic leukemia cell line HL-60. Cancer Res. 45:3090-3095. [PubMed] [Google Scholar]
- 13.Hass, R., H. Gunji, R. Datta, S. Kharbanda, A. Hartmann, R. Weichselbaum, and D. Kufe. 1992. Differentiation and retrodifferentiation of human myeloid leukemia cells is associated with reversible induction of cell cycle-regulatory genes. Cancer Res. 52:1445-1450. [PubMed] [Google Scholar]
- 14.Jiang, H., J. Lin, Z. Z. Su, F. R. Collart, E. Huberman, and P. B. Fisher. 1994. Induction of differentiation in human promyelocytic HL-60 leukemia cells activates p21, WAF1/CIP1, expression in the absence of p53. Oncogene 9:3397-3406. [PubMed] [Google Scholar]
- 15.Kingston, R. E., C. A. Bunker, and A. N. Imbalzano. 1996. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 10:905-920. [DOI] [PubMed] [Google Scholar]
- 16.Kontaraki, J., H. H. Chen, A. Riggs, and C. Bonifer. 2000. Chromatin fine structure profiles for a developmentally regulated gene: reorganization of the lysozyme locus before trans-activator binding and gene expression. Genes Dev. 14:2106-2122. [PMC free article] [PubMed] [Google Scholar]
- 17.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroeger, P., C. Stewart, T. Schaap, A. van Wijnen, J. Hirshman, S. Helms, G. Stein, and J. Stein. 1987. Proximal and distal regulatory elements that influence in vivo expression of a cell cycle-dependent human H4 histone gene. Proc. Natl. Acad. Sci. USA 84:3982-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montecino, M., J. Lian, G. Stein, and J. Stein. 1996. Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry 35:5093-5102. [DOI] [PubMed] [Google Scholar]
- 20.Montecino, M., S. Pockwinse, J. Lian, G. Stein, and J. Stein. 1994. DNase I hypersensitive sites in promoter elements associated with basal and vitamin D dependent transcription of the bone-specific osteocalcin gene. Biochemistry 33:348-353. [DOI] [PubMed] [Google Scholar]
- 21.Moreno, M. L., S. A. Chrysogelos, G. S. Stein, and J. L. Stein. 1986. Reversible changes in the nucleosomal organization of a human H4 histone gene during the cell cycle. Biochemistry 25:5364-5370. [DOI] [PubMed] [Google Scholar]
- 22.Mueller, P., P. Garrity, and B. Wold. 1997. Ligation-mediated PCR for genomic sequencing and footprinting, p. 15.5.1-15.5.26. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and J. A. Smith (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 23.O'Neill, L. P., and B. M. Turner. 1995. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 14:3946-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauli, U., S. Chrysogelos, G. Stein, J. Stein, and H. Nick. 1987. Protein-DNA interactions in vivo upstream of a cell cycle-regulated human H4 histone gene. Science 236:1308-1311. [DOI] [PubMed] [Google Scholar]
- 25.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 26.Ramsey-Ewing, A., A. J. van Wijnen, G. S. Stein, and J. L. Stein. 1994. Delineation of a human histone H4 cell cycle element in vivo: the master switch for H4 gene transcription. Proc. Natl. Acad. Sci. USA 91:4475-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice, J. C., and C. D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263-273. [DOI] [PubMed] [Google Scholar]
- 28.Rollini, P., and R. E. Fournier. 1999. Long-range chromatin reorganization of the human serpin gene cluster at 14q32.1 accompanies gene activation and extinction in microcell hybrids. Genomics 56:22-30. [DOI] [PubMed] [Google Scholar]
- 29.Rovera, G., D. Santoli, and C. Damsky. 1979. Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester. Proc. Natl. Acad. Sci. USA 76:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambucetti, L. C., D. D. Fischer, S. Zabludoff, P. O. Kwon, H. Chamberlin, N. Trogani, H. Xu, and D. Cohen. 1999. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J. Biol. Chem. 274:34940-34947. [DOI] [PubMed] [Google Scholar]
- 31.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 32.Spencer, V. A., and J. R. Davie. 1999. Role of covalent modifications of histones in regulating gene expression. Gene 240:1-12. [DOI] [PubMed] [Google Scholar]
- 33.Stein, G., J. Lian, J. Stein, R. Briggs, V. Shalhoub, K. Wright, U. Pauli, and A. van Wijnen. 1989. Altered binding of human histone gene transcription factors during the shutdown of proliferation and onset of differentiation in HL-60 cells. Proc. Natl. Acad. Sci. USA 86:1865-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein, G. S., M. Montecino, A. J. van Wijnen, J. L. Stein, and J. B. Lian. 2000. Nuclear structure-gene expression interrelationships: implications for aberrant gene expression in cancer. Cancer Res. 60:2067-2076. [PubMed] [Google Scholar]
- 35.Stein, G. S., J. L. Stein, A. J. van Wijnen, and J. B. Lian. 1996. Transcriptional control of cell cycle progression: the histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol. Int. 20:41-49. [DOI] [PubMed] [Google Scholar]
- 36.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 37.van den Ent, F. M., A. J. van Wijnen, J. B. Lian, J. L. Stein, and G. S. Stein. 1994. Cell cycle controlled histone H1, H3, and H4 genes share unusual arrangements of recognition motifs for HiNF-D supporting a coordinate promoter binding mechanism. J. Cell. Physiol. 159:515-530. [DOI] [PubMed] [Google Scholar]
- 38.van der Meijden, C. M. J., P. S. Vaughan, A. Staal, W. Albig, D. Doenecke, J. L. Stein, G. S. Stein, and A. J. van Wijnen. 1998. Selective expression of specific histone H4 genes reflects distinctions in transcription factor interactions with divergent H4 promoter elements. Biochim. Biophys. Acta 1442:82-100. [DOI] [PubMed] [Google Scholar]
- 39.van Wijnen, A. J., F. M. van den Ent, J. B. Lian, J. L. Stein, and G. S. Stein. 1992. Overlapping and CpG methylation-sensitive protein-DNA interactions at the histone H4 transcriptional cell cycle domain: distinctions between two human H4 gene promoters. Mol. Cell. Biol. 12:3273-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaughan, P. S., F. Aziz, A. J. van Wijnen, S. Wu, H. Harada, T. Taniguchi, K. J. Soprano, J. L. Stein, and G. S. Stein. 1995. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature 377:362-365. [DOI] [PubMed] [Google Scholar]
- 41.Zeng, Y. X., and W. S. el Deiry. 1996. Regulation of p21WAF1/CIP1 expression by p53-independent pathways. Oncogene 12:1557-1564. [PubMed] [Google Scholar]