Abstract
The proinflammatory cytokine interleukin-1 (IL-1) transmits a signal via several critical cytoplasmic proteins such as MyD88, IRAKs and TRAF6. Recently, serine/threonine kinase TAK1 and TAK1 binding protein 1 and 2 (TAB1/2) have been identified as molecules involved in IL-1-induced TRAF6-mediated activation of AP-1 and NF-κB via mitogen-activated protein (MAP) kinases and IκB kinases, respectively. However, their physiological functions remain to be clarified. To elucidate their roles in vivo, we generated TAB2-deficient mice. The TAB2 deficiency was embryonic lethal due to liver degeneration and apoptosis. This phenotype was similar to that of NF-κB p65-, IKKβ-, and NEMO/IKKγ-deficient mice. However, the IL-1-induced activation of NF-κB and MAP kinases was not impaired in TAB2-deficient embryonic fibroblasts. These findings demonstrate that TAB2 is essential for embryonic development through prevention of liver apoptosis but not for the IL-1 receptor-mediated signaling pathway.
Interleukin-1 (IL-1) is known to have multiple biologic activities, such as comitogenic effects on T cells and the induction of inflammatory response to infection and injury (9). The physiological events induced by IL-1 occur as a result of the activation of various molecules, including adapters, protein kinases, and transcription factors, which are relevant to the signaling cascade. When IL-1 binds to its cognate receptor, the IL-1 receptor type I (IL-1RI) forms a high-affinity complex with the IL-1R accessory protein, and an adapter protein, MyD88, and serine/threonine kinase IRAKs are recruited to the receptor complex (6, 19, 25, 39). IRAKs are activated and then dissociate from the receptor complex and interact with TRAF6, a member of the TRAF family (7). TRAF6 plays a crucial role in the IL-1-induced activation of IκB kinases (IKKs) and mitogen-activated protein kinases (MAPKs), which finally leads to the activation of transcription factors NF-κB and AP-1, respectively (2, 17, 21, 26). However, the mechanism by which TRAF6 relays its activation signal to IKKs and MAPKs remains to be clarified.
A member of the MAPK kinase kinase family, TAK1 was originally identified as a molecule involved in the TGF-β signaling pathway (40). It has recently been reported that TAK1 participates in the IL-1R-mediated signaling pathway (27). In vitro studies indicated that IL-1 activates TAK1, which in turn activates MKK6 and IKK complex, leading to the activation of MAPK and NF-κB, respectively (27). TAK1 has also been shown to associate with TRAF6 in an IL-1-dependent manner, suggesting that TAK1 is a critical regulator in the IL-1 signaling pathway (27). Furthermore, TAK1 binding protein 1 (TAB1) and TAB2 were identified as binding partners of TAK1 by yeast two-hybrid screening (32, 33). TAB1 constitutively binds to TAK1 and phosphorylates TAK1 upon stimulation with IL-1, suggesting that it functions as an activator of TAK1 (27). TAB2 has been shown to function as an adapter molecule linking TRAF6 to TAK1 (33, 34). In unstimulated cells, TAB2 localizes within the membrane fraction. When stimulated with IL-1, TAB2 translocates to the cytosol and binds to TRAF6 and TAK1, thereby regulating the activation of MAPK and NF-κB. In addition, a biochemical fractionation study has identified TRIKA1 and -2 as signaling components that have a capacity to activate the IKK complex and MAPKs in a TRAF6-dependent manner (8, 38). It is also found that TRIKA1 consists of the ubiquitin conjugating enzyme Ubc13 and the Ubc-like protein Uev1A, whereas TRIKA2 is a ternary complex of TAK1, TAB1, and TAB2.
These in vitro studies indicated that TAK1, TAB1, and TAB2 play pivotal roles in the IL-1 signaling pathway. However, their physiological functions remain to be clarified. To elucidate these functions in vivo, we generated TAB2-deficient mice. TAB2-deficient mice died in utero due to fetal liver degeneration. This phenotype is very similar to that of NF-κB p65-, IKKβ-, and NEMO/IKKγ-deficient mice (4, 18, 20, 22, 29, 31, 36). However, TAB2−/− mouse embryonic fibroblasts (MEFs) were resistant to tumor necrosis factor alpha (TNF-α)-induced apoptosis. Furthermore, TAB2−/− MEFs exhibited normal MAPK activity and NF-κB DNA binding activity in response to IL-1. These results indicate that TAB2 plays an essential role in embryonic development but not in the IL-1 or TNF-α signaling.
MATERIALS AND METHODS
Generation of TAB2-deficient mice.
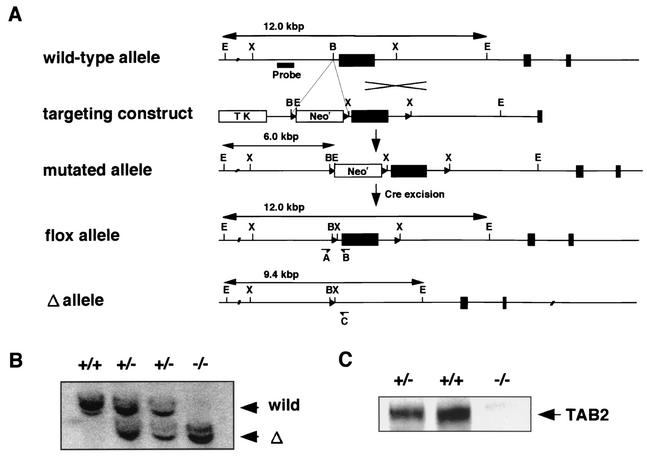
Mouse TAB2 cDNA was cloned by RT-PCR from mouse peritoneal macrophages according to the EST sequence information from the BLAST/FASTA program. Phage clones containing the murine Tab2 gene were isolated by screening a 129/SvJ genomic library (Stratagene) with the probe corresponding to the 5′ end of the mouse TAB2 cDNA. A targeting vector was designed to flank one large exon containing most of the TAB2 coding region with two loxP sites, as shown in Fig. 1. Briefly, the loxP site-flanked Neor gene fragment was inserted into the intron of the TAB2 gene. A 2.0-kb BamHI-XbaI fragment was used as the 5′ homology region; a 2.5-kb XbaI-SalI fragment, which contains most of the coding region of TAB2, was inserted between the two loxP sites; and a 6.0-kb NotI-SacII fragment was used as the 3′ homology region. The thymidine kinase gene was used for negative selection of clones with random integration. A total of 30 μg of SacII-linearized vector was electroporated into E14-1 ES cells. After positive and negative selection with G418 and ganciclovir, drug-resistant clones were picked up and screened by PCR and Southern blot analysis. Two homologous recombinant clones were then transiently transfected with 15 μg of an expression vector encoding Cre recombinase (pIC-Cre) (35) to generate the clones lacking the Neor cassette (TAB2flox/+). The obtained ES clones with the flox allele were confirmed by PCR and Southern blot analysis using a set of primers and an external probe, respectively, as shown in Fig. 1. These clones were individually microinjected into blastocysts derived from C57BL/6 mice and transferred to ICR females. Matings of chimeric male mice to C57BL/6 female mice resulted in transmission of the flox allele to the germ line. TAB2flox/+ and/or TAB2flox/flox mice were bred with CAG-cre deleter mice (30) to generate the TAB2flox/+ CAG-cre (genotype is TAB2+/−) which were then intercrossed to generate TAB2flox/− CAG-cre (genotype is TAB2−/−).
FIG. 1.
Targeted disruption of the TAB2 gene. (A) Schematic representation of the targeting strategy. A targeting construct was designed to flank one large exon of the TAB2 gene and the Neor gene with loxP sites (triangles). Cre-mediated removal was performed to generate a flox allele or Δ allele of TAB2. Abbreviations: B, BamHI; E, EcoRI; X, XbaI. Screening primers for PCR are shown as A, B, and C. (B) Southern blot analysis of EcoRI-digested genomic DNA from E11.5-derived MEFs. The probe used is indicated in panel A. (C) Immunoblot analysis of whole-cell extracts from E11.5-derived MEFs using an Ab that recognizes the N terminus of TAB2.
Expression of TAB2 and histological analysis.
TAB2 expression in mouse adult tissues was examined using mouse multiple tissue Northern blotting (Clontech). For the expression in embryo, fetuses derived at 11.5 days postcoitum were fixed with 4% paraformaldehyde and embedded in paraffin. Sections (5 μm thick) were cut and stained with hematoxylin and eosin for histological analysis. For in situ hybridization, a fragment of murine TAB2 cDNA was subcloned into pBluescript I KS(−) plasmid (Stratagene), and cRNA probes were prepared by using a DIG RNA labeling kit (Roche Diagnostics). Hybridization was performed at 50°C for 16 h, and signals were visualized with a GenPoint kit (Dako) using the horseradish peroxidase-conjugated antidigoxigenin antibody (Ab). The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was performed by using an apoptosis in situ detection kit (Wako Pure Chemical Industries), and the sections were counterstained with methyl green. Sections for immunohistochemistry were incubated with a rabbit polyclonal Ab against human Ki-67 antigen (Novocastra) at 4°C overnight. After being washed, the sections were incubated with biotin-conjugated goat anti-rabbit immunoglobulin G (Vector Laboratories), and immunoreacted cells were then visualized by the streptavidin-biotin peroxidase complex method using a Vectastain ABC Elite kit (Vector Laboratories) and diaminobenzidine tetrahydrochloride. The sections were lightly counterstained with hematoxylin.
TNF cytotoxicity.
MEFs were prepared from embryos derived at 11.5 days postcoitum according to the standard technique. They were plated at 104 cells per well onto 96-well plates 24 h prior to stimulation and treated with given amounts of TNF-α (Genzyme) or TNF-α plus cycloheximide for 24 h. Cell viability was examined using cell counting kit 8 (DOJINDO) according to the manufacturer's instructions.
Measurement of IL-6 production.
MEFs were plated at 104 cells/well onto 48-well plates 24 h prior to stimulation and treated with the stimuli for 16 h. The culture supernatants were harvested, and the concentration of IL-6 was determined using an enzyme-linked immunosorbent assay kit (R & D).
Northern blot analysis.
MEFs were treated with the stimuli for 4 h and RNA was extracted using the TRIzol reagent (Invitrogen). Ten micrograms of total RNA was developed, transferred to a nylon membrane, and hybridized with cDNA probe specific for COX-2, RANTES, IκBα, and GAPDH (glycerladehyde-3-phosphate dehydrogenase).
Gel mobility shift assay.
Preparations of nuclear extracts were performed as described previously (1). Briefly, 106 MEFs were treated with IL-1 (Genzyme), washed with cold phosphate-buffered saline and lysed with 200 μl of buffer A (0.1% NP-40, 10 mM HEPES-KOH [pH 7.8], 10 mM KCl, 0.1 mM EDTA [pH 8.0], 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], and aprotinin [2 μg/ml]). After centrifugation, pellets were lysed with buffer C (50 mM HEPES-KOH [pH 7.8], 420 mM KCl, 0.1 mM EDTA [pH 8.0], 5 mM MgCl2, 2% glycerol, 1 mM dithiothreitol, 0.5 mM PMSF, and aprotinin [2 μg/ml]) and centrifuged again, and the clear supernatants were used as nuclear extracts. A total of 10 μg of nuclear extract was incubated with an end-labeled, double-stranded oligonucleotide containing a consensus NF-κB binding site in 25 μl of binding buffer (10 mM HEPES-KOH [pH 7.8], 50 mM KCl, 1 mM EDTA [pH 8.0], 5 mM MgCl2, and 10% glycerol) for 20 min at room temperature and fractionated on a 5% polyacrylamide gel. The DNA-protein complex was visualized by autoradiography.
Abs, Western blot analysis, and in vitro kinase assay.
The antibodies (Abs) used in this study were obtained as follows: anti-phospho-JNK and p38 Abs were from Cell Signaling; Anti-JNK, p38, and IκBα Abs were from Santa Cruz. Anti-TAK1, TAB1, and TAB2 Abs were used as described previously (16, 27, 33, 34). Anti-IRAK1 Ab was generated from peptides corresponding to the C-terminal sequence of mouse IRAK1. MEFs were plated at 106 cells on 10-cm-diameter dishes 24 h prior to stimulation. Cells were starved in serum-free medium at 37°C for 2 to 3 h and stimulated with IL-1β for given periods. After stimulation, cells were washed with ice-cold phosphate-buffered saline and lysed with 100 μl of lysis buffer (1% NP-40, 20 mM Tris-Cl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, aprotinin [20 μg/ml], 10 mM NaF, 10 mM β-glycerophosphate, and 1 mM Na3VO4). After centrifugation, whole-cell extracts were mixed with 3× sodium dodecyl sulfate (SDS) sample buffer, boiled for 5 min, and resolved by SDS-polyacrylamide gel electrophoresis. Gels were transferred to polyvinylidene difluoride membranes (Amersham Pharmacia) and blocked with phosphate-buffered saline containing 5% skim milk. The membranes were incubated with various Abs, and the membrane-bound Abs were visualized with horseradish peroxidase-conjugated Ab to rabbit immunoglobulin G using enhanced chemiluminescence (DuPont). JNK in vitro kinase assay was performed as described previously (1).
RESULTS
Targeted disruption of the Tab2 gene.
To study the functional importance of TAB2 in vivo, we generated TAB2-deficient mice by gene targeting. The murine Tab2 gene was isolated from 129 mouse genomic library. The targeting vector was constructed so that one large exon, which contained most of the coding region of mouse TAB2, was placed between two loxP sites and the loxP site-flanked Neor gene was introduced into an intron of the Tab2 gene (Fig. 1A) (Materials and Methods). Correctly targeted ES cell clones were further transiently transfected with cre-expressing vector to excise the Neor gene. These ES cell clones were used to generate TAB2flox/+ mice. TAB2flox/+ mice were mated with CAG-cre deleter mice to obtain the heterozygous TAB2Δ/+ mice (= TAB2+/−) (30). The matings between these heterozygous mice were performed to generate TAB2 homozygous mice. We determined the genotype of about 90 newborn pups from these matings, but no TAB2−/− mice were observed, suggesting that the TAB2 deficiency was embryonic lethal (Table 1). TAB2 heterozygous mutant mice appeared normal and viable at birth, but about 70% of them died within a week. The reason why TAB2 heterozygous mice have less viability is not clear.
TABLE 1.
Genotype analysis of offspring from TAB2 heterozygous intercrosses
| Stage | Total no. of mice | No. of mice with genotype
|
No. of mice in resorption | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| Postnatal (∼2 weeks) | 87 | 33 (1)a | 54 (47) | 0 | |
| E11.5 | 78 | 22 | 36 | 15 | 5 |
| E12.5 | 48 | 13 | 22 | 5 (5) | 8 |
| E13.5 | 23 | 5 | 13 | 1 (1) | 4 |
The number of dead mice is indicated in parentheses.
Liver degeneration in TAB2-deficient embryos.
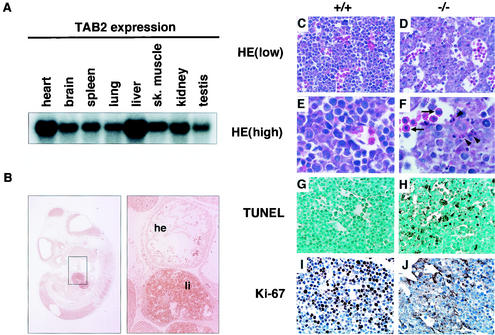
TAB2 deficiency resulted in embryonic lethality, suggesting that TAB2 plays a crucial role in embryonic development. Therefore, we examined TAB2 expression in embryo and adult tissues. In adult tissues, TAB2 mRNA was detected in all organs tested and strongly expressed in the liver and heart (Fig. 2A). We next analyzed TAB2 mRNA expression in the embryo by in situ hybridization. As shown in Fig. 2B, TAB2 mRNA was detected in all embryonic tissues at E11.5, with a strong expression in the liver. Next, to determine the stage at which TAB2 homozygous mutant mice die, timed matings were performed. TAB2−/− embryos were normal and viable at embryonic day 11.5 (E11.5) (Table 1 and data not shown). But at E12.5, severe hemorrhages were seen in the liver of TAB2−/− embryos which had already died (data not shown). Therefore, we closely examined the embryos at E12.0. The gross appearance of TAB2−/− embryos was pale and anemic (data not shown). Histological analysis showed a destructive architecture due to hypocellularity and multifocal hemorrhage in TAB2−/− fetal livers (Fig. 2C to F). In particular, hepatoblast cells, but not hematopoietic cells, contained many pyknotic and fragmented nuclei, indicating that TAB2−/− fetal livers were degenerating (Fig. 2E and F). To investigate whether this morphological abnormality in TAB2−/− fetal livers is due to apoptosis, the TUNEL assay was performed. While wild-type fetal liver contained only a few TUNEL-positive cells, there were increasing numbers of apoptotic cells in TAB2−/− fetal liver (Fig. 2G and H). In addition, on immunohistological staining with Ki-67 Ab, a proliferation marker for cells in S phase, there were significantly reduced numbers of proliferating cells in TAB2−/− fetal liver compared with those observed in wild-type liver (Fig. 2I and J). Histological analysis of other tissues such as brain and heart did not show any change in TAB2−/− embryos (data not shown). Taken together, these results indicated that the major cause of embryonic lethality in TAB2-deficient mice is increased apoptosis of hepatoblasts in the fetal liver.
FIG. 2.
Tissue distribution of TAB2 and liver degeneration of TAB2-deficient embryos. (A) Northern blot analysis of adult mouse tissues. sk., skeletal. (B) In situ hybridization analysis for the expression of TAB2 using E11.5 embryos. The panel at right represents a higher magnification of the boxed area of the panel at left. Note the strong expression of TAB2 in fetal liver. Abbreviations: li, fetal liver; he, heart. (C to F) Hematoxylin and eosin staining of transverse sections through the fetal liver of wild-type (+/+) and TAB2-deficient (−/−) embryos at E12.0. Note the hypocellularity and the abundance of pyknotic and fragmented nuclei (arrowheads) are observed in the −/− liver. Arrows indicate hematopoietic cells. (G and H) TUNEL analysis of sections of fetal liver from both embryos. Note the increasing number of positively stained cells in the −/− liver. (I and J) Staining of sections from fetal liver with an Ab to Ki-67, a proliferation marker for cells in the S phase. Note the much smaller number of proliferating cells in the −/− liver.
TAB2-deficient MEFs are resistant to TNF-induced apoptosis.
Intriguingly, the phenotype of TAB2-deficient embryos is very similar to that of NF-κB subunit p65-, IKKβ- and NEMO/IKKγ-deficient embryos (4, 18, 20, 22, 29, 31, 36). MEFs derived from these knockout mice are highly sensitive to TNF-α-induced apoptosis. To assess whether TAB2-deficient cells are prone to apoptosis, we isolated MEFs from E11.5 wild-type and TAB2−/− embryos and treated them with TNF-α. As shown in Fig. 3A, both wild-type and TAB2−/− MEFs showed resistance to TNF-α-induced apoptosis. We treated the cells with TNF-α in combination with cycloheximide. As was the case in stimulation with TNF-α alone, wild-type and TAB2−/− MEFs were similarly resistant to TNF-α-induced apoptosis. The embryonic lethality and fetal liver degeneration have been shown to be rescued by introduction of TNF-α or type I TNF-receptor deficiency in mice lacking p65 and IKKβ (3, 10, 18). Therefore, we next tried to generate double-deficient mice of TAB2 and TNF-α by intercrossing TAB2/TNF-α double heterozygous mutant mice (Table 2). However, we could not obtain any TAB2−/− TNF-α−/− pup among 66 mice analyzed, suggesting that the embryonic lethality of TAB2-deficient mice was not due to increased TNF-α-mediated apoptosis. Taken together, these results indicate that TAB2 is not critically involved in the TNF-α signaling pathway, unlike p65, IKKβ, and NEMO/IKKγ.
FIG. 3.
Normal response of TAB2-deficient MEFs to inflammatory stimuli. (A) Wild-type and TAB2-deficient MEFs were treated with the indicated concentration of TNF-α in the presence or absence of cycloheximide (CHX) for 24 h. Three independent experiments were performed in triplicate. Values are shown as the mean percentage of viable cells after treatment relative to untreated control (error bars, standard deviations). (B) Wild-type and TAB2-deficient MEFs were treated with medium alone (med), IL-1β (10 ng/ml), TNF-α (10 ng/ml), or LPS (10 μg/ml) for 16 h. After treatment, culture supernatants were harvested and levels of IL-6 were determined by enzyme-linked immunosorbent assay. Five independent experiments were performed in triplicate. Data shown are the means (error bars, standard deviations). (C) MEFs were treated with stimuli as described above for 4 h. Total RNA was extracted and subjected to Northern blot analysis using cDNA probes for COX-2, RANTES, and IκBα. The blot of GAPDH is shown as an internal control. med, medium.
TABLE 2.
Genotype analysis of offspring from TAB2+/−:TNFα+/− intercrosses
| Genotype | No. (%) of mice displaying genotype |
|---|---|
| TAB2+/+:TNFα+/+ | 4 (6.0) |
| TAB2+/+:TNFα+/− | 12 (18.2) |
| TAB2+/+:TNFα−/− | 8 (12.1) |
| TAB2+/−:TNFα+/+ | 13 (19.7) |
| TAB2+/−:TNFα+/− | 19 (28.8) |
| TAB2+/−:TNFα−/− | 10 (15.2) |
| TAB2−/−:TNFα+/+ | 0 |
| TAB2−/−:TNFα+/− | 0 |
| TAB2−/−:TNFα−/− | 0 |
| Total | 66 (100) |
Normal NF-κB and MAPK activities in response to proinflammatory stimuli in TAB2-deficient MEFs.
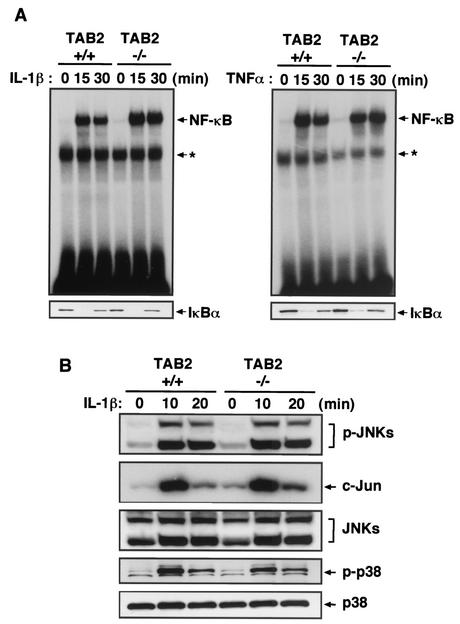
It has been reported that TAB2 is involved in the IL-1R- and Toll-like receptor (TLR)-mediated signaling pathways (33, 34). To assess whether TAB2 plays a critical role in these signaling pathways, we examined IL-6 production in response to IL-1, TNF-α, and lipopolysaccharide (LPS) in wild-type and TAB2−/− MEFs. Wild-type and TAB2−/− MEFs produced almost the same amounts of IL-6 in response to these stimuli (Fig. 3B). Furthermore, when we examined the mRNA expression of COX-2, RANTES, and IκBα, comparable levels of these gene inductions were observed between wild-type and TAB2−/− MEFs (Fig. 3C). Next, we investigated NF-κB DNA binding activity by electrophoretic mobility gel shift assay. Similar binding activities of NF-κB were detected between wild-type and TAB2−/− MEFs in response to IL-1 stimulation (Fig. 4A, top of left panel). IL-1-induced degradation of IκBα was not impaired in TAB2−/− MEFs (Fig. 4A, bottom of left panel). When stimulated with TNF-α and LPS, the activation of NF-κB and IκBα degradation occurred equally in wild-type and TAB2−/− MEFs (Fig. 4A, right panel and data not shown). We next examined activation of MAPKs in response to stimulation with IL-1. Both cells showed comparable levels of IL-1-induced phosphorylation of JNK, c-Jun, and p38 (Fig. 4B). Phosphorylation of JNK, c-Jun, and p38 in response to TNF-α and LPS was also normally induced in TAB2−/− MEFs (data not shown). Collectively, these results indicated that TAB2 does not play an essential role in the signaling pathway of IL-1, TNF-α, and LPS in MEFs.
FIG. 4.
Normal inflammatory cytokine-induced NF-κB and MAPK activity in TAB2-deficient MEFs. (A) Wild-type and TAB2-deficient MEFs were treated with IL-1β (10 ng/ml) or TNF-α (10 ng/ml) for the indicated times. NF-κB DNA binding activity was determined by electrophoretic mobility gel shift assay. Asterisks show nonspecific bands. Cytokine-induced degradation of IκBα was detected by immunoblotting with the specific Ab. (B) Phosphorylation of JNK and p38 (p-JNKs and p-p38) was detected by immunoblotting with specific Abs. Activation of c-Jun was detected by JNK in vitro kinase assay. The blots of JNKs and p38 are shown as a loading control.
Differential kinetics of phosphorylation of TAK1 and TAB1 in TAB2-deficient MEFs.
Previous reports showed that TAK1, TAB1, and TAB2 are phosphorylated after IL-1 stimulation, as detected by the delayed migration of the corresponding bands in an immunoblotting assay (16, 27, 33, 34). Therefore, we examined whether TAK1 and TAB1 are phosphorylated in response to IL-1 in TAB2−/− MEFs. IL-1-treated wild-type MEFs showed delayed migration patterns of TAK1 and TAB1. However, the band shifts of TAK1 and TAB1 following stimulation with IL-1 were significantly inhibited in TAB2−/− MEFs (Fig. 5). On the other hand, IL-1-induced phosphorylation and degradation of IRAK1 was similarly observed in wild-type and TAB2−/− MEFs (Fig. 5 and data not shown). Taken together, these results suggest that TAB2 acts downstream of IRAKs and plays an important role in the IL-1-induced activation of TAK1 and TAB1.
FIG. 5.
Differential kinetics of phosphorylations of TAK1 and TAB1 in TAB2-deficient MEFs. Wild-type and TAB2-deficient MEFs were treated with IL-1β (10 ng/ml) for the indicated times. TAK1, TAB1, TAB2, and IRAK1 were detected by immunoblotting with specific Abs. Note that the band shifts of TAK1 and TAB1 in response to IL-1 are significantly reduced in TAB2−/− MEFs. Asterisk, nonspecific bands.
DISCUSSION
In this study, we have reported the generation and analysis of TAB2-deficient mice. We demonstrated that TAB2 plays a crucial role in the embryonic development. TAB2 was originally identified as one of the TAK1 binding partners and an adapter protein which links TRAF6 to TAK1 in an IL-1-dependent manner. However, unexpectedly, TAB2 deficiency had no effect on IL-1-induced activation of NF-κB and/or MAPKs in MEFs. This result indicates that TAB2 does not play an essential role in the IL-1 signaling pathway.
TAB2 deficiency resulted in embryonic lethality, which is mainly due to severe liver degeneration during midgestation. As the expression of TAB2 mRNA was easily detected in the fetal liver and TAB2−/− fetal liver contained many apoptotic cells, TAB2 may play a critical role in the prevention of apoptosis in the fetal liver. The increased apoptosis of TAB2−/− fetal liver cells reminded us of NF-κB p65-, IKKβ-, and NEMO/IKKγ-deficient mice, all of which showed fetal liver apoptosis (4, 18, 20, 22, 29, 31, 36). It has been shown that these knockout-mouse-derived MEFs exhibited apoptosis with TNF-α treatment, and furthermore, the double mutant mice of p65/TNFRI, p65/TNF-α, and IKKβ/TNFRI could be rescued from fetal liver degeneration (3, 10, 18). These findings indicate that these molecules play a pivotal role in TNF receptor I (TNFRI)-mediated signaling and the prevention of TNF-α-induced apoptosis. In addition to these knockout mice, it has been reported that T2K-, GSK-3β-, Raf-1-, SEK1/MKK4-, and c-Jun-deficient embryos showed a similar liver degeneration during embryonic development (5, 11, 12, 13, 14, 15, 23, 28). While the critical involvement of NF-κB p65, IKKβ, and NEMO/IKKγ in the TNFR-mediated signaling pathway has clearly been shown, it remains to be elucidated whether T2K, GSK-3β, Raf-1, SEK1/MKK4, and c-Jun are important for the signaling via the TNFR. For example, whereas GSK-3β-deficient MEFs undergo apoptosis in response to TNF-α, T2K-deficient MEFs do not. On the other hand, liver degeneration of GSK-3β- and T2K-deficient mice can be circumvented by the administration of anti-TNF-α Ab or the generation of double-mutant mice with TNFR. Moreover it has been shown that TNF-α-induced NF-κB-mediated transcriptional activity was reduced in GSK-3β- and T2K-deficient MEFs. These findings suggest that these molecules are involved in TNF-α signaling by unknown mechanisms. In the case of TAB2, it does not appear to be involved in the TNFR-mediated signaling pathway, because TAB2-deficient MEFs showed a normal response to TNF-α in terms of protection against apoptosis, IL-6 production, and NF-κB DNA binding activity. In addition, introduction of TNF-α deficiency did not rescue the embryonic lethality of TAB2-deficient mice, unlike the case of mice deficient in p65 or IKKβ. In this regard, it is noteworthy that Raf-1-, SEK1/MKK4-, and c-Jun-deficient embryos died at almost the same stage as TAB2-deficient embryos. Therefore, an unknown factor(s) and its receptor(s), which play an essential role in fetal liver maturation and homeostasis, might use these molecules, including TAB2, to transmit the survival signal by an independent mechanism of the TNFR-mediated NF-κB activation.
The present study further indicates that TAB2 is not essential for IL-1 signaling in MEFs. TAB2−/− MEFs showed a normal IL-1-induced activation of NF-κB and MAPKs as well as expression of IL-6, COX-2, RANTES, and IκBα. Thus, biological activity induced by IL-1 was not affected in TAB2−/− MEFs. On the other hand, the IL-1-induced phosphorylation of TAK1 and TAB1, as shown by the delayed migration on SDS-polyacrylamide gel electrophoresis, was severely impaired in TAB2−/− MEFs. Previous reports demonstrated that the phosphorylation and activation of TAK1 and TAB1 were severely impaired in TRAF6-deficient MEFs as well as MyD88-deficient MEFs (unpublished data). These cells, unlike TAB2−/− MEFs, showed a complete loss of the IL-1-induced activation of NF-κB and MAPKs (1, 17). Although we have no evidence to clearly explain the differential phenotypes between these MEFs, there might be two possibilities. One possibility is that in the absence of TAB2, TAK1 kinase activity in response to IL-1 is completely abolished, but another MAPK kinase kinase(s) transmits the activation signal to the downstream molecules, such as JNK, p38, and/or IKK complex. Another is that the ability of TAK1 to relay the activation signal upon IL-1 stimulation remains intact or is only partially impaired in TAB2−/− MEFs. To address these points, it would be better to measure the TAK1 kinase activity in an in vitro kinase assay using MKK6 as an exogenous substrate, but we failed to do it even in wild-type MEFs. It has very recently been reported that a novel TAK1-binding protein, termed TAB3, which is closely related to TAB2 in structure, was identified in mouse, human, and Xenopus laevis (24). Therefore, TAB3 may compensate for the loss of TAB2 and support the TAK1 kinase activity to transmit the activation signal to the downstream molecules in the IL-1 signaling pathway. In addition, Vidal et al. have recently reported that dTAK1, a Drosophila melanogaster homologue of TAK1, controls activity of the Rel/NF-κB like transactivator Relish in the Imd pathway (37). Taken together, these findings make us suppose that TAK1 is associated with the IL-1R/TLR-mediated signaling pathway. Generation and analysis of TAK1-deficient mice will reveal whether mammalian TAK1 is involved in the IL-1R/TLR-mediated activation of NF-κB and MAPKs.
In conclusion, we have shown that TAB2 is required for the prevention of fetal liver apoptosis but is not essential for IL-1R-mediated signaling pathway. Our findings suggest that TAB2 participates in an unknown signaling pathway(s) other than IL-1 signaling. As TAB2 deficiency resulted in the embryonic lethality, we failed to investigate the role of TAB2 in adult animal. Conditional elimination of TAB2 will allow us to disclose unknown functions of TAB2 in the future.
Acknowledgments
We thank N. Okita, N. Iwami, and M. Kakihana for excellent technical assistance; E. Horita for secretarial assistance; and members of the Akira laboratory for their support. We also thank J. Miyazaki for CAG-cre Tg mice.
This work was supported in part by grants from Special Coordination Funds for Promoting Science and Technology; the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Japan Society for the Promotion of Science for Young Scientists; and the Virtual Research Institute of Aging of Nippon Boehringer Ingelheim.
REFERENCES
- 1.Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143-150. [DOI] [PubMed] [Google Scholar]
- 2.Baud, V., Z. G. Liu, B. Bennett, N. Suzuki, Y. Xia, and M. Karin. 1999. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 13:1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beg, A. A., and D. Baltimore. 1996. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science 274:782-784. [DOI] [PubMed] [Google Scholar]
- 4.Beg, A. A., W. C. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature 376:167-170. [DOI] [PubMed] [Google Scholar]
- 5.Bonnard, M., C. Mirtsos, S. Suzuki, K. Graham, J. Huang, M. Ng, A. Itie, A. Wakeham, A. Shahinian, W. J. Henzel, A. J. Elia, W. Shillinglaw, T. W. Mak, Z. Cao, and W. C. Yeh. 2000. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-κB-dependent gene transcription. EMBO J. 19:4976-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, Z., W. J. Henzel, and X. Gao. 1996. IRAK: a kinase associated with the interleukin-1 receptor. Science 271:1128-1131. [DOI] [PubMed] [Google Scholar]
- 7.Cao, Z., J. Xiong, M. Takeuchi, T. Kurama, and D. V. Goeddel. 1996. TRAF6 is a signal transducer for interleukin-1. Nature 383:443-446. [DOI] [PubMed] [Google Scholar]
- 8.Deng, L., C. Wang, E. Spencer, L. Yang, A. Braun, J. You, C. Slaughter, C. Pickart, and Z. J. Chen. 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351-361. [DOI] [PubMed] [Google Scholar]
- 9.Dinarello, C. A. 1996. Biologic basis for interleukin-1 in disease. Blood 87:2095-2147. [PubMed] [Google Scholar]
- 10.Doi, T. S., M. W. Marino, T. Takahashi, T. Yoshida, T. Sakakura, L. J. Old, and Y. Obata. 1999. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc. Natl. Acad. Sci. USA 96:2994-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eferl, R., M. Sibilia, F. Hilberg, A. Fuchsbichler, I. Kufferath, B. Guertl, R. Zenz, E. F. Wagner, and K. Zatloukal. 1999. Functions of c-Jun in liver and heart development. J. Cell Biol. 145:1049-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganiatsas, S., L. Kwee, Y. Fujiwara, A. Perkins, T. Ikeda, M. A. Labow, and L. I. Zon. 1998. SEK1 deficiency reveals mitogen-activated protein kinase cascade crossregulation and leads to abnormal hepatogenesis. Proc. Natl. Acad. Sci. USA 95:6881-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilberg, F., A. Aguzzi, N. Howells, and E. F. Wagner. 1993. c-jun is essential for normal mouse development and hepatogenesis. Nature 365:179-181. [DOI] [PubMed] [Google Scholar]
- 14.Hoeflich, K. P., J. Luo, E. A. Rubie, M. S. Tsao, O. Jin, and J. R. Woodgett. 2000. Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature 406:86-90. [DOI] [PubMed] [Google Scholar]
- 15.Huser, M., J. Luckett, A. Chiloeches, K. Mercer, M. Iwobi, S. Giblett, X. M. Sun, J. Brown, R. Marais, and C. Pritchard. 2001. MEK kinase activity is not necessary for Raf-1 function. EMBO J. 20:1940-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kishimoto, K., K. Matsumoto, and J. Ninomiya-Tsuji. 2000. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J. Biol. Chem. 275:7359-7364. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, N., Y. Kadono, A. Naito, K. Matsumoto, T. Yamamoto, S. Tanaka, and J. Inoue. 2001. Segregation of TRAF6-mediated signaling pathways clarifies its role in osteoclastogenesis. EMBO J. 20:1271-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Q., D. Van Antwerp, F. Mercurio, K. F. Lee, and I. M. Verma. 1999. Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science 284:321-325. [DOI] [PubMed] [Google Scholar]
- 19.Li, S., A. Strelow, E. J. Fontana, and H. Wesche. 2002. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc. Natl. Acad. Sci. USA 99:5567-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Z. W., W. Chu, Y. Hu, M. Delhase, T. Deerinck, M. Ellisman, R. Johnson, and M. Karin. 1999. The IKKβ subunit of IκB kinase (IKK) is essential for nuclear factor κB activation and prevention of apoptosis. J. Exp. Med. 189:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lomaga, M. A., W. C. Yeh, I. Sarosi, G. S. Duncan, C. Furlonger, A. Ho, S. Morony, C. Capparelli, G. Van, S. Kaufman, A. van der Heiden, A. Itie, A. Wakeham, W. Khoo, T. Sasaki, Z. Cao, J. M. Penninger, C. J. Paige, D. L. Lacey, C. R. Dunstan, W. J. Boyle, D. V. Goeddel, and T. W. Mak. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makris, C., V. L. Godfrey, G. Krahn-Senftleben, T. Takahashi, J. L. Roberts, T. Schwarz, L. Feng, R. S. Johnson, and M. Karin. 2000. Female mice heterozygous for IKK gamma/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol. Cell 5:969-979. [DOI] [PubMed] [Google Scholar]
- 23.Mikula, M., M. Schreiber, Z. Husak, L. Kucerova, J. Ruth, R. Wieser, K. Zatloukal, H. Beug, E. F. Wagner, and M. Baccarini. 2001. Embryonic lethality and fetal liver apoptosis in mice lacking the c-raf-1 gene. EMBO J. 20:1952-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz-Sanjuan, I., E. Bell, C. R. Altmann, A. Vonica, and A. H. Brivanlou. 2002. Gene profiling during neural induction in Xenopus laevis: regulation of BMP signaling by post-transcriptional mechanisms and TAB3, a novel TAK1-binding protein. Development 129:5529-5540. [DOI] [PubMed] [Google Scholar]
- 25.Muzio, M., J. Ni, P. Feng, and V. M. Dixit. 1997. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science 278:1612-1615. [DOI] [PubMed] [Google Scholar]
- 26.Naito, A., S. Azuma, S. Tanaka, T. Miyazaki, S. Takaki, K. Takatsu, K. Nakao, K. Nakamura, M. Katsuki, T. Yamamoto, and J. Inoue. 1999. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells 4:353-362. [DOI] [PubMed] [Google Scholar]
- 27.Ninomiya-Tsuji, J., K. Kishimoto, A. Hiyama, J. Inoue, Z. Cao, and K. Matsumoto. 1999. The kinase TAK1 can activate the NIK-I κB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398:252-256. [DOI] [PubMed] [Google Scholar]
- 28.Nishina, H., C. Vaz, P. Billia, M. Nghiem, T. Sasaki, J. L. De la Pompa, K. Furlonger, C. Paige, C. Hui, K. D. Fischer, H. Kishimoto, T. Iwatsubo, T. Katada, J. R. Woodgett, and J. M. Penninger. 1999. Defective liver formation and liver cell apoptosis in mice lacking the stress signaling kinase SEK1/MKK4. Development 126:505-516. [DOI] [PubMed] [Google Scholar]
- 29.Rudolph, D., W. C. Yeh, A. Wakeham, B. Rudolph, D. Nallainathan, J. Potter, A. J. Elia, and T. W. Mak. 2000. Severe liver degeneration and lack of NF-κB activation in NEMO/IKKγ-deficient mice. Genes Dev. 14:854-862. [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai, K., and J. Miyazaki. 1997. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem. Biophys. Res. Commun. 237:318-324. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt-Supprian, M., W. Bloch, G. Courtois, K. Addicks, A. Israel, K. Rajewsky, and M. Pasparakis. 2000. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol. Cell 5:981-992. [DOI] [PubMed] [Google Scholar]
- 32.Shibuya, H., K. Yamaguchi, K. Shirakabe, A. Tonegawa, Y. Gotoh, N. Ueno, K. Irie, E. Nishida, and K. Matsumoto. 1996. TAB1: an activator of the TAK1 MAPKKK in TGF-beta signal transduction. Science 272:1179-1182. [DOI] [PubMed] [Google Scholar]
- 33.Takaesu, G., S. Kishida, A. Hiyama, K. Yamaguchi, H. Shibuya, K. Irie, J. Ninomiya-Tsuji, and K. Matsumoto. 2000. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 5:649-658. [DOI] [PubMed] [Google Scholar]
- 34.Takaesu, G., J. Ninomiya-Tsuji, S. Kishida, X. Li, G. R. Stark, and K. Matsumoto. 2001. Interleukin-1 (IL-1) receptor-associated kinase leads to activation of TAK1 by inducing TAB2 translocation in the IL-1 signaling pathway. Mol. Cell. Biol. 21:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeda, K., T. Kaisho, N. Yoshida, J. Takeda, T. Kishimoto, and S. Akira. 1998. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 161:4652-4660. [PubMed] [Google Scholar]
- 36.Tanaka, M., M. E. Fuentes, K. Yamaguchi, M. H. Durnin, S. A. Dalrymple, K. L. Hardy, and D. V. Goeddel. 1999. Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity 10:421-429. [DOI] [PubMed] [Google Scholar]
- 37.Vidal, S., R. S. Khush, F. Leulier, P. Tzou, M. Nakamura, and B. Lemaitre. 2001. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-κB-dependent innate immune responses. Genes Dev. 15:1900-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, C., L. Deng, M. Hong, G. R. Akkaraju, J. Inoue, and Z. J. Chen. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412:346-351. [DOI] [PubMed] [Google Scholar]
- 39.Wesche, H., W. J. Henzel, W. Shillinglaw, S. Li, and Z. Cao. 1997. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity 7:837-847. [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi, K., K. Shirakabe, H. Shibuya, K. Irie, I. Oishi, N. Ueno, T. Taniguchi, E. Nishida, and K. Matsumoto. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-beta signal transduction. Science 270:2008-2011. [DOI] [PubMed] [Google Scholar]