Abstract
The mating-specific Gα protein of Saccharomyces cerevisiae, Gpa1, stimulates adaptation to pheromone by a mechanism independent of Gβγ sequestration. Genetic evidence suggests that Gpa1 targets the Fus3 mitogen-activated protein kinase, and it has recently been shown that the two proteins interact in cells responding to pheromone. To test the possibility that Gpa1 downregulates the mating signal by affecting the localization of Fus3, we created a Fus3-green fluorescent protein (GFP) fusion protein. In vegetative cells, Fus3-GFP was found in both the cytoplasm and the nucleus. Pheromone stimulated a measurable increase in the ratio of nuclear to cytoplasmic Fus3-GFP. In contrast, the relative level of nuclear Fus3-GFP decreased as cells recovered from pheromone arrest and did not increase when cells adapted to chronic stimulus were challenged again. Accumulation of Fus3-GFP in the nuclei of stimulated cells was also inhibited by overexpression of either wild-type Gpa1, the E364K hyperadaptive mutant form of Gpa1, or the Msg5 dually specific phosphatase. The effects of Gpa1 and Msg5 on Fus3 are partially interdependent. In a genetic screen for adaptive defective mutants, a nonsense allele of the nucleocytoplasmic transport receptor, Kap104, was identified. Truncation of the Kap104 cargo-binding domain blocked the effect of both Gpa1E364K and Msg5 on Fus3-GFP localization. Based on these results, we propose that Gpa1 and Msg5 work in concert to downregulate the mating signal and that they do so by inhibiting the pheromone-induced increase of Fus3 in the nucleus. Kap104 is required for the Gα/phosphatase-mediated effect on Fus3 localization.
When haploid yeast cells of opposite mating type are mixed, they undergo a complex mating reaction, leading to the formation of zygotes. Each mating type constitutively secretes a peptide mating pheromone that triggers cells of the opposite type to arrest in the G1 phase of the cell cycle, form projections, and induce mating-specific genes in preparation for cell and nuclear fusion. The signal to mate is transmitted across the plasma membrane by a receptor-coupled heterotrimeric G protein. Upon release from Gα (Gpa1), the Gβγ dimer (Ste4/Ste18) binds to the Ste5 scaffolding protein and stimulates a Pak kinase (Ste20). These proteins, in turn, are required for activation of the mitogen-activated protein kinase (MAPK) cascade (Ste7, Ste11, and Fus3) that ultimately communicates with regulators of transcription and cell division in the nucleus (reviewed in references 12 and 22).
In addition to the proteins that transmit the mating signal, a number of gene products have been implicated in negative regulation of the pheromone response. These include cell-type-specific proteases that degrade the pheromones (39, 40), elements that upregulate G1 cyclins (34) and elements that downregulate the pheromone receptors (29, 52), Gα (10, 11), Gβγ (27, 48, 59), the Fus3 MAPK (13, 68), and Cdc42 (60). Indirect effects on the mating pathway by global regulators of gene expression (e.g., Cdc36, Cdc39, Srm1, Mot2, and Mot3) have also been detected (5, 8, 21, 23, 31, 38, 47). These various negative regulators may serve distinct purposes. Some may ensure that the mating signal, which is propagated by constitutively expressed proteins, is not induced inappropriately. Others might provide a link to other signaling pathways. Still others may be involved in turning off the G1 arrest signal and promoting transition to S phase when the cell is chronically stimulated or immediately after zygote formation.
We are studying adaptive mechanisms that are stimulated by the pheromone-responsive Gα protein, Gpa1 (36, 62, 69). Recently, we have shown that Gpa1 binds directly to the mating-specific MAPK, Fus3 (44). Disruption of this interaction by mutation confers increased sensitivity to pheromone and a defect in Gpa1-mediated adaptation. These findings led us to ask whether Gpa1 regulates cell sensitivity to pheromone by affecting the localization of Fus3. Because MAPK signaling modules connect signaling elements located at the plasma membrane to response effectors in the nucleus, it has been presumed that, upon activation, MAPKs must be transported from the cytoplasm to the nucleus. Although some MAPKs clearly undergo signal-induced nuclear translocation (reviewed in references 26 and 50), others constitutively shuttle between the cytoplasm and nucleus and show less obvious nuclear concentration when activated (65). Hog1, the osmoregulatory MAPK of yeast, exemplifies the first class. When cells are subjected to osmotic shock, Hog1 exhibits a dramatic shift from the cytoplasm to the nucleus that is dependent on the Ran GTPase, Gsp1, and the Nmd5 importin. Conversely, Hog1 is actively exported from the nucleus by a mechanism requiring Xpo1/Crm1 as the cells adapt to the environmental change. The translocation of Hog1 is regulated by its phosphorylation state: whereas Hog1 is released from its upstream activator (Pbs2) and moved to the nucleus when phosphorylated (15, 42), the Ptp2 and Ptp3 phosphatases dephosphorylate Hog1 during adaptation to osmotic stress (25, 51). Ptp2 and Ptp3 are also thought to affect the localization of Hog1 by tethering it on opposite sides of the nuclear membrane (42). In contrast to Hog1, Fus3 constitutively shuttles between the cytoplasm and nuclei of vegetative cells and is easily detectable in both compartments. Surprisingly, there is only a slight increase in the nuclear localization of Fus3 in response to pheromone stimulation (6, 65), and there is no apparent change in the rates of Fus3 import or export (65). Here we quantify the signal-induced increase in nuclear Fus3 and show that the relative abundance of Fus3 in the nucleus and cytoplasm is correlated with sensitivity to pheromone and with the duration of the mating response. In addition, we present evidence that Gpa1 and the Msg5 phosphatase negatively regulate the mating signal by antagonizing the accumulation of Fus3 in the nucleus and that this phenomenon is dependent on the yeast homologue of transportin, Kap104.
MATERIALS AND METHODS
Molecular and microbiological techniques.
Recombinant DNA techniques were essentially as described by Sambrook et al. (54) and by Ausubel et al. (4). Dideoxy sequence reactions were performed by using the Sequenase version 2.0 kit (U.S. Biochemicals, Cleveland, Ohio).
Yeast transformations were carried out according to the method of Ito et al. (24). Yeast growth media were prepared as described by Sherman et al. (55). Amino acids were omitted as necessary to select for plasmids. For experiments requiring the induction of the GAL1 regulated genes, either cells were grown to mid-log phase in sucrose medium and galactose was added to a final concentration of 2% or the cells were cultured overnight in medium containing galactose.
Immunoblots were performed as previously described (35). Strip-purified antiserum raised against Gpa1 (62) and crude antiserum raised against Kap104 (generously provided by John Aitchison) were used as the primary antibodies.
Plasmids and strains.
All strains used in the present study were derived from strain 15Dau (MATa bar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ), which is isogenic with strain BF264-15D (49), with the exception of the gpa1 temperature-sensitive strain. The temperature-sensitive allele of GPA1 was isolated in strain 381G, and the mutant was backcrossed twice to strain 15D. The strains are listed in Table 1. The parent strain for the adaptive-defective (adaptive−) mutant hunt, DSY278, was constructed by integrating the GAL1EG28-GPA1E364K and FUS1-LEU2 transcriptional fusions at the TRP1 and HIS2 loci of 15Dau, respectively (36). Strain DSY278 was shown to contain a single copy of GAL1EG28-GPA1E364K by southern blot analysis. The A11 mutant strain was backcrossed twice before analysis. Deletion strains were constructed by transformation with disruption plasmids and PCR products containing the yeast markers URA3 and LEU2 and the bacterial markers HISG and KanMx (Table 2). Disruptions were confirmed by genomic PCR analysis and by pheromone response assays. Plasmid DSB174 (YIplac204/GAL1EG28-GPA1E364K) (62) was integrated into the TRP1 locus of the msg5Δ, ptp3Δ, and msg5Δ ptp3Δ strains to create strains TBY147, TBY141, and TBY137, respectively. Plasmid TBB105 (YIplac204/GAL1EG28-GPA1K21E R22E E364K) was integrated into the TRP1 locus of wild-type cells (strain 15Dau) to create strain TBY155. In all cases, transformants resulting from single-copy integration were identified by Southern blot. To construct strain TBY163, the msg5Δ ptp3Δ strain (HKY161) was switched to MATα by induction of a GAL1-HO plasmid and crossed to strain DSY253 (62), which contains two copies of GAL1EG28-GPA1 integrated at the LEU2 locus. After sporulation of the resulting diploid, a MATa msg5Δ ptp3Δ GAL1EG28-GPA1 segregant was identified by tetrad analysis.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| 15Dau | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ | 49 |
| DSY245 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ YIplac204/GAL1EG28-GPA1E364K | Lab stock |
| DSY253 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ YIplac128/GAL1EG28-GPA1 (2X) | Lab stock |
| DSY278 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ YIplac204/GAL1EG28-GPA1E364K FUS1-LEU2::HIS2 | 36 |
| A11 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ kap104Δ827-918 YIplac204/GAL1EG28-GPA1E364K FUS1-LEU2::HIS2 | This study |
| ZWY432 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ kap104Δ::URA3 YCplac111/kap104Δ827-918 | This study |
| IHY169 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ msg5Δ::LEU2 YIplac204/GAL1EG28-GPA1E364K FUS1-LEU2::HIS2 | This study |
| HKY137 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ msg5Δ::LEU2 | This study |
| HKY114 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ ptp3Δ::KanMx | This study |
| HKY161 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ msg5Δ::LEU2 ptp3Δ::KanMx | This study |
| IHY165 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ fus3Δ::LEU2 kss1Δ::KanMx | This study |
| IHY205 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ fus3Δ::LEU2 kss1Δ::KanMx YCplac33/FUS3K42R-GFP | This study |
| IHY217 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ fus3Δ::LEU2 kss1Δ::KanMx YIplac204/GAL1EG28-GPA1E364K YCplac33/FUS3K42R-GFP | This study |
| AEY144 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ kap104Δ::HISG YCplac111/kap104Δ827-918 | This study |
| AOY135 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ kap104Δ::HISG YIplac204/GAL1EG28-GPA1E364K YCplac111/kap104Δ827-918 | This study |
| TBY137 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ msg5Δ::LEU2 ptp3Δ::KanMx YIplac204/GAL1EG28-GPA1E364K | This study |
| TBY141 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ ptp3Δ::KanMx YIplac204/GAL1EG28-GPA1E364K | This study |
| TBY147 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ msg5Δ::LEU2 YIplac204/GAL1EG28-GPA1E364K | This study |
| TBY155 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ YIplac204/GAL1EG28-GPA1K21E R22E E364K | This study |
| TBY163 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ msg5Δ::LEU2 ptp3Δ::KanMx YIplac128/GAL1EG28-GPA1 (2×) | This study |
| MMY107 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ gpa1Δ::URA3 YIplac128/GAL1EG43-GPA1 YCplac22/GPA1K21E R22E | 44 |
| DSY419 | MATabar1Δ ade1 his2 leu2-3,112 trp1 ura3Δ gpa1ts Fus1-lacZ::URA3 | Lab stock |
TABLE 2.
Plasmids used in this study
| Plasmid | Marker | Source |
|---|---|---|
| pEL4-1 | URA3 CEN KAP104 | This study |
| kap104Δ827-918/YCplac111 | LEU2 CEN kap104Δ827-918 | This study |
| KAP104/YIplac211 | URA3 integrative KAP104 | This study |
| kap104Δ::URA3/pUC19 | URA3 integrative kap104Δ | This study |
| kap104Δ::hisG-URA3-hisG/pUC19 | URA3 integrative kap104Δ | This study |
| pYES/KAP104 | URA3 2μ GAL1-KAP104 | This study |
| pIH4.1 | URA3 CEN FUS3-GFP | This study |
| pIH4.1AA | URA3 CEN FUS3T180A Y182A-GFP | This study |
| pIH4.1KM1-1 | URA3 CEN FUS3K42R-GFP | This study |
| YCplac22/FUS3-GFP (AOB117) | TRP1 CEN FUS3-GFP | This study |
| pRS314Nab2-GFP | URA3 CEN NAB2-GFP | 14 |
| pRS316CG/MSG5-GFP (HKB101) | URA3 CEN MSG5-GFP | This study |
| YCplac111/GAL1-MSG5 (HKB100) | LEU2 CEN GAL1-MSG5 | This study |
| YCplac22/GAL1-MSG5 (AEB101) | TRP1 CEN GAL1-MSG5 | This study |
| YCplac111/Gpa1E364K (DSB117) | LEU2 CEN GPA1E364K | 61 |
| YCplac33/Gpa1E364K (HSB105) | URA3 CEN GPA1E364K | 36 |
| YCplac22/GPA1K21E R22E | TRP1 CEN GPA1K21E R22E | 44 |
| YIplac204/GAL1EG28-GPA1E364K | TRP1 GPA1E364K | 62 |
| YIplac204/GAL1EG28-GPA1K21E R22E E364K (TBB105) | TRP1 GPA1K21E R22E E364K | This study |
| YIplac128/GAL1EG28-GPA1 | LEU2 GPA1 | 62 |
| YCplac111/GAL1-PTP3 (TBB104) | LEU2 CEN GAL1-PTP3 | This study |
The plasmids used in the present study are listed in Table 2. The key details of newly constructed plasmids are described below.
(i) pEL4-1.
Plasmid pEL4-1 was recovered in the screen for complementation of the adaptive− phenotypes of strain A11. This plasmid contains the 2.75-kb KAP104 coding sequence on a 4.8-kb genomic insert.
(ii) KAP104/YIplac211.
For integrative mapping, the SstI-SphI fragment containing the coding region of KAP104 and 2.1 kb of flanking sequence was subcloned from pEL4-1 to YIplac211, an integrative URA3 vector (18).
(iii) kap104Δ::URA3/pUC19.
The SstI-SphI fragment containing the coding region of KAP104 and 2.1 kb of flanking sequence was subcloned from pEL4-1 to pUC19 to create KAP104/pUC19. The KAP104 disruption plasmid, kap104Δ::URA3/pUC19, was then generated by replacing the 1.7-kb BglII-EcoRV fragment internal to KAP104 on pUC19/KAP104 with a 1.1-kb fragment containing the URA3 gene.
(iv) kap104Δ::hisG-URA3-hisG/pUC19.
Plasmid pNKY51 (2) was digested with BamHI, and the ends were filled by reaction with Klenow. The 3.8-kb fragment containing hisG-URA3-hisG was then released by digestion with BglII and used to replace the 1.7-kb EcoRV-BglII fragment internal to the KAP104 coding region on plasmid KAP104/pUC19.
(v) pYES/KAP104.
The KAP104 coding region (from −9 to 3062 where the ATG is at position 1) was amplified by PCR with the following oligonucleotides: 5′-GCG GAT CCT GAA TAA AGA TGG CAT CGA C-3′ (which contains a BamHI site) and 5′-GGG CAT GCG CAA AAG TAT TCA AGA ATG-3′ (which contains an SphI site). Two independently generated PCR products were subcloned into pYES2.0 (Invitrogen, San Diego, Calif.) at the BamHI and SphI sites, thereby creating transcriptional fusions between KAP104 and the GAL1 promoter on the 2μ-based pYES vector.
(vi) kap104Δ827-918/YCplac111.
The C-terminal truncation allele of KAP104 was created by PCR mutagenesis by using KAP104/pUC19 as a template and the following oligonucleotides as primers: 5′-TCA GTT GGA ATC ATT GGC AAA TGC-3′ and 5′-TAA AAT TTT TTT TAC TGT ATC CTA CG-3′. The C-terminal truncation allele of KAP104 allele was then moved to YCplac111 (18) as an SstI-SphI fragment. Deletion of the 273 target bases was confirmed by sequencing.
(vii) pIH4.1. The FUS3 locus was PCR amplified from a genomic template with the following primers: 5′-GGGAATTCGACTACTGGACACTTCAGGG-3′, which anneals 1,000 bp upstream of the ATG and contains an EcoRI site, and 5′-GGCCGCGGACTAAATAATTCGTTCCAAATGAG-3′, which anneals to the 3′ end of the coding sequence and contains a SacII site. The FUS3 PCR product was then subcloned to EcoRI-SacII-cut pRS316CG (37), thus dropping out the CUP1 promoter and fusing the green fluorescent protein (GFP) coding sequence, in frame, to the 3′ end of FUS3.
(viii) pIH4.1AA (FUS3T180A Y182A-GFP) and pIH4.1KM1-1 (FUS3K42R-GFP). The mutant forms of Fus3-GFP were created by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) and plasmid pIH4.1 as a template. The primers used to create the T180A Y182A double mutation were 5′-GCAAAGCGGCATGGCCGAGGCTGTGGCCACACGTTG-3′ and 5′-CAACGTGTGGCCACAGCCTCGGCCATGCCGCTTTGC-3′. The primers used to create the K42R mutation were 5′-GGAGAAATCGTGGCAATAAGAAAGATCGAACCATTCGAT-3′ and 5′-ATCGAATGGTTCGATCTTTCTTATTGCCACGATTTCTCC-3′. The substitutions are underlined.
(ix) YCplac22/FUS3-GFP (AOB117).
The EcoRI-SacI fragment containing the FUS3-GFP translational fusion was moved from pIH4.1 to YCplac22 (18).
(x) YCplac111/GAL1-MSG5 (HKB100) and YCplac22/GAL1-MSG5 (AEB101).
The 0.680kb BamHI-EcoRI fragment containing the divergent GAL1/GAL10 promoter was moved from YCpG2 (43) to the polylinkers of YCplac22 and YCplac111 (18) to create plasmids ZWB159 and ZWB161, respectively. The MSG5 coding region (−6 to 1470) was amplified from genomic DNA with the following primers: 5′-CGCGGATCCGTGCACATGCAATTTCAC-3′, which contains a BamHI site, and 5′-AACTGCAGTTAAGGAAGAAACATCATCTG-3′, which contains a PstI site. The MSG5 PCR product was then digested with BamHI and PstI and ligated to BamHI-PstI-cut ZWB159 and ZWB161 plasmid DNA to create the GAL1-MSG5 transcriptional fusion vectors.
(xi) YCplac22/GAL1-PTP3 (TBB104).
The PTP3 coding region (−238 to 2605) was amplified from genomic DNA by using the following primers: 5′-AAGGATCCTTGCACCGGCGGATGTAACACTA-3′, which contains a BamHI site, and 5′-AACTGCAGGTTCTGGACCATGGATATTCG-3′, which contains a PstI site. The PTP3 PCR product was then digested with BamHI and PstI and ligated to the backbone of BamHI-PstI-cut AEB101 plasmid DNA, thereby changing the GAL1-MSG5 transcriptional fusion to a GAL1-PTP3 transcriptional fusion.
(xii) YIplac204/GAL1EG28-GPA1K21E R22E E364K (TBB105).
The gpa1K21E R22E E364K allele was isolated from plasmid MMB107 (YCplac22/GPA1K21E R22E E364K) (44) as a BamHI-SstI fragment and used to replace the GPA1E364K allele on plasmid DSB174 (YIplac204/GAL1EG28-GPA1E364K) (62). GAL1EG28 is ca. 18% as active as the native GAL1 promoter (20).
(xiii) pRS316CG/MSG5-GFP (HKB101).
The MSG5 locus (−381 to 1467) was PCR amplified from a genomic template by using the following primers: 5′-CCCTCGAGATGGTCCATCCTGGTAAG-3′, which contains an AvaI site, and 5′-TCCCCGCGGAGGAAGAAACATCATCTGTT-3′, which contains a SacII site. The MSG5 PCR product was then subcloned to AvaI-SacII-cut pRS316CG to create an in-frame MSG5-GFP translational fusion.
Construction of the A6 genomic library used to clone KAP104.
Yeast genomic DNA was extracted from the STE4 mutant strain A6, a derivative of strain 15Dau, and partially digested with Sau3A. The restricted DNA was fractionated by centrifugation through a 10 to 40% sucrose gradient, and 7- to 15-kb fragments were purified and ligated to YCplac33 plasmid DNA (18) digested with BamHI. The ligation mix was then used to transform Escherichia coli DH5α. A total of 54,000 transformants were recovered, of which approximately two-thirds contained an insert. The transformants were washed off the plates and grown in 38 ml of Luria broth plus ampicillin overnight at 37°C, and plasmid DNA was prepared by standard methods. Among 10 randomly picked transformants, the average insert size was 7.2 kb. A library of 33,000 plasmids each containing a genomic insert of roughly 7 kb is expected to saturate the yeast genome (54).
Pheromone response and growth assays. (i) Halo tests.
Strains were tested for pheromone-induced growth inhibition in standard halo assays as previously described (9).
(ii) Measurement of FUS1-lacZ activity.
FUS1 promoter activity was assayed in cells transformed with the centromeric reporter plasmid, pSB231 (64); β-galactosidase activity was measured as previously described (57).
(iii) Single-colony formation assay.
Strain 15Dau was transformed with two independently derived high-copy-number GAL1-KAP104 vectors, pYES/KAP104-A and pYES/KAP104-B. Transformants were grown to mid-log phase in galactose medium, and ca. 103 cells were spread onto plates containing a range of pheromone concentrations. The plates were incubated at 30°C for 6 days, and visible colonies were counted.
Genetic screens and characterization.
Backcrosses were performed as described previously (36).
Integrative mapping.
To confirm that pEL4-1 contains the gene corresponding to the adaptive− allele in strain A11, plasmid KAP104/YIplac211 was linearized with BglII, which is in the C terminus of KAP104, and used to transform a MATα derivative of strain DSY278. Uracil prototrophs were selected, and the transformants were subsequently crossed to strain A11. Strain A11 is prototrophic for leucine because its adaptive− mutation confers constitutive induction of its integrated FUS1-LEU2 reporter. The resulting diploid strains were sporulated, dissected, and scored for leucine prototrophy.
Cloning of KAP104.
The wild-type gene corresponding to the mutant allele in strain A11 was cloned by complementation. The A6 yeast genomic plasmid library was introduced into strain A11, and transformants were selected on synthetic medium lacking uracil and containing α-factor at a concentration of 9 nM. Pheromone resistant transformants were cultured on fresh medium lacking uracil and α-factor and then cured of the transforming plasmids by growth on plates containing 5-fluoroorotic acid. The growth of the cured strains was then compared to the growth of the original transformants on synthetic complete medium containing a 9 nM concentration of α-factor. Only one plasmid of 25,000 screened rendered A11 mutant cells resistant to this dose of pheromone. This plasmid, pEL4-1, was amplified and shown to normalize all of the A11 mutant phenotypes (see below). The 2.75-kb KAP104 coding sequence was located on the 4.8-kb genomic insert by creating a set of unidirectional nested deletions by using the Exo-Mung deletion kit (Stratagene).
Photomicrographs and quantification of Fus3-GFP localization.
Fluorescent and differential interference contrast (DIC) images were acquired by using a Zeiss Axioskop 2 microscope fitted with a Zeiss AxioCam digital camera and then processed by using Zeiss AxioVision software. Localization of Fus3-GFP was quantified from the digital images by using the histogram function of Adobe Photoshop 5.5. A circle of set size was used to sample the brightness of the nucleus and cytoplasm of cells from randomly chosen fields. The ratio of nuclear to cytoplasmic fluorescence was rounded to the nearest 0.1, and values corresponding to at least 25 cells for each strain and condition were plotted in histograms. All such experiments were performed at least three times.
RESULTS
Pheromone induces accumulation of Fus3 in the nucleus.
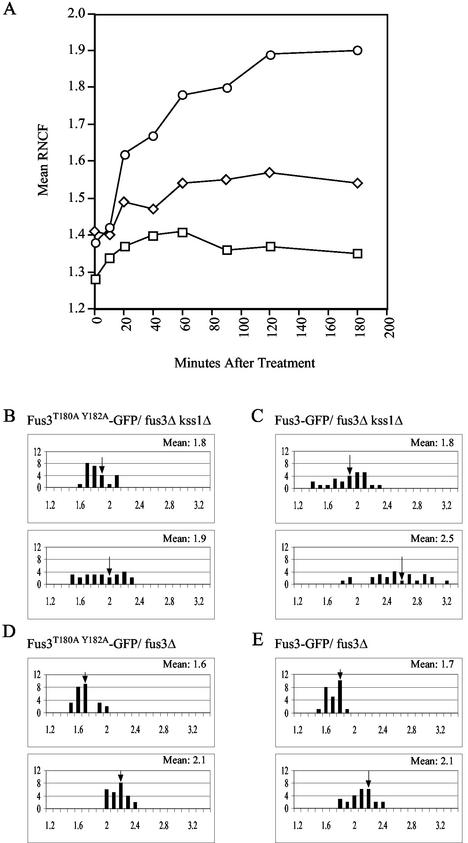
Upon activation of the mating pathway, Fus3 communicates the mating signal from the cytoplasm to the nucleus by interacting with its downstream effectors, Dig1, Dig2, and Far1 (12). Two studies have reported that the amount of Fus3 in the nucleus increases slightly when cells are treated with pheromone (6, 65). To confirm this observation and to determine whether nuclear accumulation of Fus3 is inhibited by Gpa1, we constructed an in-frame fusion between the genes encoding Fus3 and GFP. The FUS3-GFP hybrid gene proved to encode a functional MAPK, since it was able to fully rescue the response defect conferred by deletion of both FUS3 and KSS1 in halo tests (data not shown). Wild-type cells expressing FUS3-GFP from a centromeric vector were grown to mid-log phase and examined under a fluorescent microscope before and after pheromone treatment. In the vegetative culture, we observed diffuse fluorescence in both the cytoplasm and nucleoplasm. Upon stimulation with pheromone, which induces FUS3 transcription, the cells fluoresced more brightly throughout the cytoplasm and nucleoplasm but showed a proportionately greater intensification of the signal in their nuclei. In fact, the mean ratio of nuclear to cytoplasmic fluorescence (RNCF) increased from approximately 1.4 in the vegetative cells to about 1.9 in cells treated with pheromone for 3 h (Fig. 1A). A Student t test analysis of the data showed this difference to be highly significant (P < 0.0001). The mean RNCF began to rise after 10 min of treatment (P < 0.0001 for 1.42 at 10 min versus 1.62 at 20 min) and peaked about 2 h into the time course. Given that the accumulation of activated Fus3 in the nucleus must certainly precede a detectable increase in the mean RNCF, the observed nuclear accumulation of Fus3-GFP occurs early enough to contribute to the induction of mating-specific transcription and cell cycle arrest.
FIG. 1.
Pheromone induces nuclear accumulation of Fus3. Strains of the indicated genotype were transformed with the indicated GFP reporters and grown to mid-log phase. The cultures were then split and grown with or without the addition of 12 nM α-factor. Images were acquired from the untreated cultures and at various times after the addition of pheromone. The RNCF values were determined as described in Materials and Methods. In panels B to E, distributions of RNCF values are represented in histograms. The number of cells (y axis) are plotted as a function of the RNCF values (x axis). In each panel, the untreated cells are represented in the top histogram, and the cells treated for 3 h with pheromoneare represented in the bottom histogram. Arrows indicate the mean RNCF for each of the sampled populations. (A) Time course experiment showing Fus3-GFP localization in wild-type cells (○), cells overexpressing Gpa1E364K (□), and cells overexpressing Msg5 (⋄). Mean RNCF values were determined before and 10, 20, 40, 60, 90, 120, and 180 min after pheromone treatment; (B) Fus3T180A Y182A-GFP localization in fus3Δ kss1Δ cells; (C) Fus3-GFP localization in fus3Δ kss1Δ cells; (D) Fus3T180A Y182A-GFP localization in fus3Δ cells; (E) Fus3-GFP localization in fus3Δ cells.
In response to pheromone stimulation, Fus3 is phosphorylated by Ste7 on two residues, T180 and Y182, and thereby activated (17). To determine whether the pheromone-induced increase in the mean RNCF was due specifically to the nuclear accumulation of activated Fus3-GFP, we created a mutant form of the Fus3-GFP reporter, Fus3T180A Y182A-GFP, which cannot be phosphorylated by Ste7. As expected, Fus3T180A Y182A-GFP could not rescue the sterility of the fus3Δ kss1Δ strain (data not shown). Although the basal nuclear localization of the mutant reporter was normal, the Fus3T180A Y182A-GFP fus3Δ kss1Δ cells failed to exhibit a significant pheromone-induced increase in the mean RNCF (Fig. 1B) (P = 0.189 for 1.8 versus 1.9), a finding consistent with their sterile phenotype. In contrast, Fus3-GFP fus3Δ kss1Δ cells displayed a significant increase in mean RNCF after pheromone treatment (P = 0.0001 for 1.8 versus 2.5) (Fig. 1C). Interestingly, fus3Δ KSS1 cells expressing Fus3T180A Y182A-GFP responded almost normally to pheromone due to the functional redundancy of Fus3 and Kss1, and their mean RNCF increased as much as that of fus3Δ KSS1 cells expressing Fus3-GFP (compare Fig. 1D and E). This suggests that the activating phosphorylation of Fus3 is not strictly necessary for its pheromone-induced nuclear accumulation. However, it is possible that in the absence of wild-type Fus3, Fus3T180A Y182A-GFP translocates to the nucleus in complex with Kss1. Signal-induced dimerization of mammalian MAPKs is well established (26, 50), and in ERK microinjection studies Khokhlatchev et al. found that the phosphorylation of one partner in a MAPK dimer is both necessary and sufficient for nuclear translocation (28). Together, these data demonstrate that Fus3, like other MAPKs, concentrates in the nucleus in response to pathway stimulation but do not allow us to determine whether activating phosphorylation of Fus3 is essential for this phenomenon.
Adaptation to pheromone is correlated with a decrease in nuclear Fus3.
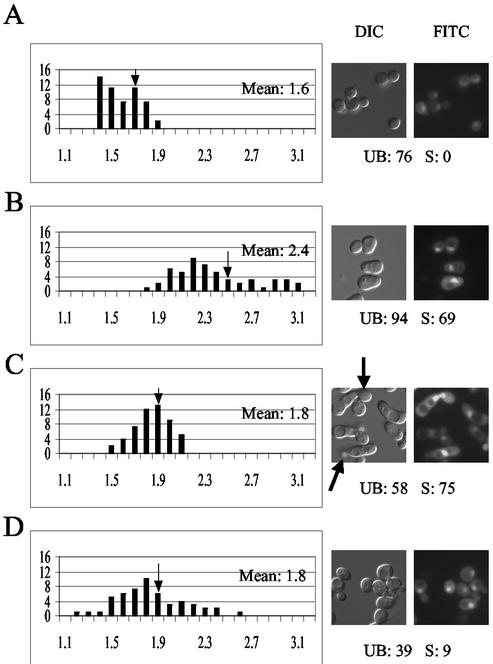
Termination of some MAPK-generated signals is correlated with decreased nuclear localization of the MAPK. For example, the Hog1 MAPK of Saccharomyces cerevisiae moves to the nucleus when cells are osmotically shocked and leaves the nucleus as the cells adapt to the new culture conditions or when they are returned to medium of normal osmolarity (15, 51). Similarly, growth factor-induced nuclear accumulation of the mammalian p42/p44 MAPK is transient, and adaptation to the stimulus correlates with reappearance of the kinase in the cytoplasm (33). To determine whether the duration of the pheromone response is correlated with the localization of Fus3, we treated mid-log-phase wild-type cells with a dose of pheromone that induces a transient response. Prior to stimulation, the mean RNCF of the culture was 1.6 (Fig. 2A). By 3 h after treatment, about 94% of the cells had arrested in G1, about 69% of the culture had formed mating projections, and the mean RNCF had risen to 2.4 (Fig. 2B). By 6 h after treatment, the culture had begun to recover. The fraction of arrested cells had decreased to 58%, more than half of the shmoos had budded and, significantly, the mean RNCF had fallen to 1.8 (Fig. 2C). An inference that can be drawn from these results is that yeast cells recover from pheromone stimulation by decreasing the level of nuclear Fus3. To test this idea, we allowed the cells to fully recover and saturate the pheromone-containing medium. At 24 h after the original stimulus, the cells were resuspended in fresh medium and challenged with pheromone again. Cells adapted to chronic pheromone treatment are expected to be unresponsive to further stimulation for up to four generations (46). By 3 h after the second treatment, only 9% of the cells had formed mating projections, and the mean RNCF remained at 1.8 (Fig. 2D). Thus, both recovery from pheromone-induced cell cycle arrest and long-term adaptation to pheromone are correlated with a decreased concentration of Fus3 in the nucleus.
FIG. 2.
Adaptation to pheromone is correlated with a decrease in nuclear Fus3. Wild-type cells transformed with Fus3-GFP were grown to mid-log phase and treated with 6 nM α-factor. Images were acquired at 0, 3, 6, and 27 h. After 24 h in the original medium, the treated culture was resuspended in fresh medium containing 6 nM α-factor, and images were taken for analysis 3 h later. RNCF values were determined as described in Materials and Methods, and the distribution of values represented in histograms. The number of cells (y axis) are plotted as a function of the RNCF values (x axis). The arrows on the histograms indicate the mean RNCF for each of the sampled populations. The arrows on the DIC images indicate cells that have responded and recovered (budded shmoos). “UB” refers to the percentage of unbudded cells. “S” refers to the percentage of shmooing cells. (A) Vegetative cells; (B) cells 3 h after treatment; (C) cells 6 h after treatment; (D) cells 27 h after first treatment and 3 h after the second treatment.
Gpa1 inhibits the pheromone-induced accumulation of Fus3 in the nucleus.
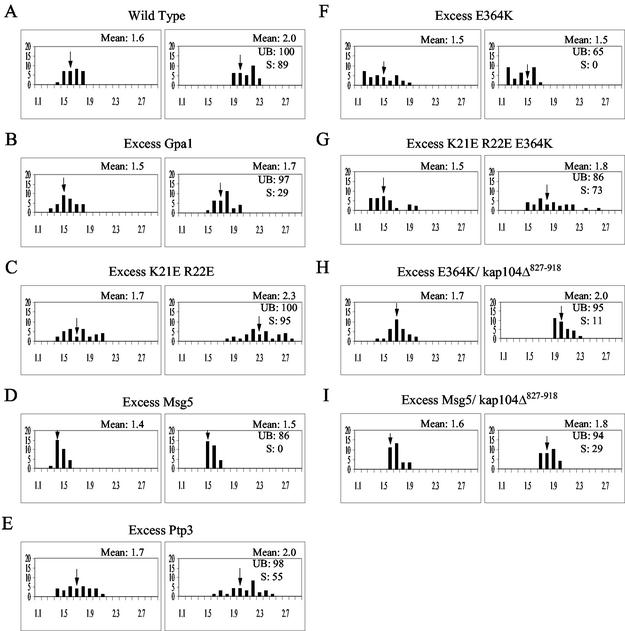
Given our recent finding that the activated form of Gpa1 binds directly to the activated form of Fus3 in vivo (44), as well as the results described above, we hypothesized that Gpa1 downregulates the mating signal by inhibiting the pheromone-induced accumulation of Fus3 in the nucleus. To test this idea, we measured the mean RNCF in cells overexpressing wild-type Gpa1, which exhibit a moderate decrease in sensitivity to pheromone (9), and in cells overexpressing the E364K hyperadaptive mutant form of Gpa1, which are completely resistant to pheromone (36). Excess Gpa1 had a moderate inhibitory effect on the pheromone-induced accumulation of Fus3 in the nucleus (Fig. 3A and B; P = 0.0011 for the induced values of 2.0 versus 1.7), whereas excess Gpa1E364K almost completely prevented it (Fig. 1A and 3F).
FIG. 3.
Gpa1 and Msg5 inhibit the pheromone-induced accumulation of Fus3 in the nucleus. MATa strains of the indicated genotype were transformed with the Fus3-GFP reporter and grown to mid-log phase in galactose medium (GAL1 promoter on). The cultures were then split and grown with or without the addition of 18 nM α-factor. Images were acquired from the untreated cultures and from the treated cultures 3 h after the addition of pheromone. The RNCF values were determined as described in Materials and Methods, and the distribution of values is represented in histograms. The number of cells (y axis) are plotted as a function of the RNCF values (x axis). In each panel, the untreated cells are represented in the histogram on the left, and the treated cells are represented in the histogram on the right. Arrows indicate the mean RNCF for each of the sampled populations. UB refers to the percentage of unbudded cells; S refers to the percentage of shmooing cells. (A) Wild-type cells; (B) GAL1EG28-GPA1 cells; (C) GAL1EG28-GPA1K21E R22 cells; (D) GAL1-MSG5 cells; (E) GAL1-PTP3 cells; (F) GAL1EG28-GPA1E364K cells; (G) GAL1EG28-GPA1K21E R22E E364K cells; (H) GAL1EG28-GPA1E364K kap104Δ::HIS YCplac111/kap104Δ827-918 cells; (I) YCplac22/GAL1-MSG5 kap104Δ::HIS YCplac111/kap104Δ827-918 cells.
To determine whether the effect of Gpa1 on Fus3 localization depends on Gpa1-Fus3 interaction, we introduced a mutation that disrupts Gpa1-Fus3 binding (K21E R22E) (44) into wild-type Gpa1 and into Gpa1E364K. In gpa1Δ cells expressing gpa1K21E R22E from a centromeric vector, the mean RNCF prior to pheromone treatment was not significantly different from that of the control cells (P = 0.1183 for the values 1.6 versus 1.7), suggesting that Gpa1 does not affect the basal localization of Fus3 (compare Fig. 3A and C). After pheromone treatment, however, the gpa1Δ gpa1K21E R22E cells showed a considerably higher than normal nuclear localization of Fus3-GFP (P = 0.0043 for the values 2.0 versus 2.3), a finding consistent with their supersensitivity (44). Moreover, cells overexpressing Gpa1K21E R22E E364K, which binds Gβγ but does not stimulate adaptation to pheromone, showed a clear shift in mean RNCF after pheromone treatment (Fig. 3G), in marked contrast to cells overexpressing Gpa1E364K (P < 0.0001 for the induced values of 1.5 versus 1.8). These data suggest that Gpa1 affects Fus3 localization by interacting directly with it.
Because Fus3 stimulates its own transcription, its sequestration by Gpa1 is expected to inhibit its expression. Preventing the increase of cytoplasmic Fus3 might, in turn, further limit the pheromone-induced accumulation of Fus3 in the nucleus. To determine whether Gpa1 can affect the localization of Fus3 without affecting Fus3 expression, we measured basal Fus3 RNCF values in sterile strains. A catalytically inactive form of the reporter, Fus3K42R-GFP, was transformed into a fus3Δ kss1Δ strain (IHY205) and into an isogenic strain that overexpresses Gpa1E364K (IHY217). Excess Gpa1E364K dramatically lowered the mean RNCF in sterile cells (P < 0.0001 for 1.4 versus 1.6), indicating that Gpa1 can inhibit the nuclear localization of Fus3 independent of its effect on Fus3 expression.
Msg5 and Ptp3 inhibit the accumulation of Fus3 in the nucleus.
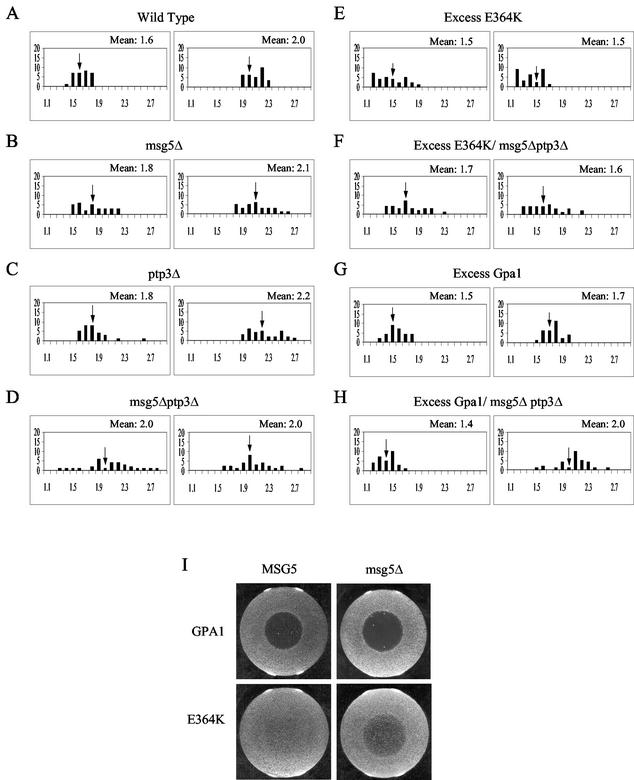
Fus3 is dephosphorylated and thereby inactivated by the pheromone-inducible, dually specific tyrosine-threonine phosphatase, Msg5, and by two tyrosine phosphatases, Ptp2 and Ptp3 (13, 68). Ptp2 is located in the nucleus (42), Ptp3 is localized to the cytoplasm (42), and Msg5-GFP is found in both compartments (D. E. Stone and H.-J. N. Kim, unpublished results). Because phosphatases are known to regulate the nucleocytoplasmic transport of some MAPKs, we asked whether the localization of Fus3 is controlled by either Msg5 or Ptp3. As shown in Fig. 4A to D, deletion of either MSG5 or PTP3 partially increased the basal mean RNCF (P = 0.0063 for 1.6 versus 1.8 [msg5Δ]; P = 0.0052 for 1.6 versus 1.8 [ptp3Δ]), and the msg5Δ ptp3Δ double deletion conferred full induction (mean RNCF = 2.0) in the absence of pheromone. Conversely, overexpression of Msg5 lowered the mean RNCF of vegetative wild-type cells expressing Fus3-GFP (Fig. 3D) (P = 0.0046 for 1.6 versus 1.4) and the mean RNCF of sterile fus3Δ kss1Δ cells expressing Fus3K42R-GFP (P < 0.0001 for 1.4 versus 1.6). In response to pheromone, wild-type cells overexpressing Msg5 did not fully arrest in G1 or shmoo, and they did not exhibit the normal increase in the relative level of nuclear Fus3-GFP (Fig. 1A [P < 0.0004 for 1.41 versus 1.57] and 3D [P = 0.022 for 1.4 versus 1.5]). Interestingly, overexpression of Ptp3 had no effect on the localization of Fus3-GFP (Fig. 3E). Thus, both Ptp3 and Msg5 appear to affect the basal localization of Fus3, whereas Msg5 may be the more important regulator in cells responding to pheromone.
FIG. 4.
Fus3 localization in cells lacking MSG5 and/or PTP3. Strains of the indicated genotype were transformed with the Fus3-GFP reporter and grown to mid-log phase in galactose medium (GAL1 promoter on). The cultures were then split and grown with or without the addition of 18 nM α-factor. Images were acquired from the untreated cultures and from the treated cultures 3 h after the addition of pheromone. The RNCF values were determined as described in Materials and Methods, and the distribution of values represented in histograms. The number of cells (y axis) are plotted as a function of the RNCF values (x axis). In each panel, the untreated cells are represented in the histogram on the left, and the treated cells are represented in the histogram on the right. Arrows indicate the mean RNCF for each of the sampled populations. (A) Wild-type cells; (B) msg5Δ cells; (C) ptp3Δ cells; (D) msg5Δ ptp3Δ cells; (E) GAL1EG28-GPA1E364K cells; (F) GAL1EG28-GPA1E364K msg5Δ ptp3Δ cells; (G) YCplac22/GAL1EG28-GPA1 cells; (H) YCplac22/GAL1EG28-GPA1 msg5Δ ptp3Δ cells. (I) Halo tests showing the epistatic effect of msg5Δ on Gpa1E364K-induced adaptation: wild-type cells (top left), msg5Δ cells (top right), YCplac111/Gpa1E364K (61) in wild-type cells (bottom left), and YCplac111/Gpa1E364K in msg5Δ cells (bottom right).
The Gpa1- and Msg5-mediated effects on Fus3 localization are partially interdependent.
Both Gpa1 and Msg5 have been implicated in adaptation to pheromone (13, 45, 62). Three observations suggest that they work together to regulate Fus3. First, both the Gα protein and the phosphatase bind to Fus3 (13, 44). Second, both proteins inhibit nuclear localization of the Fus3-GFP reporter. Third, deletion of MSG5 is partially epistatic to hyperadaptive alleles of GPA1 in halo tests (13, 69) (Fig. 4I). To further explore the functional relationship between Gpa1 and Msg5, we assessed their interdependence in affecting Fus3 localization. Deletion of MSG5 had no effect on the ability of Gpa1E364K to inhibit the pheromone-induced increase in nuclear Fus3 (data not shown). Could Ptp3 compensate for the absence of Msg5? Such functional redundancy between phosphatases has been observed in the regulation of both Fus3 and Hog1 (25, 68). Like deletion of MSG5, deletion of PTP3 compromises Gpa1E364K-mediated adaptation in halo tests (Stone and Kim, unpublished) and increases both the basal and the induced RNCF values (Fig. 4C). Although the absence of both phosphatases did not significantly compromise the inhibitory effect of Gpa1E364K on Fus3 localization (compare Fig. 4E and F) (P < 0.1321 for the induced values of 1.6 versus 1.5), the msg5Δ ptp3Δ double deletion nullified the effect of excess wild-type Gpa1 on the pheromone-induced nuclear accumulation of Fus3-GFP (compare Fig. 4G and 4H) (P = 0.0001 for the induced values of 1.7 and 2.0). These results indicate that the effect of Gpa1 on Fus3 localization is partially dependent on Msg5 and/or Ptp3.
In halo tests, Gpa1 adaptive function partially depends on Msg5; in Fus3 localization assays, Gpa1 function partially depends on Msg5 and/or Ptp3. To determine whether Msg5 adaptive function depends on Gpa1, we transformed the GAL1-MSG5 transcriptional fusion and the FUS3-GFP reporter into a gpa1 temperature-sensitive strain related to strain 15D. The temperature-sensitive form of Gpa1 is partially functional at 23°C and fully inactivated at 37°C (data not shown). Therefore, Fus3-GFP localization was assessed before and after pheromone treatment of log-phase cells cultured at the permissive temperature, and log-phase cells were shifted to the restrictive temperature at time zero. Although the pheromone-induced increase in the mean RNCF of wild-type cells cultured at high and low temperatures was not as great as that observed at 30°C, the effect of the gpa1 temperature-sensitive mutation (gpats) was clear. As shown in Table 3, loss of functional Gpa1 significantly increased the mean RNCF of cells expressing the normal level of Msg5 (when comparing cultures grown at permissive temperature, P = 0.001 for 1.6 versus 1.8 and P = 0.0014 for 1.8 versus 2.1; when comparing cultures grown at the restrictive temperature, P = 0.1754 for 1.7 versus 1.8 and P = 0.0112 for 1.9 versus 2.1). This hyperaccumulation of Fus3-GFP to the nuclei of Gpa1-deficient cells was almost completely suppressed by the overexpression of Msg5 at 23°C (P = 0.0932 for 1.5 versus 1.6 and P = 0.0911 for 1.6 versus 1.7). However, the effect of excess Msg5 on Fus3-GFP localization was significantly diminished in cells cultured at 37°C and therefore completely lacking functional Gpa1 (P < 0.0001 for 1.5 versus 1.9 and P = 0.0048 for 1.6 versus 1.8). These data indicate that the adaptive function of Msg5 is partially dependent on Gpa1.
TABLE 3.
Effect of Msg5 overexpression on Fus3-GFP localization in a gpa1 temperature-sensitive background
| Background | Effect of Msg5 overexpression (mean RNCF)
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
GAL1-MSG5 repressed at:
|
GAL1-MSG5 induced at:
|
|||||||
| 23°C
|
37°C
|
23°C
|
37°C
|
|||||
| − | +a | − | + | − | + | − | + | |
| gpa1ts | 1.8 | 2.1 | 1.8 | 2.1 | 1.6 | 1.7 | 1.9 | 1.8 |
| GPA1 | 1.6 | 1.8 | 1.7 | 1.9 | 1.5 | 1.6 | 1.5 | 1.6 |
+, 3 h after addition of 36 nM pheromone.
Together, the results of the epistasis tests suggest that Msg5 plays a role in Gpa1-mediated adaptation and, in particular, in inhibiting pheromone-induced accumulation of Fus3 in the nucleus. Ptp3 may play a similar role, or it may work in concert with Gpa1 only in the absence of Msg5.
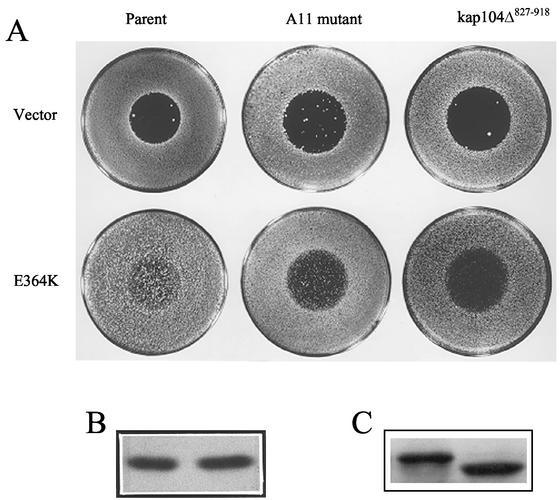
C-terminal truncation of Kap104 confers an adaptive defect and is epistatic to Gpa1E364K.
In a genetic screen designed to identify gene products required for Gpa1-mediated adaptation, we isolated a mutant strain, designated A11, that exhibited a recessive supersensitive/adaptive− phenotype. When expressed in the A11 background, Gpa1E364K was significantly compromised in its ability to downregulate the pheromone response, as indicated in halo tests (Fig. 5A) and mating-specific transcription assays (data not shown). Segregation analysis indicated that the mutant phenotype was due to mutation of a single locus. The corresponding wild-type gene was cloned by complementation of the adaptive defect, and an integrative mapping strategy was used to confirm the identity of the mutant allele and the cloned gene. The nucleotide sequence of the cloned gene matched that of KAP104 (for karyopherin protein of 104 kDa), a homologue and functional analogue of the mammalian nuclear import protein, karyopherin-β2 (transportin) (1). Transport receptors of this class bind directly to their cargo molecules and mediate unidirectional movement through the nuclear pore complex (NPC), either into or out of the nucleus. An established function of Kap104 is to import two mRNA binding proteins, Nab2 and Nab4, after they have released their cargo in the cytoplasm (1, 32).
FIG. 5.
Effect of Kap104 truncation on pheromone-induced cell cycle arrest. (A) Strains DSY278 and A11 were transformed with YCplac33 (18) (top row) or YCplac33/Gpa1E364K (36) (bottom row). Strain ZW432 (15Dau kap104Δ::URA3/kap104Δ827-918) was transformed with YCplac22 (18) (top row) or YCplac22/Gpa1E364K (62) (bottom row). Transformants were cultured and tested in glucose medium. (B) Immunoblot probed with Gpa1 antibody. Protein was extracted from strains DSY278 (left) and A11 (right), both transformed with YCplac33/Gpa1E364K. (C) Immunoblot probed with Kap104 antibody. Protein was extracted from strain DSY278 (left) and strain A11 (right).
The A11 allele of KAP104 contains a nonsense mutation and is predicted to encode a truncation product lacking the carboxy-terminal 91 residues of the 918-amino-acid Kap104 protein. However, it is possible that a nonsense suppressor intrinsic to this strain could result in the production of some full-length Kap104. The mutant phenotypes of strain A11 might therefore be due to a lower-than-normal level of full-length Kap104 rather than to its complete absence and replacement by the truncation product. To distinguish these possibilities, a deletion allele lacking the last 273 bp of the KAP104 coding region, kap104Δ827-918, was constructed. This allele encodes a shortened form of Kap104, missing the C-terminal 91 amino acids. A centromeric vector containing kap104Δ827-918 was expressed in a kap104Δ background, and these cells were compared to the isogenic wild-type and A11 mutant strains in halo tests. As shown in Fig. 5A, the halo phenotypes of the kap104Δ827-918 and A11 mutant strains were essentially the same. The two alleles conferred similar degrees of supersensitivity in cells expressing wild-type Gpa1 and had a similar detrimental effect on the hyperadaptive function of Gpa1E364K. Immunoblot analysis demonstrated that strain A11 expresses a shortened form of Kap104 at a level similar to that of the full-length protein in wild-type cells (Fig. 5C) and the normal steady-state level of Gpa1 (Fig. 5B). Therefore, the most likely explanation for the adaptive− phenotypes of strain A11 is that the C-terminal one-tenth of Kap104, which comprises about one-quarter of the cargo-binding domain, is critical for proper negative regulation of the pheromone response. Truncation of this domain is epistatic to GPA1E364K.
The genetic link between Kap104, a karyopherin, and Gpa1-mediated adaptation led us to wonder whether the adaptive− phenotype conferred by kap104Δ827-918 is due to mislocalization of Fus3. To test this possibility, we transformed the Fus3-GFP reporter into strains AEY144 (relevant genotype MATa kap104Δ827-918/kap104Δ) and AOY135 (relevant genotype MATa GAL1EG28-GPA1E364K kap104Δ827-918/kap104Δ) and quantified the relative degree of nuclear fluorescence. Although the truncation of Kap104 had no detectable effect on Fus3-GFP localization in GPA1 cells (data not shown), the kap104Δ827-918 allele largely suppressed the effect of Gpa1E364K on Fus3 localization (compare Fig. 3F and H) (P < 0.0001 for 1.5 versus 1.7, and P < 0.0001 for 1.5 versus 2.0). Thus, the loss of Gpa1E364K hyperadaptive function seen in halo tests (Fig. 5) and in culture (Fig. 3F) correlates with restoration of normal Fus3 levels in the nuclei of cells responding to pheromone. Because Gpa1 and Msg5 appear to cooperate to promote an adaptive mechanism involving Fus3 localization, we next sought to determine whether the effect of Msg5 overexpression on Fus3 localization would also be diminished in kap104Δ827-918 mutant cells. As can be seen by comparing Fig. 3D and I, this is indeed the case. The C-terminal truncation of Kap104 decreases the ability of excess Msg5 to inhibit both pheromone-induced cell cycle arrest and accumulation of Fus3-GFP in the nucleus (P = 0.003 for 1.4 versus 1.6 and P < 0.0001 for 1.5 versus 1.8). Together, these results suggest that Gpa1 and Msg5 downregulate the mating signal by antagonizing the signal-induced nuclear translocation of Fus3 and that Kap104 is required for Gα/phosphatase-mediated regulation of Fus3 localization.
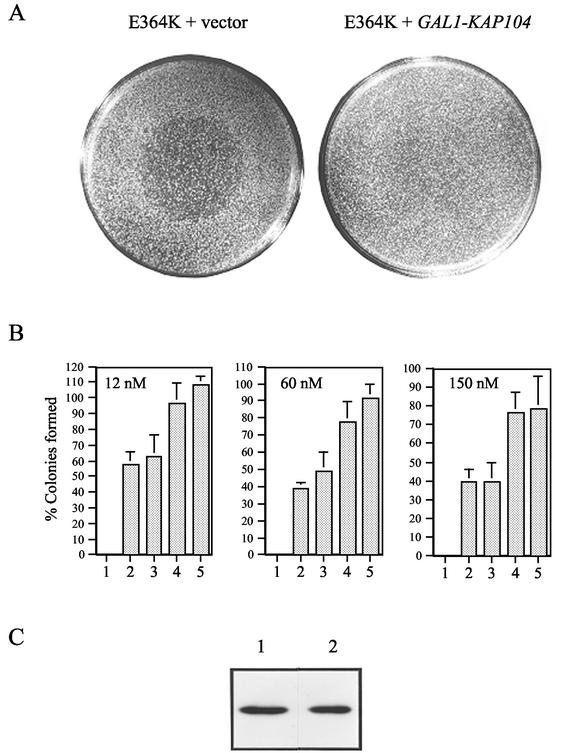
Overexpression of Kap104 inhibits growth but potentiates Gpa1-mediated adaptation.
Because kap104Δ827-918 is epistatic to GPA1E364K, we wondered whether Kap104 overexpression could enhance Gpa1E364K hyperadaptive function. The KAP104 coding region was therefore fused to the GAL1 promoter. Although induction of the GAL1-KAP104 construct in wild-type cells conferred a severe growth defect (cells overexpressing Kap104 took 7 days to form macroscopic colonies compared to the usual 2 days) and an increased sensitivity to pheromone (data not shown), excess Kap104 was nevertheless able to augment the ability of Gpa1E364K to promote growth at otherwise lethal doses of α-factor. Whereas cells expressing the wild-type level of Gpa1E364K form turbid halos, cells expressing Gpa1E364K in combination with excess Kap104 are completely resistant to pheromone and form no halo at all (Fig. 6A). The combined effect of Gpa1E364K expression and Kap104 overexpression was quantified in a single-colony formation assay (Fig. 6B). Cells expressing the wild-type level of Gpa1E364K and either the wild-type level or the GAL1-induced level of Kap104 were spread on plates containing various doses of α-factor, as well as on control plates lacking pheromone. At all three pheromone concentrations, the cells overproducing Kap104 formed significantly more colonies, increasing the plating efficiency by 64% at the lowest dose and almost doubling it at the two higher doses. Immunoblot analysis showed that the steady-state level of Gpa1 was unaffected by the induction of GAL1-KAP104 (Fig. 6C). Thus, Kap104 overexpression potentiates the adaptive function of Gpa1.
FIG. 6.
Overexpression of Kap104 potentiates Gpa1-mediated adaptation. Strain 15Dau was transformed with the vectors indicated below and the transformants were tested for their ability to grow in a range of pheromone concentrations. The transformants were cultured and assayed in galactose medium. (A) Halo tests: YCplac111/Gpa1E364K and pYES (left); YCplac111/Gpa1E364K, and pYES/KAP104 (right). (B) Single-colony formation assays. The ability to form colonies on medium containing discrete concentrations of α-factor was assayed. The numbers plotted are the percentages of cells of a given strain that formed colonies at the indicated dose. Each datum point is the mean of at least two trials with each of at least six transformants. Plasmids pYES/KAP104-A and pYES/KAP104-B were constructed by using DNA derived from two independent PCR amplifications of the KAP104 coding region. Columns: 1, pYES; 2, YCplac111/Gpa1E364K; 3, pYES and YCplac111/Gpa1E364K; 4, pYES/KAP104-A and YCplac111/Gpa1E364K; 5, pYES/KAP104-B and YCplac111/Gpa1E364. (C) Immunoblot probed with Gpa1 antibody. Protein was extracted from strain 15Dau transformed with YCplac111/Gpa1E364K (left) and with pYES/KAP104 and YCplac111/Gpa1E364K (right).
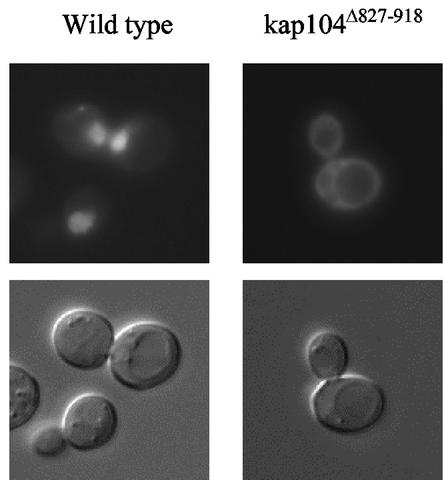
C-terminal truncation of Kap104 results in mislocalization of Nab2.
The two known substrates of Kap104 are Nab2 and Nab4. The kap104Δ827-918 form of Kap104 lacks the C-terminal one-quarter of the substrate-binding domain (HEAT domains 17 and 18) (7) and might therefore be unable to bind its cargo. To test this, we examined the localization of Nab2-GFP in strains A11 and DSY278. As expected, the Nab2 reporter was found concentrated in the nuclei of wild-type cells but spread throughout the cytoplasm of kap104Δ827-918 cells (Fig. 7).
FIG. 7.
Effect of kap104Δ827-918 on Nab2-GFP localization. Wild-type and A11 cells transformed with Nab2-GFP were examined in mid-log phase.
DISCUSSION
Because MAPK signaling cascades are used by all eukaryotic cells to control growth and differentiation, the timing and duration of MAPK responses must be tightly regulated. For MAPKs that participate in multiple pathways, cross talk must be controlled as well. Although MAPK pathways have been subjected to intense investigation, the ways in which they are regulated are not fully understood. A priori, regulatory mechanisms could involve expression, enzymatic activity, sequestration and/or localization, and stability of MAPKs. Induction of MAPK responses is thought to depend on the activation of MAPKs by their MEKs, and their subsequent accumulation in the nucleus, where they phosphorylate transcription factors and other regulatory targets. Such signal-induced localization has been demonstrated in a number of systems (26, 50). Inactivation of MAPKs partially depends on the action of specific protein phosphatases and, in some cases, the regulated export of MAPKs from the nucleus is thought to play a role in downregulation (15, 33). The yeast mating signal is mediated by a heterotrimeric G protein that communicates with a MAPK cascade composed of Ste11 (MEKK), Ste7 (MEK), and Fus3 (MAPK). The free Gβγ subunit activates the MAPK module, and both the inactive and the active forms of the Gα subunit, Gpa1, negatively regulate the response. Prior to this work, genetic evidence suggested that Gpa1 stimulates adaptation by indirectly inactivating Ste4 (Gβ) (36) and through a mechanism acting at the level of Fus3 (13, 69; A. T. Ellicott and D. E. Stone, unpublished results). In a proteomic screen for gene products that associate with Gpa1 in cells responding to pheromone, we identified Fus3 (44). Additional experiments showed that the activated form of Gpa1 binds to the activated form of Fus3 in cells responding to pheromone. This discovery led us to study the relationship between the localization of Fus3 and adaptation to pheromone. We found that the relative level of nuclear Fus3 decreases under conditions in which cells are recovering from pheromone treatment or have adapted to chronic pheromone stimulation and that both Gpa1 and the dual-specific phosphatase Msg5 negatively affect the nuclear localization of Fus3. The adaptive functions of Gpa1 and Msg5 are partially dependent on one another and on the Kap104 importin. To our knowledge, this is the first time that the Gα subunit of a heterotrimeric G protein has been shown to affect the subcellular localization of a MAPK.
Gpa1 and Msg5 downregulate the mating signal by affecting the localization of Fus3.
Because the expression of Fus3 is induced ∼5-fold by pheromone (53), it is difficult to detect signal-induced nuclear localization of Fus3 comparable to that documented for other MAPKs. Using a myc-tagged form of Fus3, Elion et al. found that the level of the kinase in the nucleus is slightly enhanced by pheromone treatment (6), and van Drogen et al. came to the same conclusion using a Fus3-GFP reporter (65). To determine whether the nuclear concentration of Fus3 rises and falls as cells respond to pheromone and adapt to it, we developed a quantitative assay that measures the relative proportion of Fus3 in the nucleus and cytoplasm. Although the RNCF values varied considerably within a given culture, four observations indicate that this measurement is sensitive to physiologically relevant changes in Fus3 localization. First, pheromone treatment of wild-type cells causes a 25 to 50% increase in the mean RNCF (Fig. 1 to 4). Statistical analysis showed this difference to be highly significant (P < 0.0001). We speculate that the variability of the RNCF values within a given culture is due to the lack of cell cycle synchrony. Second, the increase in nuclear Fus3 is easily detectable after only 20 min of pheromone treatment, suggesting that the buildup of Fus3 in the nucleus is relevant to the induction of mating response. Third, the mean RNCF of wild-type cultures decreases as the cells recover and begin to bud (Fig. 2A to C). Fourth, Fus3 does not concentrate in the nucleus when cells that have adapted to chronic stimulation are rechallenged (Fig. 2D). This observation correlates with the insensitivity of such cells to pheromone-induced cell cycle arrest and may be relevant to the ability of recovered cells to “remember” their exposure to pheromone and remain unresponsive to further stimulation for multiple generations (46).
The availability of a quantitative assay for the relative level of nuclear Fus3 enabled us to ask whether downregulation of the mating signal by Gpa1 and Msg5 is associated with changes in Fus3 localization. The data shown in Fig. 2, 3, and 4 clearly indicate that both of these proteins, when manipulated to promote hyperadaptation, inhibit the pheromone-induced accumulation of Fus3 in the nucleus, and both are dependent on the wild-type function of Kap104 to accomplish this. It is noteworthy that both Gpa1 and Msg5 can inhibit the nuclear localization of Fus3 without affecting Fus3 expression. Is the decreased concentration of nuclear Fus3 in stimulated cells the cause or the result of the mating pathway downregulation? Although this question cannot be definitively answered on the basis of correlative results, Gpa1 and Msg5 are both known to interact directly with Fus3. Moreover, mutational disruption of the Gpa1-Fus3 interaction increased pheromone sensitivity and localization of Fus3 to the nuclei of stimulated cells (Fig. 3C), implying that Gpa1 regulates the mating signal by directly binding to the MAPK. Therefore, we favor the former possibility: Gpa1 and Msg5 inhibition of Fus3 accumulation in the nucleus is an adaptive mechanism.
It is important to note that the Gpa1 and Msg5 adaptive functions are partially interdependent. Deletion of MSG5 compromises Gpa1E364K-induced recovery from pheromone arrest (Fig. 4I) and, when combined with the loss of Ptp3, another cytoplasmic phosphatase that targets Fus3, slightly lessens the effect of Gpa1E364K on Fus3 localization (Fig. 4E and F). Furthermore, the msg5Δ ptp3Δ double deletion is completely epistatic to the effect of excess wild-type Gpa1 on Fus3 localization in stimulated cells (Fig. 4G and H). Conversely, the effect of Msg5 overexpression on Fus3 localization is weakened by inactivation of Gpa1 (Table 3). Together, these results suggest that Msg5 acts with Gpa1 to downregulate the mating signal by inhibiting the nuclear localization of Fus3. Because Gpa1 is found primarily at the plasma membrane (19, 58), its effect on Fus3 localization is likely due to sequestration. Msg5 might also act as a cytoplasmic tether for Fus3, in the same way that Ptp3 serves as a cytoplasmic anchor for the Hog1 MAPK of S. cerevisiae (42). Alternatively, or in addition, Msg5 might inhibit nuclear accumulation of Fus3 by dephosphorylating it. Signal-induced nuclear localization of other MAPKs is known to be dependent on their phosphorylation state (for examples, see references 26 and 50), and the same may be true for Fus3. Consistent with this possibility, recent evidence suggests that phosphorylation of Fus3 promotes its release from the Ste5 scaffolding protein (65). Dephosphorylation of Fus3 might therefore promote its binding to Ste5.
Although the data do not yet allow us to formulate a detailed model of the Gpa1-Msg5 functional relationship, we can propose two general possibilities. Gpa1 and Msg5 may act in parallel, separately impinging on Fus3. When the kinase dissociates from one of the regulators, the other may capture it. In this scenario, Msg5 might sequester Fus3 and/or inactivate it. A distinct possibility is that Gpa1 and Msg5 act on Fus3 in a complex. Perhaps Gpa1 facilitates the docking of the phosphatase to the kinase. In this case, the primary role of Msg5 would be dephosphorylation of Fus3 rather than sequestration of it. Coimmunoprecipitation experiments might allow us to detect Gpa1-Fus3-Msg5 complexes if they exist. Whatever the mechanism, it is clear that the role of Msg5 is distinct from that of Ptp3. Whereas deletion of either phosphatase affects the basal RNCF, only overexpression of Msg5 represses the induced RNCF.
Based on photobleaching studies of cells expressing Fus3-GFP, van Drogen et al. concluded that Fus3 shuttles constitutively between the cytoplasm and nucleus and that its rate of transport is not affected by pheromone treatment (65). According to their model, Fus3 is recruited to the plasma membrane in stimulated cells by Ste5, where it is activated by Ste7. Phosphorylation of Fus3 triggers its dissociation from Ste5. Some of the activated kinase molecules then move to the nucleus, whereas some concentrate at the shmoo tip. What controls this differential localization of activated Fus3? We have shown that the activated form of Gpa1 binds to the activated form of Fus3 in cells responding to pheromone and that the Gpa1-Fus3 interaction is important both in the chemotropic sensing of pheromone and in adaptation to pheromone (44). Moreover, Gpa1 concentrates at the shmoo tips of responding cells (44). Therefore, we propose that Gpa1 captures some of the activated Fus3 molecules as they are released from Ste5, thereby targeting the kinase to its substrates at the plasma membrane and simultaneously modulating the intensity of the mating signal.
Does this mean that Gpa1 inhibits the Fus3-dependent mating responses in the nucleus while at the same time promoting mating-specific events at the plasma membrane? It is interesting to suppose that Msg5, whose expression is strongly induced by pheromone, acts as a temporal switch that alters the consequences of the Gpa1-Fus3 interaction. Early in the mating reaction, when relatively little Msg5 is present, the activated Fus3 that dissociates from Gpa1 is free to move to the nucleus. When the expression of Msg5 is fully induced (near the time of zygote formation), however, any activated Fus3 that dissociates from Gpa1 is sequestered and/or inactivated by the phosphatase. Thus, toward the end of the mating reaction (perhaps as mating partners begin to fuse or in newly formed zygotes), Gpa1 and Msg5 work together to prevent the transport of activated Fus3 to the nucleus. This relieves the cell cycle block and downregulates mating-specific gene expression when these responses are no longer appropriate. Note that in this model, the Kd for the Gpa1-Fus3 interaction determines the relevance of Msg5 to the adaptive function of Gpa1. The model correctly predicts that, in the Fus3-GFP localization assay, Gpa1E364K, which binds Fus3 considerably better than does wild-type Gpa1, is less badly compromised by deletion of MSG5 and PTP3 than is wild-type Gpa1 (Fig. 4).
An important point in the model proposed by van Drogen et al. (65) is that pheromone treatment does not alter the rates of nuclear import or export of Fus3. Nevertheless, Fus3 clearly accumulates in the nuclei of cells responding to pheromone. Most likely, this is due to decreased cytoplasmic tethering and/or increased nuclear anchoring of phosphorylated Fus3. There are a number of examples in which transcription factors retain activated MAPKs in the nuclei of yeast and mammalian cells (3, 16, 33). Although van Drogen et al. found that deletion of known Fus3 interactors had no effect on the nucleocytoplasmic distribution of Fus3 (65), their assessment was not quantitative. Differences of 0.2 in the mean RNCF of a cell population are not easily discerned, despite their significance. Alternatively, the retention factors responsible for the pheromone-induced change in Fus3 distribution may not be known.
Involvement of Kap104 in the pheromone response.
It is clear from our data that Kap104 is required for proper regulation of the pheromone response. Deletion of the carboxy-terminal 91 amino acids of Kap104, which make up about one-quarter of the presumed cargo-binding domain, confers supersensitivity to pheromone in cells expressing wild-type Gpa1 and compromises the hyperadaptive activity of Gpa1E364K and excess Msg5. These effects of the kap104Δ827-918 mutation were apparent both in pheromone-induced growth inhibition assays (Fig. 5A) and in pheromone-induced transcriptional assays (data not shown) and could not be attributed to abnormally low expression of either Gpa1 or kap104Δ827-918 (Fig. 5B and C). Furthermore, the kap104Δ827-918 mutation suppressed the effects of excess Gpa1E364K and excess Msg5 on Fus3 localization (Fig. 3H and I). Thus, the epistatic effects of kap104Δ827-918 on Msg5- and Gpa1-mediated adaptation to pheromone correlate with restoration of normal Fus3 levels in the nuclei of stimulated cells.
A second indication that Gpa1-mediated adaptation depends on Kap104 is that the hyperadaptive activity of Gpa1E364K is potentiated by Kap104 overexpression. Approximately twice as many cells expressing Gpa1E364K and excess Kap104 can overcome what are normally lethal concentrations of pheromone, as can control cells expressing Gpa1E364K and the wild-type level of Kap104 (Fig. 6A and B), despite the decreased growth rate and increased sensitivity to pheromone conferred by Kap104 overexpression. Immunoblot analysis demonstrated that this enhanced resistance to pheromone is not due to an increase in the steady-state level of Gpa1 (Fig. 6C). Thus, changes in both the structure and expression of Kap104 affect the adaptive signaling mechanism(s) stimulated by Gpa1: overexpression of Kap104 enhances recovery, whereas truncation of Kap104 confers an adaptive defect.
How does Kap104 affect Fus3 localization? One possibility is that Kap104 imports a protein that acts as a nuclear tether for Fus3, although this is purely speculative. Another possibility is that Kap104 directly exports Fus3. This is unlikely because there is no precedent for a Ran-dependent karyopherin protein acting as both an importer and an exporter. Based on the structural and biochemical evidence, interaction of a given karyopherin with Ran-GTP in the nucleus is thought to stimulate either cargo release (for an importer) or cargo binding (for an exporter) (see references 7 and 41 and references therein). Kap104 is an import receptor. It is concentrated on the cytosolic side of nuclear pore complexes, where it is thought to interact with the RNA-binding proteins Nab2 and Nab4 (1). Thus far, Nab2 and Nab4 are the only known Kap104 cargo proteins. Moreover, Fus3 does not contain a region similar to the Kap104-binding domain found in Nab2 (56, 63) or a stretch of residues like the M9 sequence of hnRNP A1 (rich in glycine, serine, and asparagine) that mediates the interaction of human transportin with its cargo (67). If Kap104 is not itself the Fus3 export receptor, then perhaps it recycles the unknown transporter. After cargo release, all transport receptors must move back or be moved back through the NPCs to their point of origin. This is true for both importers and exporters. In at least one case, recycling of one transporter depends on the action of another: karyopherin α (importin) is returned to the cytoplasm by the exporter, CAS1/Cse1 (30). A deficiency in recycling the Fus3 exporter could lead to increased levels of Fus3 in the nucleus, just as we observed when kap104Δ827-918 cells overexpressing Gpa1E364K or Msg5 were challenged with pheromone (Fig. 3). Of course, this explanation assumes that Fus3 is escorted across the nuclear membrane by specific transport receptors, and this has not been established. Fus3 may bind directly to nuclear pore proteins, as does Erk2 (66).
A third way in which Kap104 might affect the pheromone response pathway is by ensuring the proper expression of a key regulator. Because Kap104 is essential for the reimport of Nab2 and Nab4 and thus indirectly responsible for the export of mature mRNAs, loss of Kap104 function would be expected to adversely affect gene expression. Given that Nab2-GFP is mislocalized in kap104Δ827-918 cells (Fig. 7), it would be surprising if the mutant cells expressed all genes normally. What genes might be required to limit the amount of nuclear Fus3? We have found that Gpa1 and Msg5 are likely to play a role in controlling the localization of Fus3. However, the steady-state level of Gpa1 is not reduced in mutant strain A11 (Fig. 5B), and the signal intensity of an Msg5-GFP reporter appears to be unaffected by the truncation of Kap104 (data not shown). The phenotypes conferred by kap104Δ827-918 could also be due to a decrease in the level of an unknown Fus3 exporter, an increase in the level of a Fus3 nuclear tether or, less directly, by loss of transcription factors such as Mot2 or Mot3, which are required for downregulation of the mating signal (5, 8, 21, 23, 31, 38, 47). Although the mechanism is unclear, our data indicate that Kap104 is required for proper regulation of the pheromone response and in particular for the Gpa1/Msg5-mediated effect on Fus3 localization.
Acknowledgments
E.B. and I.M.H. contributed equally to this study.
We thank John Aitchison for the Kap104 antibody and helpful discussions, Sue Liebman and Kent Duncan for plasmids, and Metodi Metodiev for suggesting the Fus3K42R-GFP experiment.
This work was supported by American Cancer Society Research grant RPG-94-034-06-MBC and by National Science Foundation grant MCB-0111397.
REFERENCES
- 1.Aitchison, J. D., G. Blobel, and M. P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624-627. [DOI] [PubMed] [Google Scholar]
- 2.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alepuz, P. M., A. Jovanovic, V. Reiser, and G. Ammerer. 2001. Stress-induced MAP kinase Hog1 is part of transcription activation complexes. Mol. Cell 7:767-777. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Cade, R. M., and B. Errede. 1994. MOT2 encodes a negative regulator of gene expression that affects basal expression of pheromone-responsive genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3139-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, K.-Y., J. E. Kranz, S. K. Mahanty, K.-S. Park, and E. A. Elion. 1999. Characterization of Fus3 localization: active Fus3 localizes in complexes of varying size and specific activity. Mol. Biol. Cell 10:1553-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chook, Y. M., and G. Blobel. 1999. Structure of the nuclear transport complex karyopherin-β2Ran-GppNHp. Nature 399:230-237. [DOI] [PubMed] [Google Scholar]
- 8.Clark, K. L., and G. F. Sprague. 1989. Yeast pheromone response pathway: characterization of a suppressor that restores mating to receptorless mutants. Mol. Cell. Biol. 9:2682-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, G. M., D. E. Stone, and S. I. Reed. 1990. Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol. Cell. Biol. 10:510-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohlman, H. G., D. Apaniesk, Y. Chen, J. Song, and D. Nusskern. 1995. Inhibition of G-protein signaling by mutants with dominant gain-of-function mutations in Sst2p, a pheromone desensitization factor in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:3635-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dohlman, H. G., J. Song, D. Ma, W. E. Courchesne, and J. Thorner. 1996. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, genetic interaction, and physical association with Gpa1 (G protein α subunit). Mol. Cell. Biol. 16:5194-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohlman, H. G., and J. W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703-754. [DOI] [PubMed] [Google Scholar]
- 13.Doi, K., A. Gartner, G. Ammerer, B. Errede, H. Shinkawa, K. Sugimoto, and K. Matsumoto. 1994. MSG5, a novel protein phosphatase promotes adaptation to pheromone response in S. cerevisiae. EMBO J. 13:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan, K., J. G. Umen, and C. Guthrie. 2000. A putative ubiquitin ligase required for efficient mRNA export differentially affects hnRNP transport. Curr. Biol. 10:687-696. [DOI] [PubMed] [Google Scholar]
- 15.Ferrigno, P., F. Posas, D. Koepp, H. Saito, and P. A. Silver. 1998. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin β homologs NMD5 and XPO1. EMBO J. 17:5606-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaits, F., G. Degols, K. Shiozaki, and P. Russell. 1998. Phosphorylation and association with the transcription factor Atf1 regulate localization of Spc1/Sty1 stress-activated kinase in fission yeast. Genes Dev. 12:1464-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartner, A., K. Nasmyth, and G. Ammerer. 1992. Signal transduction in Saccharomyces cerevisiae requires tyrosine and threonine phosphorylation of FUS3 and KSS1. Genes Dev. 6:1280-1292. [DOI] [PubMed] [Google Scholar]
- 18.Gietz, R. D., and A. Sugino. 1988. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 19.Gillen, K. M., M. Pausch, and H. G. Dohlman. 1998. N-terminal domain of Gpa1 (G protein alpha subunit) is sufficient for plasma membrane targeting in yeast Saccharomyces cerevisiae. J. Cell Sci. 111:3235-3244. [DOI] [PubMed] [Google Scholar]
- 20.Giniger, E., and M. Ptashne. 1988. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc. Natl. Acad. Sci. USA 85:382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grishin, A. V., M. Rothenberg, M. A. Downs, and K. J. Blumer. 1998. Mot3, a Zn finger transcription factor that modulates gene expression and attenuates mating pheromone signaling in Saccharomyces cerevisiae. Genetics 149:879-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP Kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irie, K., K. Yamaguchi, K. Kawase, and K. Matsumoto. 1994. The yeast MOT2 gene encodes a putative zinc finger protein that serves as a global negative regulator affecting expression of several categories of genes, including mating-pheromone-responsive genes. Mol. Cell. Biol. 14:3150-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacoby, T., H. Flanagan, A. Faykin, A. G. Seto, C. Mattison, and I. Ota. 1997. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J. Biol. Chem. 272:17749-17755. [DOI] [PubMed] [Google Scholar]
- 26.Kaffman, A., and E. K. O'Shea. 1999. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 15:291-339. [DOI] [PubMed] [Google Scholar]
- 27.Kao, L.-R., J. Peterson, R. Ji, L. Bender, and A. Bender. 1996. Interactions betweeen the ankyrin repeat-containing protein Akr1p and the pheromone response pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:168-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khokhlatchev, A. V., B. Canagarajah, J. Wilsbacher, M. Robinson, M. Atkinson, E. Goldsmith, and M. H. Cobb. 1998. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93:605-615. [DOI] [PubMed] [Google Scholar]
- 29.Konopka, J. B., D. D. Jenness, and L. H. Hartwell. 1988. The C-terminus of the Saccharomyces cerevisiae α-pheromone receptor mediates an adaptive response to pheromone. Cell 54:609-620. [DOI] [PubMed] [Google Scholar]
- 30.Kutay, U., F. R. Bischoff, S. Kostka, R. Kraft, and D. Gorlich. 1997. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90:1061-1071. [DOI] [PubMed] [Google Scholar]
- 31.Leberer, E., D. Dignard, D. Harcus, M. Whiteway, and D. Y. Thomas. 1994. Molecular characterization of SIG1, a Saccharomyces cerevisiae gene involved in negative regulation of G-protein-mediated signal transduction. EMBO J. 13:3050-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, D. C. Y., and J. D. Aitchison. 1999. Kap104p-mediated nuclear import. J. Biol. Chem. 274:29031-29037. [DOI] [PubMed] [Google Scholar]
- 33.Lenormand, P., J. M. Brondello, A. Brunet, and J. Pouyssegur. 1998. Growth factor-induced p42/p44 MAPK nuclear translocation and retention requires both MAPK activation and neosynthesis of nuclear anchoring protiens. J. Cell Biol. 142:625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leza, M. A., and E. A. Elion. 1999. POG1, a novel yeast gene, promotes recovery from pheromone arrest via the G1 cyclin CLN2. Genetics 151:531-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, E., M. J. Cismowski, and D. E. Stone. 1998. Phosphorylation of the pheromone responsive Gβ protein of Saccharomyces cerevisiae does not affect its mating-specific signaling function. Mol. Gen. Genet. 258:608-618. [DOI] [PubMed] [Google Scholar]
- 36.Li, E., E. Meldrum, H. Stratton, and D. E. Stone. 1998. Substitutions in the pheromone responsive Gβ protein of Saccharomyces cerevisiae confer a defect in recovery from pheromone treatment. Genetics 148:947-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, J.-J., and S. Lindquist. 1999. Oligopeptide-repeat expansions modulate “protein only” inheritance in yeast. Nature 400:573-576. [DOI] [PubMed] [Google Scholar]
- 38.Lopes, M. D. B., J.-Y. Ho, and S. I. Reed. 1990. Mutations in cell division cycle genes CDC36 and CDC39 activate the Saccharomyces cerevisiae mating pheromone response pathway. Mol. Cell. Biol. 10:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKay, V. L., S. K. Welch, M. Y. Insley, T. R. Manney, J. Holley, G. C. Saaru, and M. L. Parker. 1988. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc. Natl. Acad. Sci. USA 85:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcus, S., C.-B. Xue, F. Nadier, and J. M. Becker. 1991. Degradation of a-factor by a Saccharomyces cerevisiae α-mating-type specific endopeptidase: evidence for a role in recovery of cells from G1 arrest. Mol. Cell. Biol. 11:1030-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattaj, I. W., and E. Conti. 1999. Snail mail to the nucleus. Nature 399:208-210. [DOI] [PubMed] [Google Scholar]
- 42.Mattison, C. P., and I. M. Ota. 2000. Two protein tyrosine phosphatases, Ptp2 and Ptp3, modulate the subcelluar localization of the Hog1 MAP kinase in yeast. Genes Dev. 14:1229-1235. [PMC free article] [PubMed] [Google Scholar]
- 43.Mendenhall, M. D., H. E. Richardson, and S. I. Reed. 1988. Dominant negative protein kinase mutations that confer a G1 arrest phenotype. Proc. Natl. Acad. Sci. USA 85:4426-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metodiev, M. V., D. Matheos, M. D. Rose, and D. E. Stone. 2002. Regulation of MAPK function by direct interaction with the mating-specific Gα in yeast. Science 296:1483-1486. [DOI] [PubMed] [Google Scholar]
- 45.Miyajima, I., K. Arai, and K. Matsumoto. 1989. GPA1val-50 mutation in the mating-factor signaling pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:2289-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, S. 1984. Yeast cells recover from mating pheromone α factor-induced division arrest by desensitization in the absence of α factor destruction. J. Biol. Chem. 259:1004-1010. [PubMed] [Google Scholar]
- 47.Neiman, A. M., F. Chang, K. Komachi, and I. Herskowitz. 1990. CDC36 and CDC39 are negative elements in the signal transduction pathway of yeast. Cell Regul. 1:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pryciak, P. M., and L. H. Hartwell. 1996. AKR1 encodes a candidate effector of the Gβγ complex in the Saccharomyces cerevisiae pheromone response pathway and contributes to control of both cell shape and signal transduction. Mol. Cell. Biol. 16:2614-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed, S. I., J. A. Hadwiger, and A. T. Lorincz. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA 82:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reiser, V., G. Ammerer, and H. Ruis. 1999. Nucleocytoplasmic traffic of MAP kinases. Gene Expr. 7:247-254. [PMC free article] [PubMed] [Google Scholar]
- 51.Reiser, V., H. Ruis, and G. Ammerer. 1999. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10:1147-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reneke, J. E., K. J. Blumer, W. E. Courchesne, and J. Thorner. 1988. The carboxy-terminal segment of the yeast α-factor receptor is a regulatory domain. Cell 55:221-234. [DOI] [PubMed] [Google Scholar]
- 53.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennet, Y. D. D. He, H. Y. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 55.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Siomi, M. C., M. Fromont, J.-C. Rain, L. Wan, F. Weng, P. Legrain, and G. Dreyfuss. 1998. Functional conservation of the transportin nuclear import pathway in divergent organisms. Mol. Cell. Biol. 18:4141-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slater, M. R., and E. A. Craig. 1987. Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1906-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song, J., J. Hirschman, K. Gunn, and H. B. Dohlman. 1996. Regulation of membrane and subunit interactions by N-myristoylation of a G protein α subunit in yeast. J. Biol. Chem. 271:20273-20283. [DOI] [PubMed] [Google Scholar]
- 59.Spain, B. H., D. Koo, M. Ramakrishnan, B. Dzudzor, and J. Colicelli. 1995. Truncated forms of a novel yeast protein suppress the lethality of a G protein α subunit deficiency by interacting with the β subunit. J. Biol. Chem. 270:25435-25444. [DOI] [PubMed] [Google Scholar]
- 60.Stevenson, B. J., B. Ferguson, C. D. Virgilio, E. Bi, J. R. Pringle, G. Ammerer, and G. F. Sprague. 1995. Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 9:2949-2963. [DOI] [PubMed] [Google Scholar]
- 61.Stone, D. E., and S. I. Reed. 1990. G protein mutations that alter the pheromone response in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:4439-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stratton, H. F., J. Zhou, S. I. Reed, and D. E. Stone. 1996. The mating-specific Gα protein of Saccharomyces cerevisiae downregulates the mating signal by a mechanism that is dependent on pheromone and independent of Gβγ sequestration. Mol. Cell. Biol. 16:6325-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Truant, R., R. A. Fridell, R. E. Benson, H. Bogerd, and B. R. Cullen. 1998. Identification and functional characterization of a novel nuclear localization signal present in the yeast Nab2 poly(A)+ RNA binding protein. Mol. Cell. Biol. 18:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trueheart, J., J. D. Boeke, and G. R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Drogen, F., V. M. Stucke, G. Jorritsma, and M. Peter. 2001. MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 3:1051-1059. [DOI] [PubMed] [Google Scholar]
- 66.Whitehurst, A. W., J. L. Wilsbacher, Y. You, K. Luby-Phelps, M. S. Moore, and M. H. Cobb. 2002. ERK2 enters the nucleus by a carrier-independent mechanism. Proc. Natl. Acad. Sci. USA 99:7496-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wozniak, R. W., M. P. Rout, and J. D. Aitchison. 1998. Karyopherins and kissing cousins. Trends Cell Biol. 8:184-188. [DOI] [PubMed] [Google Scholar]
- 68.Zhan, X.-L., R. J. Deschenes, and K.-L. Guan. 1997. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 11:1690-1702. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, J., M. Arora, and D. E. Stone. 1999. The yeast pheromone-responsive Gα protein stimulates recovery from chronic pheromone treatment by two mechanisms that are activated at distinct levels of stimulus. Cell Biochem. Biophys. 30:193-212. [DOI] [PubMed] [Google Scholar]