Abstract
Proteins containing PDZ domains are involved in a large number of biological functions, including protein scaffolding, organization of ion channels, and signal transduction. We recently identified a novel PDZ domain-containing protein, PDZK1, that is selectively expressed in normal tissues, where it is associated and colocalized with MAP17, a small 17-kDa membrane-associated protein; cMOAT, an organic anion transporter implicated in multidrug resistance; and the type IIa Na/Pi cotransporter. The protein cluster formed by PDZK1, MAP17, and cMOAT is upregulated in a significant number of human carcinomas originating in the colon, breast, lung, and kidney. In order to better define the function of PDZK1 in the protein cluster and its potential role in the organization of ion channels, we generated a PDZK1 knockout mouse. While PDZK1-deficient mice developed normally, did not display any gross phenotypic abnormalities, and were fecund, lack of PDZK1 resulted in modulation of expression of selective ion channels in the kidney, as well as increased serum cholesterol levels. However, no significant redistribution of proteins known to interact with PDZK1, such as MAP17, cMOAT, and the type IIa Na/Pi cotransporter, was observed. The absence of a more significant phenotype in PDZK1-deficient mice may be due to functional compensation by other PDZ domain-containing proteins, which could be instrumental in determining the location of interacting proteins such as ion channels and other membrane-associated proteins in defined areas of the plasma membrane.
PDZK1, a recently described protein containing four PDZ domains, belongs to a cluster of proteins including MAP17 and cMOAT (7-10). All three proteins are upregulated in human carcinomas arising in the kidney, lung, colon, and breast. Although the exact function of PDZK1 is unknown, it has been postulated that it plays a role in multidrug resistance through its interaction with the organic anion transporter cMOAT, also known as MRP2, the multidrug resistance-associated protein (9, 11, 12, 14, 24). More recently, PDZK1 has been found to interact with the type IIa Na/Pi cotransporter and therefore may participate in the apical sorting of ion channels (4). Furthermore, PDZK1 is overexpressed in estrogen receptor-positive breast carcinomas compared to estrogen-negative tumors, suggesting a role for PDZK1 in tissue response to β-estradiol (3). PDZ domains were originally recognized as structural motifs in the mammalian postsynaptic density protein PSD-95 (1), the Drosophila disk large tumor suppressor Dlg (26), and the tight junction protein ZO-1 (25). Such domains, typically 80 to 120 amino acids, bind to well-defined consensus sequences and have been described in a number of proteins associated with specialized areas of the plasma membrane (2, 22). PDZ domain-containing proteins are involved in synaptic organization, control of cell proliferation, and cell differentiation (1, 6, 13, 20, 26). Some of these proteins contain several PDZ domains and, as a result, promote the clustering of a small group of proteins and organize complex biological functions, such as signal transduction (19, 21). Little is known about the function of PDZ domain-containing proteins during development and about the dispensability of such proteins. We report here the generation and characterization a PDZK1 knockout mouse and demonstrate that disruption of the gene for PDZK1 is not associated with abnormal growth and development or redistribution of interacting proteins. However, disruption of the gene for PDZK1 results in downregulation of selective ion channels and membrane-associated protein gene expression, together with increased serum cholesterol levels, in knockout mice.
MATERIALS AND METHODS
Cloning of the mouse PDZK1 cDNA.
A mouse kidney 5′ stretch plus cDNA (Clontech, Palo Alto, Calif.) was hybridized with a [32P]dCTP-labeled BglII DNA fragment containing the complete sequence of the human PDZK1 cDNA. A full-length recombinant cDNA clone was sequenced by using oligonucleotide primers (Integrated DNA Technologies, Coralville, Iowa) and a 373 automated DNA sequencer from Applied Biosystems (Branchburg, N.J.).
Isolation and mapping of the mouse gene for PDZK1.
Two pairs of oligonucleotide primers derived from the mouse PDZK1 cDNA sequence (Integrated DNA Technologies) were used to screen a 129SvJ BAC genomic library (Genome System, St. Louis, Mo.), from which two clones were obtained. Plasmid DNA was prepared by using a Qiagen kit for large constructs (Qiagen, Valencia, Calif.). Sequencing was conducted by starting with oligonucleotide primers derived from the cDNA clone sequence, followed by oligonucleotides derived from the introns. A map of the gene for PDZK1 was generated by using the sequencing data (Fig. 1A).
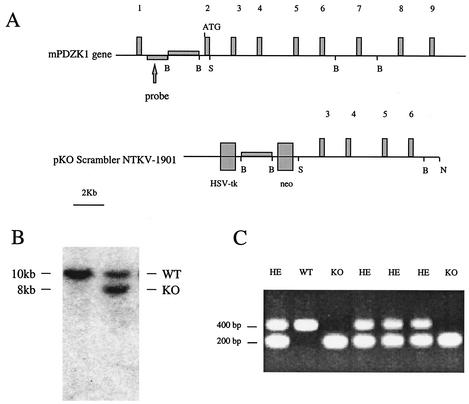
FIG. 1.
(A) Structure of the mouse gene for PDZK1 containing 9 exons. The start site of the protein is indicated in exon 2, as is the area of the gene used as a probe to screen ES cell clones for homologous recombination. The positions of restriction sites used to construct the targeting vector (B, BamHI; S, SalI; N, NotI) are also indicated. The plasmid pKO Scrambler NTKV-1901 was used to construct the targeting vector. (B) Representative Southern blot including a wild-type and a targeted ES cell clone hybridized with the external probe after digestion with BglII and XhoI. The wild-type band is approximately 10 kb, while the recombined allele is shortened to approximately 8 kb. (C) Representative PCR genotyping of tail DNA. The wild-type (WT) allele produces a band at approximately 400 bp, while the mutant allele produces a band at approximately 200 bp. Homozygous normal (WT) mice show only one band at 400 bp, and heterozygous mice (HE) show both 400- and 200-bp bands, while knockout (KO) mice show only the 200-bp band.
Construction of the targeting vector.
A 2.7-kb BamHI DNA fragment upstream from the translation start codon of the gene for PDZK1 was isolated from the BAC genomic clone and inserted into the BglII site of the pKO Scrambler NTKV-1901 plasmid (Stratagene, La Jolla, Calif.). An 8-kb SalI-BamHI DNA fragment from the BAC clone corresponding to the sequences extending from the second intron to the sixth intron were inserted between the SalI and BamHI sites of plasmid pBluescript in order to add a NotI restriction site. The 8-kb SalI-NotI DNA fragment was inserted into pKO Scrambler NTKV-1901 already containing the 5′ region of the gene for PDZK1 in order to complete the construction of the targeting vector (Fig. 1A). The construct was verified by DNA sequencing by using specific primers. The recombinant plasmid was linearized with NotI prior to transfection of ES cells.
Transfection of ES cells.
Transfections of AB2.2-Prime ES cells (Mouse Kit; Stratagene) were performed with a Bio-Rad Gene Pulser using a 0.4-cm electrode gap cuvette (Bio-Rad, Hercules, Calif.) at 230 V and 500 μF of capacitance in a 0.9-ml volume of phosphate-buffered saline containing 10 × 106 cells and 15 μg of linearized targeting vector. Transfected ES cells were immediately plated on neomycin-resistant, mitotically inactivated mouse embryonic fibroblasts in Dulbecco modified Eagle medium containing 14% fetal bovine serum and supplemented with 2 mM l-glutamine, 0.1 mM β-mercaptoethanol, 50 U of penicillin G per ml, and 50 μg of streptomycin sulfate per ml. G418, at a concentration of 300 μg/ml, was added 24 h following the transfection to select for neomycin-resistant clones. The selection was carried out for 11 days, after which single colonies were picked by using pipette tips and transferred into two sets of 96-well plate replicas, one set for expanding the clones to be frozen and the other set for DNA analysis.
Southern blot analysis.
After the cells reached confluence, genomic DNA was extracted from ES cell clones for DNA analysis in accordance with the manufacturer's (Stratagene) protocol. After purification, the genomic DNA was digested with BglII and XhoI and run on 1% agarose gels containing 0.5 μg of ethidium bromide per ml in TAE buffer with appropriate size markers, examined under UV light, and transferred to Biodyne filters (Pall Filter, Glen Cove, N.Y.). After blotting, filters were UV cross-linked by using a Stratalinker (Stratagene) and hybridized by using a [32P]dCTP-labeled 1.6-kb DNA fragment (probe) located on the gene for PDZK1 upstream from the area of the gene used to make the targeting vector (Fig. 1A). Following the hybridization, filters were exposed to Kodak XAR film at −80°C (Fig. 1B).
Chimeras.
Two ES cell clones that had undergone homologous recombination of the gene for PDZK1 with the targeting vector were injected into C57BL/6 blastocysts at 10 to 15 cells per injection. Embryos were transferred into pseudopregnant mice at 5 to 10 blastocysts per uterine horn. Coat color chimeras were mated to either C57BL/6 or 129SvEv mice.
Genotyping of offspring.
Tail DNA was prepared by using a Qiagen DNA extraction kit in accordance with the manufacturer's protocol. Three oligonucleotide primers were designed to be used in a single PCR allowing the distinction among wild-type, heterozygous, and knockout mice. The primer sequences were the following: wild type, 5′-GACACCATTGATCCTGAGCACCCTG-3′; common, 5′-CATGCAATTGGATGCGTGGGTC-3′; knockout, 5′-CTCTATGGCTTCTGAGGCGGAAAG-3′. Amplicons generated by PCR were 426 bp for the wild-type gene and 212 bp for the knockout gene; heterozygous mice showed both amplicons (Fig. 1C).
Western blot analysis and immunoperoxidase studies.
Affinity-purified antibodies against PDZK1 and MAP17 have been previously characterized (8, 10). A polyclonal antibody was prepared against the 30-mer carboxy-terminal peptide of the cMOAT protein sequence (GSPEELLQIPGPFYFMAKEAGIENVNSTKF; Genemed Biotechnologies, Inc., South San Francisco, Calif.) (24). A cysteine was added to the sequence at its NH2 terminus in order to conjugate the peptide to keyhole limpet hemocyanin. Rabbits were immunized, and the antibody was purified by affinity chromatography using the same peptide bound to a SulfoLink column (Pierce, Rockford, Ill.). The type IIa Na/Pi cotransporter antibody was a gift from Jurg Biber (University of Zurich, Zurich, Switzerland). The anti-actin antibody was purchased from Sigma (St. Louis, Mo.).
For Western blot analysis, protein extracts prepared form PDZK1 knockout and wild-type adult mice were electrophoresed on sodium dodecyl sulfate-polyacrylamide gels, transferred to nitrocellulose, and incubated with the anti-PDZK1, anti-cMOAT, or anti-actin antibody and subsequently with anti-chicken or anti-rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase (GIBCO BRL, Gaithersburg, Md.). These incubations were followed by the ECL chemiluminescence reaction using the Super Signal West Pico Luminal reagents (Pierce).
For immunoperoxidase studies, tissues harvested from PDZK1 knockout and wild-type adult mice were fixed for 4 h in 4% paraformaldehyde in phosphate-buffered saline, pH 7.4, at 4°C and then transferred to 30% sucrose in phosphate-buffered saline, pH 7.4, overnight at 4°C. Tissues were then frozen in OCT compound (Miles Diagnostics, Elkhart, Ind.) and stored at −80°C. Immunoperoxidase studies were performed on 6-μm fixed-frozen tissue sections using either the affinity-purified primary antibody against PDZK1, MAP17, cMOAT, or the type IIa Na/Pi cotransporter. Normal chicken IgY or rabbit IgG was used as a negative control. The sections were then incubated with biotinylated anti-chicken or anti-rabbit IgG using a 1/200 dilution (Vector, Burlingame, Calif.) and subsequently treated with the Vectastain ABC reagents (Vector) and diaminobenzidine (Research Genetics, Inc., Huntsville, Ala.) in accordance with the manufacturers' protocols.
RNA isolation.
Whole kidneys were surgically removed from PDZK1 knockout and wild-type adult mice, frozen on dry ice, and stored at −80°C until use. Frozen tissue samples were simultaneously disrupted and homogenized for up to 60 s by using a Polytron (Kinematica, Lucerne, Switzerland) in lysis buffer RLT (Qiagen) containing 1.45 M β-mercaptoethanol. Total RNA was further purified in accordance with the manufacturer's protocol for the Qiagen RNeasy Mini Kit.
RT.
Total RNA was reverse transcribed by using the ABI Prism 7700 Sequence Detection System and TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, Calif.). Each reaction mixture contained 1× reverse transcription (RT) buffer containing 5.5 mM MgCl2, 500 μM each deoxynucleoside triphosphate, 2.5 μM random hexamer, 0.4 U of RNase inhibitor, 1.25 U of multiscribe reverse transcriptase/μl, 0.1 μg of total RNA, and RNase-free distilled H2O (Sigma) to a total volume of 10 μl per reaction mixture. The RT reaction cycle parameters were 10 min at 25°C, 30 min at 48°C, and 5 min at 95°C.
SYBR green real-time quantitative PCR.
Oligonucleotide primers were designed by using Primer Express software version 1.5, on the basis of gene sequences obtained from the National Center for Biotechnology Information database. All reactions were performed with an ABI Prism 7700 Sequence Detection System (Applied Biosystems). Reactions were carried out in duplicate in a 50-μl volume containing reverse-transcribed cDNAs, 25 μl of 2× SYBR Green Master Mix, and each forward and reverse primer at a concentration of 50 nM. Conditions for all SYBR Green PCRs were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 95°C for 10 s and 60°C for 1 min. Data were gathered and analyzed by using the SDS 1.9 software on a G4 Power Macintosh computer (Apple Computer, Cupertino, Calif.). Results were normalized with respect to glyceraldehyde-3-phosphate dehydrogenase expression. Ct values were exported into a Microsoft Excel worksheet for calculation of gene expression in accordance with the delta delta Ct method (Applied Biosystems). Levels of gene expression in knockout mice were represented with respect to those in wild-type animals as knockout/wild-type ratios. P values were calculated for all of the genes investigated.
Serum and urine chemistries.
Blood samples were collected from PDZK1 knockout and wild-type adult mice by cardiac puncture and subsequently centrifuged to recover sera, which were analyzed by the Massachusetts Institutes of Technology Department of Comparative Medicine Laboratory (Boston, Mass.). Urine samples were analyzed by the Clinical Laboratory at Beth Israel-Deaconess Medical Center.
RESULTS AND DISCUSSION
Protein sequences derived from human and mouse PDZK1 cDNAs were compared by using the Lasergene software. The analysis revealed 78.2% homology between the human and mouse protein sequences and that the PDZ protein interaction motifs were conserved between the two species (GenBank database accession no. AF012281 and AF220100, respectively).
Sequence analysis using specific oligonucleotides revealed that the mouse gene for PDZK1 contains nine exons, the first one upstream from the protein initiation codon (Fig. 1A).
The pKO scrambler NTKV-1901 vector containing approximately 10 kb of the mouse gene for PDZK1 interrupted by a pgkneo cassette was used as a targeting vector and transfected into low-passage ES cells (Stratagene) (Fig. 1A). Seven hundred colonies were isolated after 10 days of selection with G418. Twenty clones showed homologous recombination by Southern blot analysis hybridized with a 1.6-kb probe located upstream from the sequences used to build the targeting vector on the mouse gene for PDZK1 (Fig. 1A). Wild-type clones showed a single hybridization band at about 10 kb, while clones with homologous recombination showed an additional band at 8 kb (Fig. 1B). Two independent clones were injected into C57BL/6 blastocysts. Chimeras derived from both clones transmitted the targeted allele to the germ line. Offspring were genotyped by using a three-oligonucleotide primer PCR (Fig. 1C). Crossing of heterozygous mice produced viable homozygous offspring in both the C57BL/6 and pure 129SvEV backgrounds.
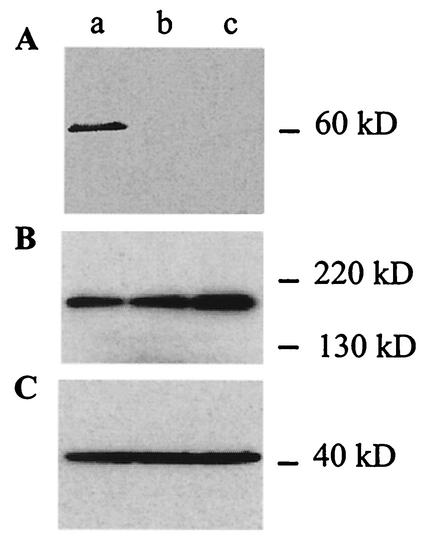
Real-time PCR performed comparing the expression of PDZK1 in kidney and liver from wild-type and knockout mice revealed that the mRNA for PDZK1 is absent in knockout mice (data not shown). Western blot analysis confirmed the absence of PDZK1 expression in knockout mouse kidney samples (Fig. 2A), while cMOAT and actin were detected in both wild-type and knockout mouse kidney samples (Fig. 2B and C). Immunoperoxidase studies using a PDZK1 polyclonal antibody showed the typical localization of PDZK1 associated with the brush border of kidney proximal tubular epithelial cells in wild-type mice, while no staining was observed in knockout counterparts (Fig. 3) (10).
FIG. 2.
Western blot analysis of wild-type mouse kidney (lane a) and PDZK1 knockout mice derived from two independent ES cell clones (lanes b and c) incubated with affinity-purified PDZK1 (A), cMOAT (B), and actin (C) antibodies. A single band at approximately 60 kDa corresponding to PDZK1 is detected in wild-type mouse kidney, while it is absent in knockout mouse kidney samples (A), while a single band at approximately 190 kDa corresponding to cMOAT (B) and a single band at approximately 40 kDa corresponding to actin (C) are detected in wild-type and knockout kidney samples.
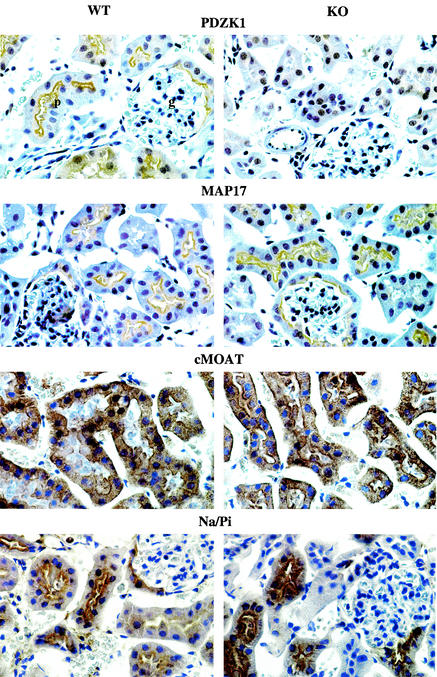
FIG. 3.
Immunoperoxidase staining of renal cortex from wild-type (WT) and knockout (KO) mice with PDZK1, MAP17, cMOAT, and the type IIa Na/Pi cotransporter. Note the typical staining pattern obtained for all four proteins associated with the apical border of proximal tubular epithelial cells in wild-type mice (p, proximal tubule; g, glomerulus). The staining of PDZK1 is absent in knockout mice, while there is no difference between wild-type and knockout mice in the staining or distribution of MAP17, cMOAT, and the type IIa Na/Pi cotransporter. Magnification, ×300.
Mice homozygous for the targeted gene were observed for more than a year. They developed normally and showed no obvious alteration in gross morphology, weight, size, and fecundity compared to wild-type mice. Breeding of mice heterozygous for the targeted gene for PDZK1 produced the expected Mendelian ratio of knockout animals of 25%. No change in the histology of multiple organs of both groups was observed (data not shown).
Immunoperoxidase studies were performed by using MAP17, cMOAT, and type IIa Na/Pi cotransporter antibodies on wild-type and knockout mouse kidneys (Fig. 3). No difference in signal intensity or localization of all three proteins was seen. In wild-type mice, PDZK1 was associated with the apical brush border of proximal tubular epithelial cells, as previously described (10). No staining for PDZK1 was seen in knockout mice. However, MAP17, cMOAT, and the type IIa Na/Pi cotransporter displayed the same characteristic distribution in wild-type and knockout mice.
RT-PCR experiments were conducted with total mRNA extracted from adult PDZK1 knockout and wild-type mouse kidneys. The levels of expression of ion channels and proteins known to be predominantly expressed in proximal tubular cells of the renal cortex were investigated, including the proteins known to interact with PDZK1, namely, MAP17, cMOAT, and the type IIa Na/Pi cotransporter (4, 9, 23). The results are summarized in Table 1. There was a significant downregulation of cationic amino acid transporter 2 (CAT-2), aquaporin I, and the postsynaptic 43-kDa protein in knockout mice compared to their wild-type counterparts (P < 0.05). All of the other ion channels and proteins investigated, including the molecules known to interact with PDZK1, did not show significant changes in the level of gene expression between PDZK1 knockout and wild-type mice (P > 0.05).
TABLE 1.
mRNA quantification in wild-type and PDZK1 knockout mouse kidney
| mRNA | Fold change (knockout/wild-type mice) |
|---|---|
| MAP17 | 0.95 ± 0.20 |
| cMOAT | 1.10 ± 0.12 |
| MDR1 | 1.15 ± 0.18 |
| Sodium-dicarboxylate transporter | 0.99 ± 0.13 |
| Organic anion-transporting polypeptide 5 | 1.28 ± 0.25 |
| Brain-derived voltage-dependent ion channel | 1.02 ± 0.16 |
| Synaptic glycoprotein | 0.87 ± 0.12 |
| Postsynaptic 43-kDa protein | 0.67 ± 0.08a |
| Sodium-potassium ATPase | 0.80 ± 0.13 |
| Sodium-sulfate cotransporter | 0.88 ± 0.16 |
| H+-peptide transporter | 0.98 ± 0.18 |
| Aquaporin 1 | 0.53 ± 0.18a |
| CAT-2 | 0.38 ± 0.14a |
| Brain K+ channel protein 1 | 1.00 ± 0.16 |
| Glycine transporter 1 | 1.06 ± 0.09 |
| Type II sodium-phosphate cotransporter | 1.05 ± 0.07 |
| Sodium-glucose cotransporter | 1.00 ± 0.24 |
P < 0.05.
We have no evidence from our own experiments or published reports that any of the three modulated proteins interact with PDZK1. Aquaporin 1, CAT-2, and the postsynaptic 43-kDa protein have different functions. Aquaporin 1 is one of the 10 aquaporins known to transport solute-free water across cell membranes. In the kidney, it plays a role in concentrating urine (5). CAT-2 is a member of a family of closely related cationic amino acid transporters responsible for the transport of arginine and lysine in mammalian cells and plays a major role in nitric oxide metabolism (18). The 43-kDa postsynaptic protein is colocalized in a 1:1 ratio in the postsynaptic membrane with the acetylcholine receptor and has been proposed to play a role in the organization of the acetylcholine receptor (27). The role of the postsynaptic 43-kDa protein in the kidney is unknown.
Since PDZK1 is expressed predominantly in the kidney and liver, serum and urine chemistry profiles were obtained to identify any abnormalities of renal and hepatic functions resulting from the targeted disruption of the gene for PDZK1. Serum analysis revealed a significant increase in the cholesterol levels in knockout mice compared to those in wild-type mice (Table 2). All other levels in serum, including those of markers of liver and renal function, were similar in both groups. Urinalysis did not reveal any significant difference between knockout and wild-type mice (Table 3).
TABLE 2.
Comparison of serum chemistry profiles of wild-type and PDZK1 knockout mice
| Parameter | Wild-type mice | Knockout mice |
|---|---|---|
| Alkaline phosphatase (IU/liter) | 55.0 ± 6.0 | 68.5 ± 5.2 |
| Alanine transaminase (IU/liter) | 29.0 ± 1.5 | 26.5 ± 3.0 |
| Apartate aminotransferase (IU/liter) | 77.0 ± 2.9 | 101.5 ± 17.8 |
| Gamma-glutamyl transpeptidase (IU/liter) | 2.3 ± 1.5 | 0.5 ± 0.5 |
| Albumin (g/dl) | 2.9 ± 0.1 | 2.8 ± 0.1 |
| Total protein (g/dl) | 5.2 ± 0.1 | 4.8 ± 0.3 |
| Globulin (g/dl) | 2.3 ± 0.2 | 2.0 ± 0.2 |
| Total bilirubin (mg/dl) | 0.1 ± 0.0 | 0.2 ± 0.0 |
| Direct bilirubin (mg/dl) | 0.1 ± 0.0 | 0.1 ± 0.0 |
| Blood urea nitrogen (mg/dl) | 27.0 ± 1.5 | 32.3 ± 1.6 |
| Creatinine (mg/dl) | 0.1 ± 0.0 | 0.1 ± 0.0 |
| Cholesterol (mg/dl) | 89.3 ± 16.7 | 160.8 ± 8.3a |
| Glucose (mg/dl) | 230.0 ± 15.3 | 226.3 ± 11.7 |
| Calcium (mg/dl) | 9.5 ± 0.7 | 9.1 ± 0.1 |
| Phosphorus (mg/dl) | 5.6 ± 0.5 | 5.6 ± 0.4 |
| Total CO2 (mEq/liter) | 18.7 ± 2.6 | 18.3 ± 1.7 |
| Chloride (mEq/liter) | 104.0 ± 1.7 | 103.8 ± 0.5 |
| Potassium (mEq/liter) | 4.3 ± 0.4 | 4.9 ± 0.2 |
| Sodium (mEq/liter) | 142.0 ± 0.6 | 142.5 ± 1.3 |
| Albumin/globulin ratio | 1.3 ± 0.2 | 1.4 ± 0.1 |
| Na/K ratio | 33.3 ± 2.8 | 29.3 ± 1.4 |
| Indirect bilirubin (mg/dl) | 0.0 ± 0.0 | 0.1 ± 0.0 |
| Anion gap (mEq/liter) | 26.0 ± 2.5 | 25.5 ± 1.0 |
P < 0.05.
TABLE 3.
Comparison of urine chemistry profiles of wild-type and PDZK1 knockout mice
| Parameter | Wild-type mice | Knockout mice |
|---|---|---|
| Creatinine (mg/dl) | 45.0 ± 3.05 | 46.4 ± 10.0 |
| Sodium (mEq/liter) | 112.0 ± 14.5 | 116.0 ± 15.6 |
| Potassium (mEq/liter) | 364.0 ± 27.2 | 364.0 ± 43.3 |
| Chloride (mEq/liter) | 232.0 ± 18.4 | 229.0 ± 35.4 |
| Phosphorus (mEq/liter) | 170.0 ± 15.0 | 149.0 ± 19.6 |
The increase in serum cholesterol in PDZK1 knockout mice compared to their wild-type counterparts was a surprise, since there is no known evidence that PDZK1 plays a role in cholesterol metabolism. However, this is an interesting finding that warrants further investigation.
Frequently, phenotypic changes associated with the deletion of a given gene are more limited than expected, as demonstrated in two other PDZ protein knockout mice, PSD-93 and PSD-95 (15, 16). It is possible that a functional compensatory process involving other PDZ domain-containing proteins takes place in PDZK1 knockout mice, explaining, at least in part, the modest changes observed, particularly concerning the absence of redistribution of proteins known to interact with PDZK1, as demonstrated in renal tissue samples. Such compensatory mechanisms have been described in MALS/Veli (mammalian LIN-7/vertebrate homolog of LIN-7), a member of a complex of three PDZ proteins, LIN-2/7/10, involved in the basolateral localization of a receptor tyrosine kinase in Caenorhabditis elegans. Knocking out MALS-1 or MALS-2 did not reveal any significant phenotype, while knocking out both MALS-1 and -2 resulted in a dramatic upregulation of MALS-3 (17). The generation of knockout mice for proteins interacting with PDZK1, as well as double knockouts, may provide additional tools with which to understand the role of PDZK1 in the apical sorting of ion channels and multidrug resistance.
Acknowledgments
We are grateful to Jurg Biber for the gift of the type IIa Na/Pi cotransporter antibody, Joel Lawitts for blastocyst injection and generating the chimeras, and Pushpa Srivastava for performing the DNA sequence analysis.
This work was supported by grants from the Beth Israel Hospital Pathology Foundation, Inc.; the Elsa U. Pardee Foundation; and the Nell and Nancy Fund, a cancer prevention fund supported by AFLAC Incorporated and The Pine Mountain Benevolent Association.
REFERENCES
- 1.Cho, K.-O., C. A. Hunt, and M. B. Kennedy. 1992. The rat brain postsynaptic density fraction contains a homolog of the drosophila discs-large tumor suppressor protein. Neuron 9:929-942. [DOI] [PubMed] [Google Scholar]
- 2.Doyle, D. A., A. Lee, J. Lewis, E. Kim, M. Sheng, and R. MacKinnon. 1996. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85:1067-1076. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh, M. G., D. A. Thompson, and R. J. Weigel. 2000. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 60:6367-6375. [PubMed] [Google Scholar]
- 4.Gisler, S. M., I. Stagljar, M. Traebert, D. Bacic, J. Biber, and H. Murer. 2001. Interaction of the type IIa NA/Pi cotransporter with PDZ proteins. J. Biol. Chem. 276:9206-9213. [DOI] [PubMed] [Google Scholar]
- 5.Jenq, W., D. R. Cooper, P. Bittle, and G. Ramirez. 1999. Aquaporin-1 expression in proximal tubule epithelial cells of human kidney is regulated by hyperosmolarity and contrast agents. Biochem. Biophys. Res. Commun. 256:240-248. [DOI] [PubMed] [Google Scholar]
- 6.Kim, E., M. Niethammer, A. Rothschild, Y. N. Jan, and M. Sheng. 1995. Clustering of shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 378:85-88. [DOI] [PubMed] [Google Scholar]
- 7.Kocher, O., P. Cheresh, L. F. Brown, and S. W. Lee. 1995. Identification of a novel gene, selectively up-regulated in human carcinomas, using the differential display technique. Clin. Cancer Res. 1:1209-1215. [PubMed] [Google Scholar]
- 8.Kocher, O., P. Cheresh, and S. W. Lee. 1996. Identification and partial characterization of a novel membrane-associated protein (MAP17) up-regulated in human carcinomas and modulating cell replication and tumor growth. Am. J. Pathol. 149:493-500. [PMC free article] [PubMed] [Google Scholar]
- 9.Kocher, O., N. Comella, A. Gilchrist, R. Pal, K. Tognazzi, L. F. Brown, and J. H. M. Knoll. 1999. PDZK1, a novel PDZ domain-containing protein up-regulated in carcinomas and mapped to chromosome 1q21, interacts with cMOAT (MRP2), the multidrug resistance-associated protein. Lab. Investig. 79:1161-1170. [PubMed] [Google Scholar]
- 10.Kocher, O., N. Comella, K. Tognazzi, and L. F. Brown. 1998. Identification and partial characterization of PDZK1: a novel protein containing PDZ interaction domains. Lab. Investig. 78:117-125. [PubMed] [Google Scholar]
- 11.Koike, K., T. Kawabe, T. Tanaka, S. Toh, T. Uchiumi, M. Wada, S. Akiyama, M. Ono, and M. Kuwano. 1997. A canalicular multispecific organic anion transporter (cMOAT) antisense cDNA enhances drug sensitivity in human hepatic cancer cells. Cancer Res. 57:5475-5479. [PubMed] [Google Scholar]
- 12.Kool, M., M. de Haas, G. L. Scheffer, R. J. Scheper, M. J. T. van Eijk, J. A. Juijn, F. Baas, and P. Borst. 1997. Analysis of expression of cMOAT (MRP2), MRP3, MRP4, and MRP5, homologues of the multidrug resistance-associated protein gene (MRP1), in human cancer cell lines. Cancer Res. 57:3537-3547. [PubMed] [Google Scholar]
- 13.Li, M., Y. N. Jan, and L. Y. Jan. 1992. Specification of subunit assembly by the hydrophilic amino-terminal domain of the shaker potassium channel. Science 257:1225-1230. [DOI] [PubMed] [Google Scholar]
- 14.Loe, D. W., R. G. Deeley, and S. P. C. Cole. 1996. Biology of the multidrug resistance-associated protein, MRP. Eur. J. Cancer 32A:945-957. [DOI] [PubMed] [Google Scholar]
- 15.McGee, A. W., J. R. Topinka, K. Hashimoto, R. S. Petralia, S. Kakizawa, F. Kauer, A. Aguilera-Moreno, R. J. Wenthold, M. Kano, and D. S. Bredt. 2001. PSD-93 knock-out mice reveal that neuronal MAGUKs are not required for development of function of parallel fiber synapses in cerebellum. J. Neurosci. 21:3085-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Migaud, M., P. Charlesworth, M. Dempster, L. C. Webster, A. M. Watabe, M. Makhinson, Y. He, M. F. Ramsay, R. G. M. Morris, J. H. Morrison, T. J. O'Dell, and S. G. N. Grant. 1998. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature 396:433-439. [DOI] [PubMed] [Google Scholar]
- 17.Misawa, H., Y. Kawasaki, J. Mellor, N. Sweeney, J. Kiwon, R. A. Nicoll, and D. S. Bredt. 2000. Contrasting localizations of MALS/LIN-7 PDZ proteins in brain and molecular compensation in knockout mice. J. Biol. Chem. 276:9264-9272. [DOI] [PubMed] [Google Scholar]
- 18.Nicholson, B., C. K. Manner, J. Kleeman, and C. L. MacLoed. 2001. Sustained nitric oxide production in macrophages requires the arginine transporter CAT2. J. Biol. Chem. 276:15881-15885. [DOI] [PubMed] [Google Scholar]
- 19.Pawson, T., and J. D. Scott. 1997. Signaling through scaffold, anchoring, and adaptor proteins. Science 278:2075-2080. [DOI] [PubMed] [Google Scholar]
- 20.Poulat, F., P. de Santa Barbara, M. Desclozeaux, S. Soullier, B. Moniot, N. Bonneaud, B. Boizet, and P. Berta. 1997. The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J. Biol. Chem. 272:7167-7172. [DOI] [PubMed] [Google Scholar]
- 21.Ranganathan, R., and E. M. Ross. 1997. PDZ domain proteins: scaffolds for signaling complexes. Curr. Biol. 7:R770-R773. [DOI] [PubMed] [Google Scholar]
- 22.Songyang, Z., A. S. Fanning, C. Fu, J. Xu, S. M. Marfatia, A. H. Chishti, A. Crompton, A. C. Chan, J. M. Anderson, and L. C. Cantley. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275:73-77. [DOI] [PubMed] [Google Scholar]
- 23.Takenaka, M., E. Imai, T. Kaneko, T. Ito, T. Moriyama, A. Yamauchi, M. Hori, S. Kawamoto, and K. Okubo. 1998. Isolation of genes identified in mouse renal proximal tubule by comparing different gene expression profiles. Kidney Int. 53:562-572. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi, K., M. Wada, K. Kohno, T. Nakamura, T. Kawabe, M. Kawakami, K. Kagotani, K. Okumura, S. Akiyama, and M. Kuwano. 1996. A human canalicular multispecific organic anion transporter (cMOAT) gene is overexpressed in cisplatin-resistant human cancer cell lines with decreased drug accumulation. Cancer Res. 56:4124-4129. [PubMed] [Google Scholar]
- 25.Willott, E., M. S. Balda, A. S. Fanning, B. Jameson, C. Van Itallie, and J. M. Anderson. 1993. The tight junction protein ZO-1 is homologous to the Drosophila discs-large tumor suppressor protein of septate junctions. Proc. Natl. Acad. Sci. USA 90:7834-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woods, D. F., and P. J. Bryant. 1991. The discs-large tumor suppressor gene of drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell 66:451-464. [DOI] [PubMed] [Google Scholar]
- 27.Yoshihara, C. M., and Z. W. Hall. 1993. Increased expression of the 43-kD protein disrupts acetylcholine receptor clustering in myotubes. J. Cell Biol. 122:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]