Abstract
Recombination plays a central role in the repair of broken chromosomes in all eukaryotes. We carried out a systematic study of mitotic recombination. Using several assays, we established the chronological sequence of events necessary to repair a single double-strand break. Once a chromosome is broken, yeast cells become immediately committed to recombinational repair. Recombination is completed within an hour and exhibits two kinetic gaps. By using this kinetic framework we also characterized the role played by several proteins in the recombinational process. In the absence of Rad52, the broken chromosome ends, both 5′ and 3′, are rapidly degraded. This is not due to the inability to recombine, since the 3′ single-stranded DNA ends are stable in a strain lacking donor sequences. Rad57 is required for two consecutive strand exchange reactions. Surprisingly, we found that the Srs2 helicase also plays an early positive role in the recombination process.
The process of recombination plays an essential role during meiosis and in DNA repair during vegetative growth. Double-strand breaks (DSBs) arise frequently as a consequence of exposure to external insults or as a direct result of natural cell metabolism. Recombinational repair of DSBs is important in solving collapsed replication forks during DNA replication (20). If left unrepaired, DSBs result in broken chromosomes, genetic alterations, or cell death. Repair of DSBs (DSBR) takes place in eukaryotes by two competing processes: nonhomologous end joining and homologous recombination. In yeast cells, homologous recombination is the prevalent mechanism used (reviewed in reference 36).
In a classic model for DSB-initiated recombination (53) (Fig. 1A), single-stranded degradation of the broken DNA molecule generates protruding 3′-OH ends that can invade homologous regions, creating a D-loop. The invasion process yields regions of heteroduplex DNA (hDNA) that may contain mismatches. The invading 3′ end is then used to prime DNA synthesis. Eventually, the displaced donor strand pairs with single-stranded DNA (ssDNA) from the opposing end of the DSB, also serving as a template for DNA synthesis. Ligation results in the formation of a structure containing two Holliday junctions, which can be resolved to yield either crossover or noncrossover products. Mismatch repair of the hDNA may result in gene conversion events (Fig. 1A, left).
FIG. 1.
(A) Three alternative models of DSB-initiated recombination resulting in gene conversion. (Panel 1) DSBR model. The DSBR model involves invasion of a homologous template by the two broken ends and formation of an intermediate containing two Holliday junctions. After mismatch repair, resolution leads to gene conversion. (Panel 2) One-ended SDSA model. A single broken end invades the homologous sequence and primes DNA synthesis. Newly synthesized DNA reanneals and ligates to the opposite broken arm. A second round of mismatch repair results in gene conversion. (Panel 3) Double-ended SDSA model (DE-SDSA). Invasion is carried out by the two broken arms, followed by DNA synthesis and annealing of the two newly synthesized DNA. Only one round of hDNA repair is necessary for gene conversion. B and R represent the BamHI and EcoRI restriction site polymorphisms, respectively. (B) Schematic representation of our experimental system. Open rectangles represent the ura3 alleles on chromosomes II and V. A black box represents the HOcs; a gray box depicts the inactive HOcs-inc flanked by the BamHI (B) and EcoRI (R) restriction sites. Transfer of the cells to galactose-containing medium results in gene conversion.
An alternative model for gene conversion, termed the synthesis-dependent strand-annealing (SDSA) model (32), proposes that after DSB formation and resection, a single 3′ single-stranded end invades the intact homologous template. DNA synthesis is followed by reannealing of the newly synthesized DNA with the opposite broken arm. In the basic version of this model (Fig. 1A, center), only gene conversion, and not crossover, can be obtained, although variations allowing crossing-over have been also proposed (reviewed in reference 36).
Homologous recombination is catalyzed by a number of proteins encoded mostly by genes of the RAD52 epistasis group (36, 51). RAD52 seems to play a very central role, affecting almost all of the types of known recombination events (11, 36, 39). In vitro Rad52p exhibits strand-annealing activity (31, 49) and has been shown to stimulate the strand exchange activity of Rad51p, the functional homolog of the bacterial RecA protein (45, 51). Rad55 and Rad57 stimulate the in vitro strand exchange activity of Rad51p (50). The role played by other proteins in recombination is less clear. Srs2 is a DNA helicase with 3′-5′ polarity (41). Mutations in SRS2 result in an increased rate of spontaneous gene conversion (6, 23, 42). In addition, a role for Srs2p in the removal of terminal heterologies during recombination has been proposed (37). Mutations in SRS2 are lethal in the absence of some recombination genes, such as RAD54 and SGS1 (7, 35). It is thought that this is due to the accumulation of lethal recombination intermediates. Accordingly, this lethality can be alleviated by eliminating recombination altogether or by disabling checkpoint functions (7, 18).
Despite the importance of recombination in cell survival and the existence of detailed models, a thorough dissection of the actual mechanism of mitotic recombination has remained elusive, mainly due to the low rate at which spontaneous recombination occurs. Most laboratories commonly use genetic systems that are based on the selection of rare recombinants among a cell population. Although convenient, this procedure introduces selective biases on the types of events analyzed, whereas the fate of the majority of the cells remains invisible. To overcome these limitations, yeast strains have been developed in which recombination can be induced in each cell of a population. Recombination is initiated by a DSB at a unique genomic location (8, 12, 14-16, 38, 52, 58). In the strains developed by our laboratory, virtually 100% of the cells in each culture uniformly participate in the recombination event. This gives us the opportunity to dissect the complex course of the DSBR process.
Here, we use such strains to time and order the different steps in the repair of a single broken chromosome during vegetative growth. We then dissect the role played by different proteins in the recombination process.
MATERIALS AND METHODS
Strains.
All of the yeast strains used in the present study are isogenic derivatives of strain MK205 (MATa-inc ura3-HOcs ade3::GALHO ade2-1 leu2-3,112 his3-11,15 trp1-1 can1-100), a derivative of OI29 (16). The ura3-HOcs allele on chromosome V was created by inserting a 39-bp oligonucleotide at the NcoI site of the 1.2-kb BamHI fragment containing the URA3 gene and inserting it into the genome. In this strain a DSB can be created on chromosome V, and it carries no donor sequences for repair by homologous recombination. In the isogenic strain MK203, a 1.2-kb ura3-Hocs-inc gene carrying BamHI and EcoRI polymorphisms was inserted at a HpaI site within LYS2 sequences, as described previously (16).
Deletions of the RAD52, RAD57, and SRS2 genes were created by one- or two-step transplacement by using plasmids pSM20 and pSM51 (43) and plasmid pTZradH (2). All chromosomal configurations were verified by Southern blot analysis after transformation.
Media and growth conditions.
Saccharomyces cerevisiae strains were grown at 30°C, unless specified otherwise. Standard YEP medium (1% yeast extract, 2% Bacto Peptone) supplemented with 3% glycerol (YEPGly), 2% galactose (YEPGal), or 2% dextrose (YEPD) was used for nonselective growth. We added 1.8% Bacto Agar for solid media.
Repair efficiency measurement.
Each strain was streaked onto YEPGly plates. Individual colonies were resuspended in water, appropriately diluted, and plated on YEPD and YEPGal plates. Colonies were counted after 3 days of incubation at 30°C.
Induction experiments.
Single colonies were resuspended in rich YEPGly medium, grown to logarithmic phase, centrifuged, and resuspended in YEPGal. At timely intervals, samples were plated on YEPD plates to score viability and commitment to gene conversion (CGC [see below]), and DNA was extracted and subjected to the different assays. At least three independent experiments were carried out for each strain analyzed. Although slight variations in the time of appearance of DSBs were sometimes seen, the relative timing of the different steps analyzed was constant within experiments.
Southern blot analysis.
Southern blotting was carried out as described previously (16).
Nondenaturing slot blots.
DNA was either directly spotted onto nylon Hybond+ filters or denatured first by incubation for 15 min in the presence of 0.2 N NaOH. Hybridization and exposure were carried out as in Southern blot analysis. The results obtained for the denatured samples were used to calibrate those obtained without denaturation.
PCR assays.
Portions (5 ng) of genomic DNA were amplified in each sample. For quantitative measurements of intact chromosome V, samples were removed at cycle 18. Otherwise, reactions were allowed to proceed to cycle 35. Taq polymerase was used in standard reaction conditions. The compositions of individual primers are available upon request.
Survival.
Survival was assayed by plating samples of cells grown on liquid YEPGal onto YEPD plates at different times during a DSB induction experiment. Colonies were counted after 3 days incubation.
CGC.
For each independent time point, 20 individual colonies were subjected to either Southern blot analysis or PCR to confirm the transfer of information in individual colonies. The PCRs consisted of 1 cycle of 1 min at 98°C, followed by 25 cycles at standard conditions. The DNA thus obtained was then digested with BamHI or EcoRI and subjected to electrophoresis.
Quantitation of results.
Southern blots and ethidium bromide-stained agarose gels were quantified by using the TINA (version 2.1) and NIHimage (version 1.62) computer programs.
Calculation of cumulative curves.
Noncumulative curves were converted to cumulative ones by a modification of the method described in reference 34. T50 values were algebraically interpolated and represent the time at which 50% of the cells have completed a particular assayed step.
RESULTS
The haploid strains of the yeast S. cerevisiae used in the present study bear two copies of the URA3 gene; one of them, located on chromosome V, carries the recognition site for the yeast HO site-specific endonuclease (33) inserted as a short oligonucleotide (ura3-HOcs). The second copy, located on chromosome II, carries a similar site containing a single-base-pair mutation that prevents recognition by the endonuclease (ura3-HOcs-inc). In addition, the ura3 alleles differ at two restriction sites, located to the left (BamHI) and to the right (EcoRI) of the HOcs-inc insertion; these polymorphisms are used to monitor the transfer of information between the chromosomes. In these strains, the HO gene is under the transcriptional control of the GAL1 promoter. Upon transfer of the cells to galactose-containing medium, the HO endonuclease is produced at high levels. The enzyme creates a DSB in 100% of the cell population. The broken chromosomes are then repaired by a mechanism that copies the HOcs-inc information, together with the flanking markers, resulting in a gene conversion event. During the repair, the donor chromosome remains unchanged (Fig. 1B). In the present study, we analyze strains carrying 1.2 kb of shared homology. In these strains repair is carried out only by conversion events not associated with crossing-over, making the physical analysis simpler (16). The region around the break is identical to that of the unbroken allele, except for the two polymorphic sites and the Hocs-inc mutation, so as to avoid production of nonhomologous tails that may affect the recombinational repair (37, 48). There is no genetic selection for recombinational products; instead, repair is monitored in the entire cell population. During the course of the experiment, cell viability remains high in the wild-type strain, MK203, which constitutes our standard.
We have used different assays to monitor specific steps during the recombination event.
(i) Generation of DSBs.
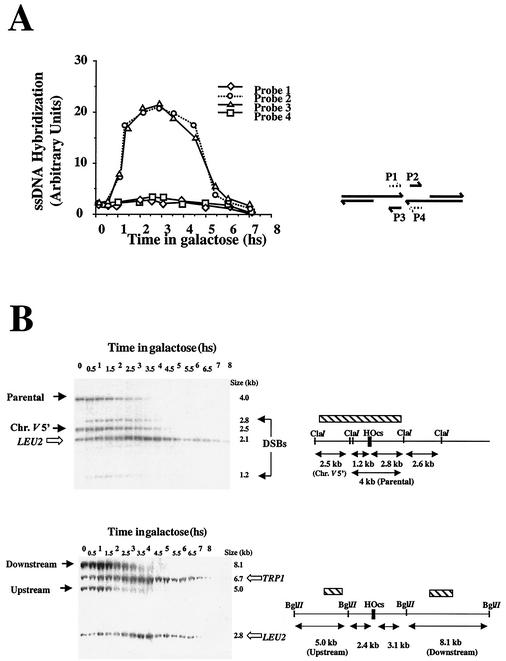
The kinetics of DSB formation and repair of strain MK203 was monitored by Southern blot analysis of DNA extracted at different times after transfer to galactose-containing medium (16) (Fig. 2A). Within 30 min, two additional bands appeared, representing the broken chromosome arms. The intensity of these bands increased with time, reaching a maximal level 2 h after transfer to galactose (t = 2 h). By t = 5 h, the DSBs are no longer detected by Southern analysis. The band representing intact chromosome V decreases in intensity and then reappears with kinetics that inversely correlate with those of DSB formation. Interestingly, in comparison to other chromosomes, the total amount of chromosome V (intact as well as broken) transiently diminishes in our Southern blots (“ratio V/III”). Chromosome V reaches its lowest level at t = 3 h, and is restored to original levels by t = 7 h (Fig. 2A and C). Processing of intermediates and complete repair of the DSB occurs during this time interval. The recombination intermediates are not resolved in our Southern blot analysis but can be monitored by other assays (see below). Quantitative PCR assays (Fig. 2B) corroborate the kinetics inferred by Southern blot analysis.
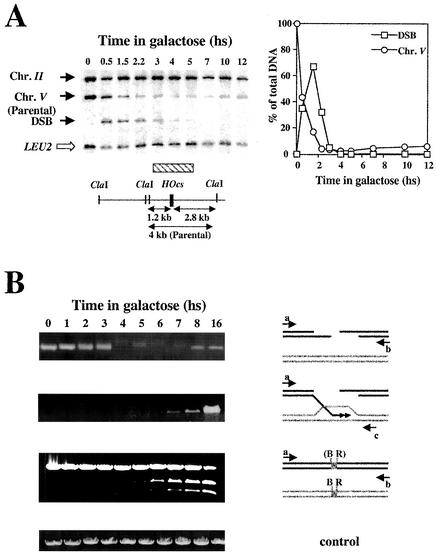
FIG. 2.
(A) Southern blot analysis of DNA extracted from MK203 at timely intervals. The DNA was digested with ClaI and probed with a fragment of chromosome V carrying the URA3 gene (hatched box) and with LEU2 sequences that served as a loading standard. (B) Quantitative PCR measures the relative amounts of intact chromosome V. Pictured below is a quantitative control reaction with primers complementary to a region on chromosome VIII. (C) Quantitation of Southern blot analysis. “Percent of broken chr. V” refers to the levels of the 2.8 and 1.2 kb fragments resulting from DSB formation. “Ratio of chr. V/chr. III” refers to the ratio between the 4-kb fragment representing unbroken chr. V sequences and the LEU2 band. hs, hour(s).
(ii) ssDNA resection.
All current models of recombination (reviewed in reference 36) (Fig. 1A) propose that the DNA immediately adjacent to the DSBs is resected 5′ to 3′. DNA resection has been extensively analyzed, primarily at the MAT locus, and the rate of resection has been found to be under complex genetic control (21, 55, 59).
We used nondenaturative slot blot analysis to monitor the ssDNA intermediate. RNA probes complementary to each of the four strands that border the DSB were used to hybridize blots in which only DNA present as ssDNA in vivo can be detected. Upon transfer of the cells to galactose, only the two RNA probes complementary to the DNA strands with 3′ ends detected exposed ssDNA (Fig. 3A). The accumulation of ssDNA follows DSB induction with a 30-min delay (compare Fig. 2C and 3A). In our regular Southern blots, which analyze double-stranded DNA (dsDNA), the exposed ssDNA ends are not detected; instead, we observe a steady decrease in the relative intensity of bands that represent sequences flanking the DSB site. This disappearance is very short lived in MK203 since DNA synthesis eventually overcomes ssDNA degradation. Resection could be monitored in the cell population by carrying out Southern blot analyses with various restriction enzymes and probes at different distances from the DSB. The decrease in intensity of a band is due to the resection of the dsDNA into ssDNA molecules, which are no longer detected in these Southern blots. The decrease in intensity of bands thus allows us to measure the relative proportion of resected DNA. These results confirmed that the resection from both ends extends symmetrically within the cell population. In different experiments, the maximal length of chromosome V resected in MK203 before resynthesis varied from 3.0 to 3.5 kb.
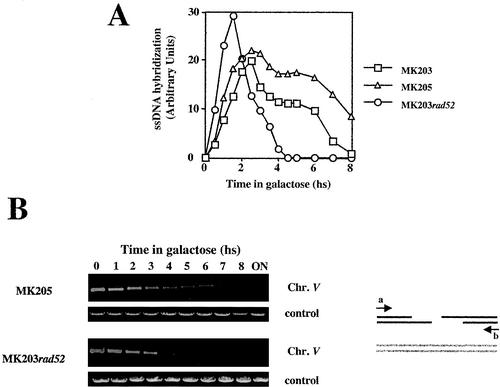
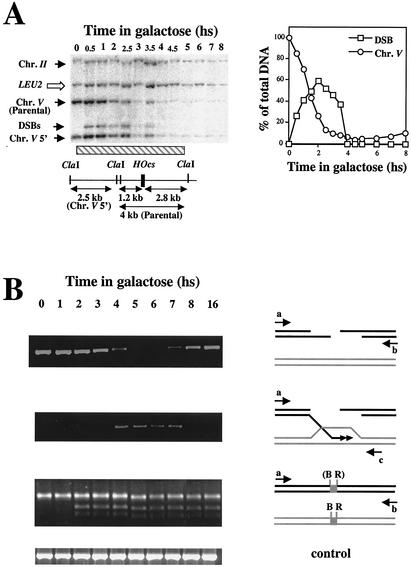
FIG. 3.
(A) Accumulation of ssDNA intermediates: results from nondenaturative slot blots with four RNA probes is plotted as a function of time. (B) Analysis of ssDNA resection in strain MK205. Southern blot analysis of DNA digested with ClaI and BglII are shown. Additional probes homologous to the LEU2 and TRP1 sequences were used as loading controls. hs, hour(s).
DNA resection can be analyzed more easily in the isogenic strain MK205. In this strain, no ectopic homologous sequences are present. Resection occurs at the same rate as that of MK203 but can be monitored for longer periods of time by using probes at different positions along chromosome V. Figure 3B shows two examples of such an analysis.
Resection of the 2.5-kb ClaI fragment, located 1.2 kb upstream of the DSB (“chr.V 5′”) is initially detected in both MK203 and MK205 at t = 1 h (see also Fig. 2A). In MK205, resection continues without reverting to DNA synthesis, and by t = 4.5 h this band can no longer be detected. The BglII fragment upstream of the DSB, located 2.4 kb away, begins to disappear 1.5 h after transfer to galactose. A fragment 3.1 kb downstream decreases in intensity from t = 2 h and cannot be detected by t = 5 h. When results from all of the Southern blot analyses are compiled, we can calculate the rate of resection in our strains. This rate is 35 to 50 nucleotides/min, a rate that is slightly slower than previous estimates (5, 21, 47). The differences may be technical or due to the different strain background used.
(iii) Invasion and DNA synthesis.
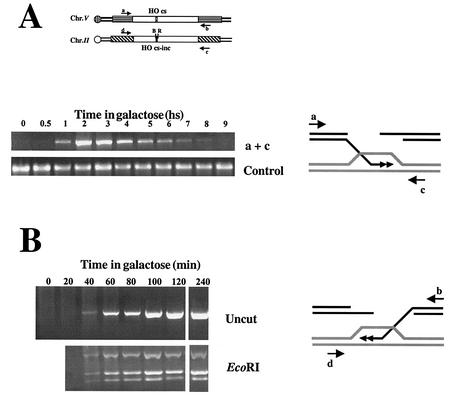
In most recombination models, invasion of the homologous duplex and DNA synthesis follows resection. We monitored this step by using a PCR invasion-extension assay (59). Two primers are used in each PCR: one is complementary to sequences on chromosome V immediately adjacent to the region of homology, and the second anneals to sequences on chromosome II, flanking the homology border. In cells not expressing the HO endonuclease, no PCR product is detected since the primers locate to different chromosomes. Once chromosome V is broken, the activated strand invades the homologous sequences on chromosome II to initiate new DNA synthesis. The extended 3′ end thus becomes covalently linked to sequences on chromosome II. This intermediate was detected by our PCR invasion-extension assay, quantified, and characterized (Fig. 4A and B).
FIG. 4.
(A) Invasion and DNA synthesis assay. Primers adjacent to homology on chromosomes V and II amplify the polymerized invading intermediate. (B) hDNA repair is already maximal at the first time point at which the invasion-extension intermediate can be amplified. (C) Gene conversion assay. Equal amounts of PCR products obtained with primers a and b were digested with BamHI and subjected to gel electrophoresis. (D) Asymmetric repair during gene conversion. Equal amounts of PCR products obtained with primers b and d (upper panels) or primers a and c (lower panels) were digested with either EcoRI or BamHI. hs, hour(s).
Polymerized invasion intermediates can be seen as early as 1 h after DSB induction, only 30 min after the initial detection of the broken chromosome. Invasion levels peak ca. 3 h after DSB induction and then slowly diminish. Although broken chromosome V is no longer detected by 5 h after induction, some invasion-extension intermediates can still be detected at t = 8 h. Similar results were obtained with primers a+c or b+d (Fig. 4 and data not shown).
The primers used in this assay are located immediately adjacent to the region of shared homology between chromosomes II and V, outside the predicted regions of hDNA. Thus, ca. 500 and 800 bp of newly synthesized DNA are detected during invasion from the right and left arms, respectively. DNA synthesis does not proceed much further: PCR carried out with a primer located 1,300 bp from the DSB showed 20% of the level seen with primer c (800 bp from the broken ends). Another primer located 300 bp further (1,600 bp from the DSB) detected only about 1% of the original level.
(iv) Transfer of information.
In our strains, gene conversion is manifested by the transfer of two polymorphic restriction enzyme sites. We can monitor the kinetics and extent of gene conversion in the cell population by measuring the relative amounts of intact chromosome V containing either restriction site (Fig. 4C).
Transfer of the BamHI site to chromosome V reaches detectable levels 2 h after induction of the DSBs; however, a majority of the cells do not undergo gene conversion until 4 h after induction (Fig. 5A). This peak correlates with the timing of maximal invasion-extension. Thus, gene conversion appears to be strongly linked to the invasion step. Evidence that the two stages are linked is apparent by digesting the PCR products obtained in our invasion-extension assay with EcoRI or BamHI. Strikingly, even in the first PCR products detected the polymorphic sites are already seen (Fig. 4B), indicating a kinetic coupling between DNA synthesis and heteroduplex repair.
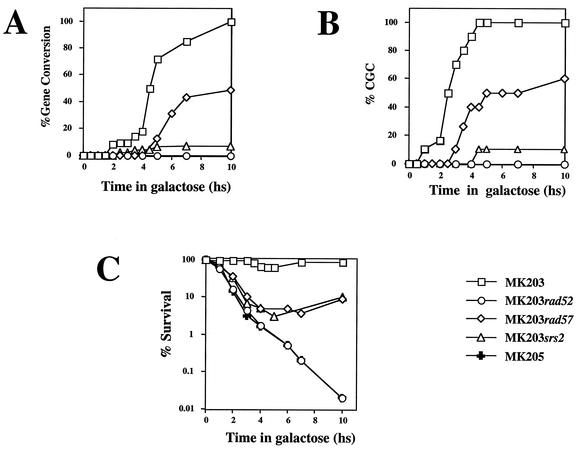
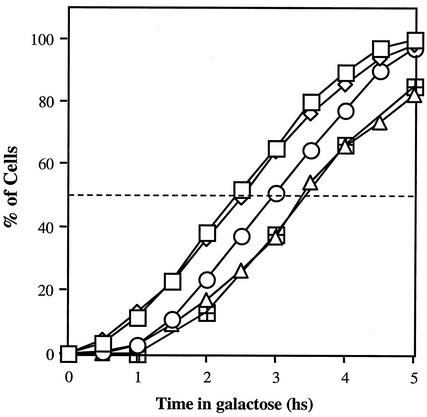
FIG. 5.
(A) Kinetics of gene conversion, as measured by the PCR gene conversion assay. (B) CCG. (C) Survival after DSB induction. hs, hour(s).
Current models dictate the formation of hDNA that is corrected to result in either a conversion event or a restoration of the parental sequences. If correction is unbiased, the models predict a maximal level of gene conversion of 50%. We detect higher levels of information transfer to the invading strand (90 and 70% for BamHI and EcoRI, respectively; Fig. 4D). Gene conversion exceeding 50% implies directionality in the repair process: the invading strand is usually the one repaired.
A simple explanation for the coupling and directionality is that a short stretch of the invading DNA is degraded (e.g., by the editing functions of the DNA polymerase) before engaging in DNA synthesis. We favor this model because it has previously been shown that gene conversion in our system is not affected by mutations in the mismatch repair genes (15), whereas mutations in the editing domain of polymerase delta do affect gene conversion (Y. Aylon and M. Kupiec, unpublished data; see also reference 37).
(v) CGC.
Plating of timely aliquots from a DSB induction experiment onto glucose-containing medium (YEPD) monitors the proportion of viable (colony-forming) cells in the population at a given time. Viability remains high for strain MK203 throughout the experiments (Table 1 and Fig. 5C). Viable colonies can be analyzed by PCR to evaluate the relative levels of gene conversion (scored by the presence or absence of the two polymorphic restriction sites). Since the HO endonuclease is very short-lived, transfer to YEPD plates almost immediately halts further production of DSBs (17). Since there is no selective pressure on cells to carry out gene conversion in order to survive on glucose-containing plates, any mechanism that restores the integrity of the genome enables the formation of a viable colony. Only cells that have irrevocably begun the repair process by a homologous recombination mechanism develop into colonies showing gene conversion on YEPD plates. We call this phenomenon CGC. CGC precedes actual gene conversion of the cell culture by at least 90 min (Fig. 5A and B). For example, at t = 4 h only 20% of the cells have transferred the polymorphic sites; however, when plated on YEPD at this time point, the vast majority of the population (>90%) complete the repair by gene conversion (Fig. 5B).
TABLE 1.
Survival of different strains on YEPGal (constitutive expression of the HO endonuclease) versus YEPD (no expression) plates
| Strain (temp [°C]) | % Survival ± SDa |
|---|---|
| MK203 | 88.3 ± 8.1 |
| MK205 | 0.01 ± 0.005 |
| MK203rad52 | 0.03 ± 0.01 |
| MK203rad57 (20) | 0.02 ± 0.02 |
| MK203rad57 (30) | 5.7 ± 1.8 |
| MK203srs2 | 11.0 ± 1.5 |
That is, the mean CFU on YEPGal/CFU on YEPD.
Genetic control of recombination. (i) Is lack of homology the same as lack of homologous recombination proteins?
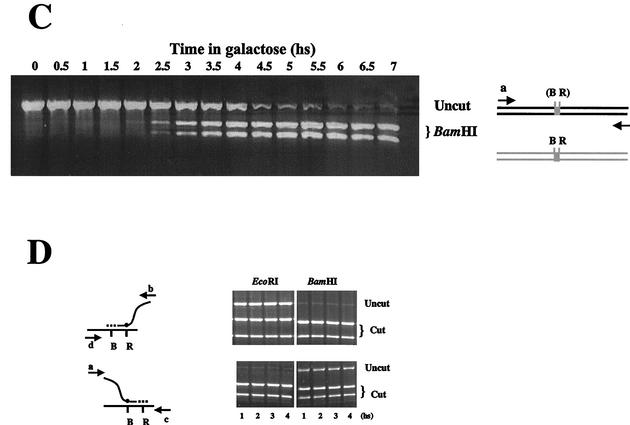
Strain MK205 carries the HO recognition site (HOcs) on chromosome V, as MK203, but has no homologous sequences on chromosome II. The isogenic strain MK203rad52 carries 1.2 kb of ectopic homology but lacks the RAD52 gene. In MK205, recombination is prevented by the absence of homologous sequences; in MK203rad52, recombination is prevented by the lack of recombinational machinery. Both strains show extremely low viability in response to a DSB (Table 1 and Fig. 5C), indicating that they are unable to repair the broken chromosome. However, they exhibit very different kinetics of chromosome V processing. Resection of the broken ends occurs in MK203 and MK205, both Rad+ strains, with similar kinetics irrespective of the presence of potential donor sequences in the genome but is strikingly different in strains lacking RAD52 (Fig. 6A). The MK203rad52 strain accumulates resected DNA very rapidly, but ssDNA levels diminish as early as 1.5 h after the peak of DSB formation and are undetectable by t = 4.5 h. Intact chromosome V cannot be detected by PCR around that time (Fig. 6B). Thus, after an initial burst of resection, dsDNA degradation of the broken arms of chromosome V takes place. In MK205, the levels of ssDNA remain high for an extended period after DSB induction (Fig. 6A).
FIG. 6.
(A) ssDNA resection in strains MK203, MK203rad52, and MK205. (B) Quantitative PCR of remaining chromosome V in strains MK205 and MK203rad52. hs, hour(s).
(ii) Role of RAD57.
The Rad57 protein also functions in the RAD52 pathway. Together with Rad55p, it stimulates Rad51p-mediated recombination, probably by facilitating the loading of Rad51p onto the single-stranded tails (50). Mutations in the RAD55 and RAD57 genes have been previously shown to act in a temperature-dependent fashion (9, 25, 40). Similar phenotypes could be seen in our strains: at 20°C, MK203rad57 strains were indistinguishable from MK203rad52, and survival on YEPGal plates was extremely low. At 30°C, however, survival of rad57 cells plated on YEPGal increased to 5.7% (Table 1).
DSB induction experiments were performed with strain MK203rad57 at 30°C. Southern blot analysis shows that the DSBs appear with kinetics similar to those of the control MK203 (compare Fig. 7 and 2). In contrast to MK203, however, DSBs and parental chromosome V bands rapidly disappear. The kinetics of chromosome V disappearance are similar to those of the MK203rad52 strain, except that at later times the band representing intact chromosome V reappears and accumulates to a low level (Fig. 7A). PCR assays that measure the proportion of intact chromosome V confirm these results (Fig. 7B).
FIG. 7.
(A) Southern blot analysis of DNA extracted from MK203rad57 at timely intervals. (B) Analysis of recombination intermediates in strain MK203rad57 PCR assays for intact chromosome V, invasion plus DNA synthesis, and gene conversion. hs, hour(s).
Detection of invading molecules in MK203rad57 is delayed 5 h compared to that of those in wild-type strains (6 h versus 1 h after induction; Fig. 7B). In addition, the invading intermediates accumulate over time and reach extremely high levels after overnight incubation. We conclude that MK203rad57 cells at 30°C are dually defective. They are deficient at a stage that precedes ssDNA invasion-extension, and although eventually able to complete this process they are also defective in the resolution of the polymerized invasion intermediates. MK203rad57 also shows delayed and reduced kinetics of gene conversion compared to MK203 (Fig. 5A and 7B). Even after prolonged exposure to galactose, most of the cell population has not transferred the DNA polymorphisms to intact chromosome V. Our results are consistent with a defect of rad57 strains in carrying out the strand-transfer reactions of recombination.
(iii) Role of SRS2.
The SRS2 gene encodes an helicase with a poorly understood role in recombination. srs2 mutants exhibit a spontaneous hyperrecombinational phenotype, primarily for gene conversion events (6, 23). Although Srs2 is usually assumed to channel repair to a nonrecombinogenic pathway, there are instances in which it plays a prorecombinogenic role (6). We therefore tested whether srs2 mutants were able to repair a broken chromosome. MK203srs2 showed reduced viability on YEPGal plates (Table 1), indicating a defect in DSBR.
DSB induction experiments were performed with strain MK203srs2. DSBs are created with kinetics identical to those of wild-type strains; however, subsequent processing of the ends dramatically differs. Despite the presence of homology in the genome, sequences adjacent to the DSB are extensively resected, reminiscent of strains with no homologous regions, such as MK205 (Fig. 8A). At very late times, a small proportion (ca. 10%) of chromosome V is resynthesized and recovered as intact chromosome. Although successful polymerized invasion molecules appear with normal kinetics, they are detected in drastically reduced amounts compared to those of the wild type (Fig. 8B). Initial transfer of polymorphisms is detected as in wild-type strains, but gene conversion and repair reaches merely 10% of wild-type levels (Fig. 5A and 8B). Prolonged induction of DSB formation does not result in accumulation of recombination intermediates or products. We conclude that, in DSB-initiated recombination assays, srs2 shows a hyporecombinational phenotype.
FIG. 8.
(A) Southern blot analysis of DNA extracted from MK203srs2 at timely intervals. The LEU2 fragment shown is derived from the srs2::LEU2 allele. (B) Analysis of recombination intermediates in strain MK203srs2 PCR assays for intact chromosome V, invasion plus DNA synthesis, and gene conversion. For the gene conversion analysis, equal amounts of PCR products obtained with primers a and b were digested with BamHI and subjected to gel electrophoresis. hs, hour(s).
Although srs2 and rad57 strains exhibit comparable survival in our assays, they show differences in CGC. When strains are plated on YEPD 4 h after transfer to galactose, virtually all MK203 cells and ca. 40% of the surviving MK203rad57 cells repair DSBs by gene conversion. At this time no srs2 cells are committed to gene conversion (Fig. 5B). Even after 10 h of continuous DSB induction, only 10% of the surviving MK203srs2 cells exhibit converted chromosomes. This is in marked contrast to the full conversion of MK203 and 60% conversion of surviving MK203rad57 cells at this time point. Thus, srs2 shows a strong defect in CGC. The low survival of srs2 cells is independent of the repair mechanism used. Accordingly, only 10% of the srs2 cells exhibit a gene conversion event, as measured in the cell population by PCR (which detects the event irrespectively of viability). Among survivors on YEPD plates, the same proportion is observed.
The rapid decrease in cell viability of rad57 and srs2 strains initially follows kinetics similar to that of rad52 and MK205. After t = 3 h rad57 and srs2 recover somewhat, in accordance with their capability to carry out some recombinational repair (Fig. 5C).
DISCUSSION
Much of our understanding of the mechanisms of recombination comes from studies carried out in yeast cells. Tetrad analysis data obtained throughout the years fit quite well with the DSBR model of recombination (46, 53). Although meiotic and mitotic recombination share important elements of the repair process, results have accumulated suggesting that they may fit different models. Using various physical assays, the different steps in the meiotic recombination process have been characterized and ordered in time. These studies have helped to confirm most features of DSBR, while detecting minor inconsistencies that call for modification and refinement of the model (3, 4, 13, 34). In the present study, we initiate an analogous systematic study of mitotic recombination.
Cumulative curves depicting the percentage of cells that have completed each stage can be helpful in ordering the successive steps of the repair process (34). When all of the curves obtained in our studies are plotted together, a consistent view of the recombination process becomes apparent (Fig. 9). To better describe the different steps in the recombination process, we define T50 as the time at which 50% of the population has completed each step. DSB formation (measured by Southern blot analysis) matches perfectly with the reduction in the levels of intact chromosome V, as detected by quantitative PCR. The calculated T50 of chromosome break is 2.45 h for the Southern blot assay and 2.50 h for the PCR assay. Accumulation of a population exhibiting ssDNA ends follows, with a delay of 30 min (T50 = 3.0 h). Invasion and DNA synthesis coincide with gene conversion (T50 = 3.43 and 3.46 h, respectively) and take place 1 h after the appearance of the DSB. In other words, the recombination process can be divided into two kinetic phases. Immediately after the DSB is created, a 30-min gap is seen before the appearance of the ssDNA intermediate. After its creation, a second gap of similar length is observed, before invasion-extension can be detected. Once this step is carried out, gene conversion is completed without any further delay. It is important to note that the PCR assay that measures invasion and DNA synthesis does not require ligation of the newly synthesized molecules. In contrast, the PCR assay that measures transfer of information can detect only molecules that have religated the original break on chromosome V. The fact that the two curves are almost identical implies that the ligation reaction that rejoins the broken arms immediately follows DNA synthesis.
FIG. 9.
Comparison of the kinetics of accumulation of the different intermediates in the recombination process. Symbols: □, DSB (Southern blot analysis); ◊, chromosome break (PCR); ○, ssDNA filament; ▵, invasion; ⊞, gene conversion. hs, hour(s).
Each cell requires ca. 1 h to repair the broken chromosome. This process includes a search for homology, physical intertwining of donor and recipient DNA molecules, DNA synthesis, ligation of the synthesized DNA, and transfer of information between the chromosomes. Our estimate is remarkably similar to the one obtained for mating-type switch, despite the inherent differences between the processes and strains used (59).
Double Holliday junction molecules (DHJMs), as predicted by the DSBR model, are easily detected in meiotic cells (44). We saw no evidence for such structures in our DNA samples. Although lack of detection constitutes negative evidence, we believe that these structures are not major intermediates of mitotic recombination in MK203. Our results are most consistent with an SDSA type of recombination, which does not form DHJMs and does not create crossover molecules. We have previously shown that a minimal length of homology (ca. 1.6 kb) is required in order to observe a coupling between gene conversion and crossing over. This minimal homology length probably provides the stability required for DHJM formation (16).
What is the significance of the two gaps observed in the kinetics of recombination? The first delay, between chromosome breakage and the appearance of the active ssDNA intermediate, may be required for the induction and recruitment of recombination proteins. A large protein complex must probably assemble before the broken ends are processed. Evidence for the binding of several proteins in vivo to broken DNA ends has been gathered recently (24, 26, 29, 54, 56). In addition, the creation of the DSB elicits a checkpoint response (27). Recent models propose that several proteins must be recruited to the broken ends for the activation of the checkpoint pathway (19, 28). The kinetic delay observed may allow time for assembly of either of these complexes.
Recruitment of the protein complexes activates the broken chromosomal arms. A 5′-to-3′ resection takes place, creating an ssDNA intermediate. This activated filament is pivotal in all homology-dependent pathways; it is believed to be directly involved in homology search and annealing. In addition, it serves as a primer for DNA synthesis. The resection is probably carried out by multiple exonucleases (30, 55). In the absence of Rad52p, resection is carried out, but it exhibits faster kinetics (Fig. 6A). Thus, Rad52p may moderate ssDNA resection, perhaps providing the time necessary to coordinate between the activation of filament and the homology search.
After the activated filament has accumulated, a second delay takes place before polymerized invasion intermediates are produced. This period of time may be required to complete a genome-wide homology search. Very little is known about the mechanism that scans the genome for potential homology. The ssDNA filament is probably sheathed in Rad51 protein, forming a structure that can carry out a strand transfer reaction. It is not clear, however, whether this Rad51 filament is the probe that searches for homology, since some recombinational processes can take place in the absence of Rad51p (36). Rad52p seems to play a more central role and is more likely to be involved in the actual homology recognition through its annealing activity (31).
The activated filament invades the homologous region and primes DNA synthesis by using donor sequences as a template. In meiotic recombination, hDNA is formed at this stage and remains stable until late in the process, before being repaired (4). Contrary to what is seen in meiotic cells, few hDNA molecules were detected in our assays, even at early time points. Repair of the heteroduplex seems to take place concomitantly with invasion and DNA synthesis. We detected a similar frequency of invasion-extension by both ends. The slight asymmetry seen between conversion of the BamHI and the EcoRI sites is due to differential repair of hDNA. Our results are thus consistent with models in which invasion by the left or the right arms occurs independently, but with equal frequency in the population, and also with two-ended models. In one-ended models, the invading strand, after carrying out DNA synthesis, reanneals to the opposite, unmodified arm (Fig. 1A, center). This necessitates two independent rounds of hDNA repair in order to create a gene conversion event: the invading strand must be corrected during invasion and then always serves as the donor of information during reannealing. This seems unlikely. In addition, the rapid transition between the invasion and religation steps precludes a second round of homology search to find the unmodified broken arm. Rather, the kinetic link between the two processes suggests a close proximity of the reannealing strands.
Previously, a model (double-ended SDSA) was proposed in which invasion is carried out by both arms, followed by untangling and annealing of the newly synthesized strands to each other (16) (Fig. 1A, right). In this case, no second round of homology search or hDNA repair is required. Rather, the two arms are in close proximity and may even have participated together in the homology search. The results presented in the present study provide further support for this model.
Our detailed kinetic dissection has enabled characterization of other features of recombination. We have defined CGC as the step in the repair process at which the cells are obligated to complete DSBR by gene conversion. CGC markedly precedes actual conversion. What is the molecular nature of CGC? The T50 of CGC perfectly coincides with the creation of the DSB (2.50 h). Therefore, immediately after DSB formation, a molecular event takes place that leads irreversibly to gene conversion. This event precedes the creation of the activated ssDNA filament and takes place long before invasion of the homologous duplex. The biochemical nature of this event is not currently known, but it may involve modification of the broken ends or be related to the recruitment of checkpoint or DNA processing proteins. The lack of CGC in srs2 mutants correlates well with a putative role for the Srs2 protein in either of these two functions.
We have used our assays to pinpoint the execution point of different proteins with a role in recombination. Rad52 may play a role during homology search (36). Since 5′-to-3′ resection of the broken ends takes place in MK203rad52 strains, resection can be carried out independently of the homology search. In rad52 strains, ssDNA accumulates quickly and is rapidly degraded. Thus, Rad52 also plays a structural role, protecting both DNA strands from exonucleolytic degradation or stabilizing the Rad51 filament. Accordingly, recent in vitro studies have shown that human Rad52 protein can form multimeric complexes able to bind to DNA ends (56). To learn whether the protein plays a role at subsequent steps, conditional mutants should be analyzed.
The Rad55 and Rad57 proteins form a stable heterodimer that stimulates in vitro the strand exchange activity of Rad51p (50). Consistently, rad55 strains showed results identical to those of rad57 in our assays (data not shown). In these mutants the coupling between invasion-extension and reannealing observed in the wild type is lost. rad57 cells show a severe delay in performing the invasion and DNA synthesis stage. Eventually, invasion intermediates appear and accumulate. Only a small percentage of the cell population successfully proceeds to the next step, which requires reannealing and ligation of the arms. We conclude that Rad57p is required twice during recombination: initially, to carry out invasion-extension, and subsequently, to complete gene conversion. Rad57p may be needed to promote Rad51p-mediated strand exchange during invasion and to facilitate the untangling of the newly synthesized strands required to complete gene conversion. A higher-order protein complex is probably active in these processes, since at low temperatures rad57 strains fail to carry out even the first process. At 20°C, the broken ends of MK203rad57 are rapidly degraded, as in rad52 strains. These results suggest that under these conditions a functional Rad52 protein is not sufficient to protect the Rad51 filament from degradation.
Most of the rad57 cells die upon induction of a DSB, but among survivors, a majority has undergone gene conversion, indicating that cells completing the repair by conversion have higher chances of surviving. However, not all of the cells that undergo gene conversion survive: in PCR assays, we detected a higher level of gene conversion than that of overall survival (Fig. 5A and 7B). A window of opportunity available to complete the recombination process seems to exist. Cells that miss this window are irreversibly committed to death, even after completion of the repair.
SRS2 plays a complex and still poorly understood role in DNA repair and recombination. In numerous assays, mutations in this gene cause an hyper-recombinational phenotype (reviewed in references 7 and 18). We have examined the kinetics of DSBR in a strain deleted for the SRS2 gene. After DSB formation, DNA processing occurs with wild-type kinetics, but extensive resection is observed. Invasion and DNA synthesis also takes place with normal kinetics but only in a small fraction of the population (∼10%). The transfer of polymorphic sites occurs also in about 10% of the DNA molecules. These results are consistent with a small subpopulation of cells able to carry out the entire recombinational repair normally, whereas the rest of the population continues to resect the DNA as if no homology was detected in the cell. Perhaps detection of homology and/or strand invasion is required to limit resection.
Many different roles have been proposed for Srs2 in recombination. Most of them involve the processing of postinvasion intermediates (1, 7, 18). Our results show that Srs2 is required at an earlier stage, since most of the population exhibits defects already at the resection step. It should be noted that no heterologous sequences have to be removed from the broken ends in MK203 to allow repair by recombination. Thus, the effect of the srs2 mutation cannot be related to the removal of terminal heterologies (37). Rather, srs2 mutants exhibit a general defect in DSBR that may be exacerbated by the presence of heterologous ends.
Srs2 may be involved in facilitating the processes of homology recognition or strand transfer carried out by the Rad52 and Rad51 proteins. In the absence of the Srs2 helicase activity, the majority of DNA molecules cannot complete the invasion-extension reaction and behave as if no ectopic homology was present in the cell. Since srs2 mutants exhibit a spontaneous hyperrecombinational phenotype, this may not be the only role for the Srs2 protein. Under different circumstances (for example, when recombination is initiated by a single-strand nick), the Srs2 helicase activity may revert the invasion process, thus preventing the formation of recombination intermediates. The fact that an srs2 rad52 haploid mutant is viable (35) also implies that DSBs are not the primary substrate for Srs2 activity.
An alternative (nonexclusive) explanation for the function of Srs2 is that it plays a role in the coordination between DSBR and cellular processes monitored by checkpoint mechanisms (18, 22). According to this idea, the rapid loss of viability observed in srs2 strains is due to lack of coordination between repair and cellular functions. For example, the unloading of specific proteins may require the helicase activity of Srs2 (41). In agreement with this idea, a recent study has shown that Srs2 is required for cell recovery after checkpoint arrest (57).
srs2 mutants are very defective in CGC: only 10% of the surviving colonies on YEPD showed a gene conversion event. In other words, survival on YEPD in srs2 strains seems to be independent of the ability to repair the break by gene conversion. This is strikingly different from the results obtained in all of the other recombination-defective or checkpoint-defective strains that we have studied (Fig. 5B and data not shown). The defect in CGC observed implies that, in the absence of the helicase, alternative repair mechanisms can be used to reconstitute the broken chromosome. The nature of these mechanisms remains a mystery, since srs2 mutants have been found to be defective in sister chromatid recombination (6) and in nonhomologous end joining (10).
Although rad57 and srs2 mutants are defective at different stages of the recombination process, they both exhibit comparable loss of viability. In fact, rad57, srs2, and also rad52 and MK205 exhibit rapid kinetics of cell death that are remarkably similar (Fig. 5C). Thus, all strains defective in recombinational repair experience an early event that commits them to death independently of the nature of their recombinational defects. This event could be related to the induction of checkpoint functions. Accordingly, the efficient processing of DSB ends has been directly linked to appropriate checkpoint regulation (21). Further studies are required to understand why these cells are unable to utilize alternative repair pathways, such as end joining, to improve survival.
In summary, we have carried out a systematic study of the repair of a broken chromosome. Our results have allowed the ordering of the different stages of recombination in the temporal scale, and they provide knowledge about the relative length and order of the stages. Our results clearly differentiate between lack of homology and lack of recombination machinery, since in rad52 mutants both the 5′ and the 3′ ssDNA ends are rapidly degraded, whereas they are stable in a strain lacking potential recombination partners. We have also characterized the role of Rad57 and Srs2 in the framework established. Finally, we have defined a novel phenomenon, CGC: wild-type cells are committed to repair by recombination as soon as the break appears and before the ends are processed into ssDNA filaments. We have shown that srs2 mutants are defective in CGC.
Acknowledgments
This work was supported by a grant of the Israel Science Foundation to M.K. Y.A. acknowledges the generous support of the Constantiner Institute for Molecular Genetics.
We thank Rivka Steinlauf for excellent technical help and all members of the Kupiec lab for support and ideas.
Y.A. and B.L. contributed equally to this study.
REFERENCES
- 1.Aboussekhra, A., R. Chanet, A. Adjiri, and F. Fabre. 1992. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12:3224-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aboussekhra, A., R. Chanet, Z. Zgaga, C. Cassier-Chauvat, M. Heude, and F. Fabre. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair: characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17:7211-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allers, T., and M. Lichten. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106:47-57. [DOI] [PubMed] [Google Scholar]
- 4.Allers, T., and M. Lichten. 2001. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8:225-231. [DOI] [PubMed] [Google Scholar]
- 5.Fishman-Lobell, J., and J. E. Haber. 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258:480-484. [DOI] [PubMed] [Google Scholar]
- 6.Friedl, A. A., B. Liefshitz, R. Steinlauf, and M. Kupiec. 2001. Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat. Res. 486:137-146. [DOI] [PubMed] [Google Scholar]
- 7.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 8.Haber, J. E. 1995. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays 17:609-620. [DOI] [PubMed] [Google Scholar]
- 9.Hays, S. L., A. A. Firmenich, and P. Berg. 1995. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. USA 92:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegde, V., and H. Klein. 2000. Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 28:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiom, K. 1999. DNA repair: Rad52, the means to an end. Curr. Biol. 9:R446-R448. [DOI] [PubMed] [Google Scholar]
- 12.Holmes, A., and J. E. Haber. 1999. Physical monitoring of HO-induced homologous recombination. Methods Mol. Biol. 113:403-415. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, N., and N. Kleckner. 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106:59-70. [DOI] [PubMed] [Google Scholar]
- 14.Inbar, O., and M. Kupiec. 1999. Homology search and choice of homologous partner during mitotic recombination. Mol. Cell. Biol. 19:4134-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inbar, O., and M. Kupiec. 2000. Recombination between divergent sequences leads to cell death in a mismatch-repair-independent manner. Curr. Genet. 38:23-32. [DOI] [PubMed] [Google Scholar]
- 16.Inbar, O., B. Liefshitz, G. Bitan, and M. Kupiec. 2000. The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 275:30833-30838. [DOI] [PubMed] [Google Scholar]
- 17.Kaplun, L., Y. Ivantsiv, D. Kornitzer, and D. Raveh. 2000. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc. Natl. Acad. Sci. USA 97:10077-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein, H. L. 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 20.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94:399-409. [DOI] [PubMed] [Google Scholar]
- 22.Liberi, G., I. Chiolo, A. Pellicioli, M. Lopes, P. Plevani, M. Muzi-Falconi, and M. Foiani. 2000. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 19:5027-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liefshitz, B., R. Steinlauf, A. Friedl, F. Eckardt-Schupp, and M. Kupiec. 1998. Genetic interactions between mutants of the “error-prone” repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 407:135-145. [DOI] [PubMed] [Google Scholar]
- 24.Lisby, M., R. Rothstein, and U. H. Mortensen. 2001. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA 98:8276-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lovett, S. T., and R. K. Mortimer. 1987. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength, and mating type. Genetics 116:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, S. G., T. Laroche, N. Suka, M. Grunstein, and S. M. Gasser. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97:621-633. [DOI] [PubMed] [Google Scholar]
- 27.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 28.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills, K. D., D. A. Sinclair, and L. Guarente. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97:609-620. [DOI] [PubMed] [Google Scholar]
- 30.Moreau, S., E. A. Morgan, and L. S. Symington. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1, and Rad27 nucleases in DNA metabolism. Genetics 159:1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortensen, U. H., C. Bendixen, I. Sunjevaric, and R. Rothstein. 1996. DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl. Acad. Sci. USA 93:10729-10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nassif, N., J. Penney, S. Pal, W. R. Engels, and G. B. Gloor. 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14:1613-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nickoloff, J. A., J. D. Singer, and F. Heffron. 1990. In vivo analysis of the Saccharomyces cerevisiae HO nuclease recognition site by site-directed mutagenesis. Mol. Cell. Biol. 10:1174-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Padmore, R., L. Cao, and N. Kleckner. 1991. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 66:1239-1256. [DOI] [PubMed] [Google Scholar]
- 35.Palladino, F., and H. L. Klein. 1992. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132:23-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parket, A., O. Inbar, and M. Kupiec. 1995. Recombination of Ty elements in yeast can be induced by a double-strand break. Genetics 140:67-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastink, A., and P. H. Lohman. 1999. Repair and consequences of double-strand breaks in DNA. Mutat. Res. 428:141-156. [DOI] [PubMed] [Google Scholar]
- 40.Rattray, A. J., and L. S. Symington. 1995. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics 139:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rong, L., and H. L. Klein. 1993. Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:1252-1259. [PubMed] [Google Scholar]
- 42.Rong, L., F. Palladino, A. Aguilera, and H. L. Klein. 1991. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schild, D. 1995. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics 140:115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwacha, A., and N. Kleckner. 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83:783-791. [DOI] [PubMed] [Google Scholar]
- 45.Song, B., and P. Sung. 2000. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J. Biol. Chem. 275:15895-15904. [DOI] [PubMed] [Google Scholar]
- 46.Stahl, F. 1996. Meiotic recombination in yeast: coronation of the double-strand-break repair model. Cell 87:965-968. [DOI] [PubMed] [Google Scholar]
- 47.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugawara, N., F. Paques, M. Colaiacovo, and J. E. Haber. 1997. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc. Natl. Acad. Sci. USA 94:9214-9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiyama, T., J. H. New, and S. C. Kowalczykowski. 1998. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA 95:6049-6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sung, P. 1997. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 11:1111-1121. [DOI] [PubMed] [Google Scholar]
- 51.Sung, P., K. M. Trujillo, and S. Van Komen. 2000. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 451:257-275. [DOI] [PubMed] [Google Scholar]
- 52.Sweetser, D. B., H. Hough, J. F. Whelden, M. Arbuckle, and J. A. Nickoloff. 1994. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol. Cell. Biol. 14:3863-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 54.Teo, S. H., and S. P. Jackson. 2000. Lif1p targets the DNA ligase Lig4p to sites of DNA double-strand breaks. Curr. Biol. 10:165-168. [DOI] [PubMed] [Google Scholar]
- 55.Tsubouchi, H., and H. Ogawa. 1998. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol. Cell. Biol. 18:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Dyck, E., A. Z. Stasiak, A. Stasiak, and S. C. West. 1999. Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398:728-731. [DOI] [PubMed] [Google Scholar]
- 57.Vaze, M. B., A. Pellicioli, S. E. Lee, G. Ira, G. Liberi, A. Arbel-Eden, M. Foiani, and J. E. Haber. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10:373-385. [DOI] [PubMed] [Google Scholar]
- 58.Weng, Y., S. L. Barton, J. W. Cho, and J. A. Nickoloff. 2001. Marker structure and recombination substrate environment influence conversion preference of broken and unbroken alleles in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:461-468. [DOI] [PubMed] [Google Scholar]
- 59.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]