Abstract
The NF-κB/Rel family of transcription factors participates in the control of a wide array of genes, including genes involved in embryonic development and regulation of immune, inflammation, and stress responses. In most cells, inhibitory IκB proteins sequester NF-κB/Rel in the cytoplasm. Cellular stimulation results in the degradation of IκB and modification of NF-κB/Rel proteins, allowing NF-κB/Rel to translocate to the nucleus and act on its target genes. Calmodulin (CaM) is a highly conserved, ubiquitously expressed Ca2+ binding protein that serves as a key mediator of intracellular Ca2+ signals. Here we report that two members of the NF-κB/Rel family, c-Rel and RelA, interact directly with Ca2+-loaded CaM. The interaction with CaM is greatly enhanced by cell stimulation, and this enhancement is blocked by addition of IκB. c-Rel and RelA interact with CaM through a similar sequence near the nuclear localization signal. Compared to the wild-type protein, CaM binding-deficient mutants of c-Rel exhibit increases in both nuclear accumulation and transcriptional activity on the interleukin 2 and granulocyte macrophage colony-stimulating factor promoters in the presence of a Ca2+ signal. Conversely, for RelA neither nuclear accumulation nor transcriptional activity on these promoters is increased by mutation of the sequence interacting with CaM. Our results suggest that CaM binds c-Rel and RelA after their release from IκB and can inhibit nuclear import of c-Rel while letting RelA translocate to the nucleus and act on its target genes. CaM can therefore differentially regulate the activation of NF-κB/Rel proteins following stimulation.
NF-κB/Rel proteins are a family of transcription factors that are expressed in virtually all cell types and regulate a wide range of genes (50). NF-κB/Rel proteins have been shown to play important roles in the regulation of immune, inflammation, and stress responses, embryonic development, growth control, and apoptosis (2, 15, 18, 19, 50). The mammalian NF-κB/Rel family consists of five proteins, namely NF-κB1 (p50/p105), NF-κB2 (p52/p100), c-Rel, RelA (p65), and RelB. They are characterized by an N-terminal Rel homology domain (7) that mediates DNA binding, dimerization, and IκB binding and also harbors a nuclear localization signal (NLS). Sequences outside the Rel domain are less conserved, and in the case of c-Rel, RelA, and RelB they contain transactivation domains.
NF-κB/Rel proteins are regulated primarily at the level of subcellular localization. In most cells, NF-κB/Rel is complexed with an inhibitory IκB protein. This interaction masks the NLS of NF-κB/Rel, and consequently the NF-κB/Rel-IκB complex resides in the cytoplasm. NF-κB/Rel is known to be activated by over 150 different stimuli such as, for example, inflammatory cytokines, mitogens, oxidative stress, UV and gamma radiation, viruses, and bacterial lipopolysaccharide (50). NF-κB/Rel-activating stimuli initiate a cascade of events that lead to the phosphorylation, ubiquitination, and subsequent degradation of the inhibitory IκB protein (27, 31, 32). Loss of IκB reveals the NLS of NF-κB/Rel, allowing NF-κB/Rel to be directed to the nucleus, where it can act on its target genes. The initiating step in NF-κB/Rel activation is the phosphorylation of IκB, which in most cases is mediated by a large kinase complex termed IKK (17, 27, 31, 32, 41).
In addition to regulation by inhibitory proteins, NF-κB/Rel activity is also influenced at several other levels. NF-κB/Rel proteins are themselves phosphorylated upon cellular stimulation (4, 17, 34, 37, 40, 45, 46, 56, 57, 66, 67). Stimulation-induced phosphorylation of the transactivation domains of RelA and c-Rel enhances their ability to activate transcription. NF-κB1 has also been shown to become phosphorylated in response to stimulation, resulting in a more stable interaction with DNA (37). In addition, there is evidence that the cAMP-dependent protein kinase PKA regulates c-Rel activity (34, 45). The outcome of activation of NF-κB/Rel proteins also depends on their interaction with other proteins. NF-κB/Rel proteins synergize with many other transcription factors and coactivators in the process of transcriptional activation (16, 51-53, 60, 68).
Calmodulin (CaM) is a small, acidic, highly conserved protein that is ubiquitously expressed. CaM is a key mediator of signals by the secondary messenger Ca2+, and it has been shown to be an essential regulator of cell cycle progression, cell motility and contraction, ion homeostasis, and other fundamental cellular processes (65). CaM is also involved in the regulation of transcription (6, 12, 13, 22), not only indirectly through CaM-dependent kinases and phosphatases, but also directly through interaction with transcription factors (10, 21, 47, 61).
Here we report that two members of the NF-κB/Rel family, c-Rel and RelA, interact directly and Ca2+ specifically with CaM. NF-κB/Rel-activating stimuli enhance the interaction with CaM, and this enhancement is blocked by the addition of IκBα. Compared to wild-type c-Rel, CaM binding-deficient mutants exhibit an increased nuclear accumulation and transcriptional activity on the granulocyte macrophage colony stimulating factor (GM-CSF) and interleukin 2 (IL-2) promoters in the presence of a Ca2+ signal. Our results suggest that CaM can inhibit transport of c-Rel, but not RelA, to the nucleus and thereby differentially regulates the activation of NF-κB/Rel proteins following cell stimulation. We therefore propose a novel role for CaM as a direct link between Ca2+ signals and the regulation of NF-κB/Rel.
MATERIALS AND METHODS
Plasmids.
The RelA, c-Rel, and NF-κB1 (p50) pRc/CMV expression plasmids used for in vitro translations and transient transfections have been described previously (38). For expression in Escherichia coli, the RelA cDNA was subcloned into pET-20b+His, a derivative of pET-20b+ (Novagen) containing an N-terminal His6 tag (49). The IκBα cDNA (38) was also subcloned into pET-20b+His. Mutated derivatives of RelA and c-Rel were made by using standard cloning and PCR techniques. The enhancer and promoter of GM-CSF (8) were obtained from genomic Jurkat cell DNA by PCR amplification and cloned into the BglII and HindIII sites, respectively, of the pGL2-Basic reporter plasmid (Promega). Existing BglII and HindIII sites of the 716-bp enhancer and the 0.6-kb promoter segment, respectively, were used, except at the 3′ end of the promoter, where the HindIII site was created with the 5′-GAGAAGCTTTAGCCTTTCTCTCTGTG-3′ primer. The reporter plasmid pGL2-IL-2 contains the enhancer and the minimal promoter of IL-2. The ClaI-HindIII fragment of pIL2CAT (14) was inserted into the Ecl136II and HindIII sites of pGL2-Basic after the ClaI site had been blunted. The hCMV-βgal normalization plasmid has been described previously (35).
Protein preparations.
The DNA- and CaM-binding nuclear proteins (shown in Fig. 1A) were purified as previously described (10), except that the final E-box DNA-Sepharose column purification step was omitted. RelA, c-Rel, and NF-κB1 were in vitro transcribed and translated by using the TNT coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. His6-tagged RelA and derivatives were overexpressed in E. coli BL21(DE3)pLysS, cells were lysed by sonication in 10 mM Tris-HCl (pH 8.0) and centrifuged, and the RelA-containing pellet was solubilized in 20 mM Tris-HCl (pH 8.0)-0.5 M NaCl-5 mM imidazole-6 M guanidine-HCl. RelA was then purified by using nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) under denaturing conditions, replacing guanidine-HCl with 8 M urea in the washing and elution steps. The purified RelA was renatured by dilution to approximately 100 μg/ml and dialysis against 20 mM HEPES (pH 8.0)-0.2 mM EDTA-1 mM dithiothreitol (DTT)-0.05% Triton X-100-0.5 M NaCl-10% glycerol with gradually decreasing concentrations of urea. EDTA was omitted in the last dialysis. His6-tagged IκBα was overexpressed in E. coli BL21(DE3)pLysS, cells were lysed by sonication in 20 mM Tris-HCl (pH 8.0)-0.5 M NaCl, and IκBα was purified from the soluble fraction by Ni-NTA agarose chromatography according to the manufacturer's instructions. The purified preparation was dialyzed against 20 mM HEPES (pH 8.0)-0.05% Triton X-100-0.1 M NaCl-10% glycerol-2 mM DTT.
FIG. 1.
NF-κB/Rel proteins bind CaM. (A) A calf thymus nuclear protein preparation purified by a series of chromatographic steps including CaM-Sepharose was analyzed by EMSA with a κB sequence used as a probe. A 100-fold molar excess of the same DNA sequence (s) or a non-κB sequence (ns) was added as competitor where indicated. In the first lane (co), no protein was added to the binding reaction. (B) RelA, c-Rel, or NF-κB1 (p50) expressed in vitro in a reticulocyte lysate was incubated with CaM-Sepharose in the presence of Ca2+ (+) or EDTA (−), eluted with 1 M NaCl plus 2 mM EDTA, and analyzed by SDS-PAGE and autoradiography. Preloads (lane 1) represent 40% (RelA and NFκB1) and 20% (c-Rel) of the protein amount incubated with CaM-Sepharose or protein A-Sepharose. RelA and c-Rel binding was inhibited by prebinding a fourfold molar excess of a CaM-binding peptide, amino acids 290 to 309 of CaM kinase II, to the CaM-Sepharose (lane 4), and RelA and c-Rel did not bind to protein A-Sepharose (lanes 5 and 6).
EMSA.
A 32P-labeled immunoglobulin κ light chain enhancer κB sequence, 5′-GTCAGAGGGGACTTTCCGAGAGGTA-3′, was used as a probe in all electrophoretic mobility shift assays (EMSAs). The EMSA of DNA- and CaM-binding nuclear proteins (Fig. 1A) was as described previously (64). In vitro-translated RelA and c-Rel or the control rabbit reticulocyte lysate was incubated with the DNA probe in 10 mM HEPES (pH 8.0)-5 mM DTT-10% glycerol-0.05% Triton X-100-50 mM NaCl for 10 min at room temperature. The binding reactions were electrophoresed on 6% polyacrylamide Tris-borate-EDTA (RelA) or Tris-glycine (c-Rel) gels at 4°C. Where indicated, proteins were preincubated with 1 ng of E. coli-produced and purified IκBα per μl for 10 min at room temperature before addition of DNA.
Cell culture and transient transfections.
Jurkat T cells and DG75 B cells were maintained in RPMI supplemented with 5% fetal calf serum and antibiotics. Cells were transiently transfected by electroporation as described previously (24), except that the amounts of expression plasmid in the DG75 transfections were 2.5 μg for the experiment illustrated in Fig. 6C and 15 μg for the one illustrated in Fig. 3D. The transfected cells were grown for 24 h. Where indicated, cells were stimulated before being harvested. PMA (Sigma) was used at a concentration of 25 ng/ml, ionomycin (Calbiochem) at 1 μg/ml, and human recombinant tumor necrosis factor alpha (TNF-α) (Sigma) at 10 ng/ml. Luciferase activity was measured with the Luciferase assay system (Promega) and normalized to β-galactosidase activity from the cotransfected hCMV-βgal plasmid.
FIG. 6.
The nuclear accumulation of c-Rel, but not of RelA, is negatively regulated by its CaM binding site. (A) Jurkat cells were transfected with expression plasmids for c-Rel or the point-mutated derivatives as indicated on the left. Twenty-four hours after transfection, cells were stimulated for 30 min with PMA (P), PMA plus ionomycin (P+I), or TNF-α (Τ). Cytoplasmic (c) and nuclear (n) extracts were prepared and analyzed by Western blotting with the anti-c-Rel C antibody. As a control of the purity of the cytoplasmic and nuclear extracts, the localization of tubulin (cytoplasmic) and that of histone H1 (nuclear) were determined by Western blotting. Shown are tubulin and histone H1 Western blots with extracts from cells transfected with wild-type c-Rel plasmid. (B) Quantitation of the results shown in panel A and two other corresponding Western blot experiments. The ratios of nuclear and cytoplasmic contents of c-Rel ± standard errors of the means (SEM) are shown. (C) DG75 cells were transfected with c-Rel or c-Rel HB− expression plasmids and were harvested 24 h later. Analysis of cytoplasmic (c) and nuclear (n) extracts was performed as described for panel A. (D) Jurkat cells were transfected with RelA or RelA H− expression plasmids. After 24 h, cells were stimulated as for panel A. Cytoplasmic (c) and nuclear (n) extracts were prepared, and RelA proteins were detected by Western blotting performed with the anti-p65 C-20 antibody. (E) Quantitation of the results shown in panel D and two other corresponding Western blot experiments. The ratios of nuclear and cytoplasmic contents of RelA ± SEM are shown.
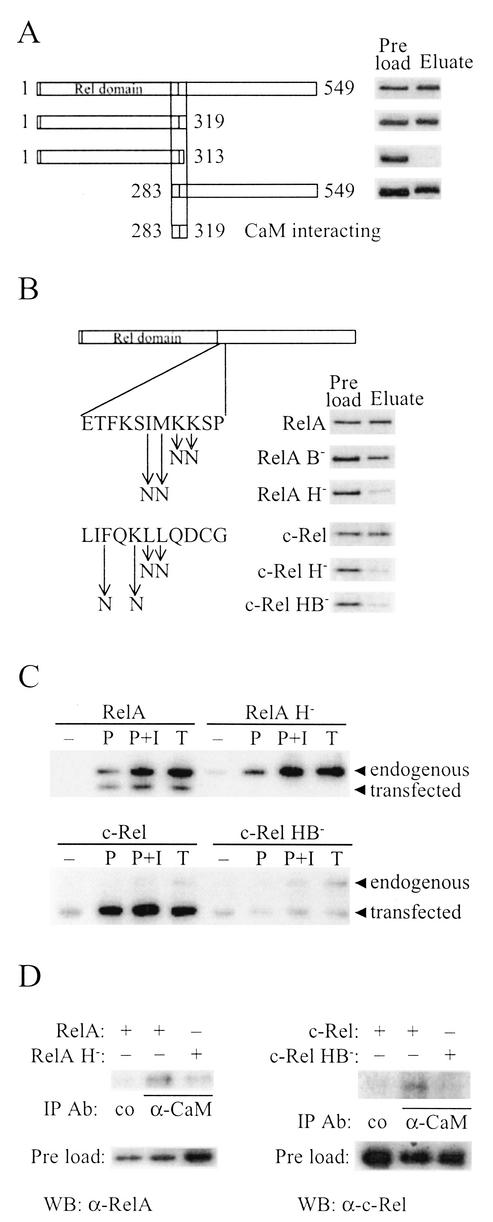
FIG. 3.
Identification of the CaM binding sequences of RelA and c-Rel. (A) E. coli-produced and purified full-length (amino acids 1 to 549) and the schematically drawn deletion derivatives of RelA were assayed for CaM-Sepharose binding in the presence of Ca2+ as described for Fig. 1B, except that proteins were visualized by silver staining. (B) E. coli-produced RelA or in vitro-translated c-Rel and derivatives with the point mutations in the CaM binding sequences shown to the left were assayed for CaM-Sepharose binding in thepresence of Ca2+ as described for Fig. 1B. RelA was visualized by silver staining and c-Rel by autoradiography. (C) Jurkat cells transiently transfected with vectors encoding RelA, RelA H−, c-Rel, or c-Rel HB− were unstimulated (−) or stimulated with PMA (P), PMA plus ionomycin (P+I), or TNF-α (T) for 20 min (RelA) or 1 h (c-Rel) prior to harvest. Whole-cell extracts were then incubated with CaM affinity resin, and bound proteins were eluted and separated by SDS-PAGE. Endogenous RelA and c-Rel and transiently expressed proteins from transfected cDNAs were detected by Western blot analysis. (D) DG75 cells were transiently transfected with vectors encoding the indicated NF-κB/Rel proteins, and whole-cell extracts were subjected to immunoprecipitation with a mouse monoclonal α-CaM antibody or a control (co) mouse monoclonal α-Flag M2 antibody. Coimmunoprecipitated RelA or c-Rel was detected by Western blot analysis. Preload lanes contain an equal volume of each extract.
Calmodulin binding assays.
E. coli-produced and in vitro-translated proteins were incubated with CaM-Sepharose (10) or protein A-Sepharose 4 Fast Flow (Pharmacia) in CaM binding buffer (25 mM HEPES [pH 7.3], 0.1 M NaCl, 10% glycerol, 0.05% Triton X-100 with 2 mM CaCl2 or EDTA) for 1 h at 4°C. After a thorough washing, bound proteins were eluted with CaM binding buffer containing 1 M NaCl and 2 mM EDTA and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Where indicated, the CaM-Sepharose was prebound with a fourfold molar excess of a CaM-binding peptide corresponding to amino acids 290 to 309 of CaM kinase II (Sigma). Cell extracts were adjusted to the above binding conditions, except that Triton X-100 was adjusted to 0.1% and 2 mM CaCl2 was included, and then they were incubated with CaM affinity resin (Stratagene) in the presence or absence of 100 ng of recombinant IκBα/μl for 1 h at 4°C. After a thorough washing, bound proteins were eluted by boiling in sample buffer and analyzed by Western blotting.
Extract preparations.
For whole-cell extracts used in CaM binding assays, cells were lysed in CaM binding buffer containing 0.1% Triton X-100 and 2 mM CaCl2 and supplemented with Complete EDTA-free protease inhibitor cocktail tablets (Roche Diagnostics GmbH), 1 mM Na3VO4, 20 mM β-glycerophosphate, and 10 mM sodium pyrophosphate. Cytoplasmic and nuclear extracts were prepared as described previously (3), except that Complete EDTA-free protease inhibitor cocktail tablets were included in the buffers. Buffers for preparing cytoplasmic and nuclear extracts for CaM binding assays were further supplemented with 1 mM Na3VO4, 20 mM β-glycerophosphate, and 10 mM sodium pyrophosphate.
Immunoprecipitation assays.
Cells were lysed in CaM IP buffer (25 mM Tris [pH 7.5], 150 mM NaCl, 2 mM CaCl2, 1 mM DTT, 1% Triton X-100, and Complete EDTA-free protease inhibitor cocktail), and the extracts were incubated together with 2 to 4 μg of α-CaM (catalogue no. 05-173; Upstate Biotechnology) or α-Flag M2 (Sigma) antibodies for 2 h at 4°C. Ten to 20 μl of protein G-Sepharose was added, and the mixture was incubated for one additional hour at 4°C. The beads were washed 3 to 10 times with 1 ml of CaM IP buffer. Antibody-bound complexes were eluted by boiling in sample buffer. The preload represents 1/350 (c-Rel) and 1/500 (RelA) of the lysates.
Western blot analysis.
Cell extracts (10 to 35 μg), eluates from CaM binding resin, or immunoprecipitations were separated by SDS-PAGE followed by Western blot analysis by using 0.4 to 1 μg of anti-c-Rel C per ml, 0.4 to 1 μg of anti-p65 C-20 per ml, 1 μg of anti-histone H1 FL-219 per ml (all from Santa Cruz Biotechnology), or 1/15,000 anti-α-tubulin (clone B-5-1-2; Sigma) antibody. Detection was performed with SuperSignal chemiluminescent substrate (Pierce).
RESULTS
RelA and c-Rel interact with CaM.
In a search for DNA binding proteins that can interact with CaM, we purified a calf thymus nuclear protein preparation by using a series of Sepharose columns that select for proteins with DNA binding properties followed by a CaM-Sepharose column (see Materials and Methods). Proteins binding to the CaM-Sepharose in the presence of Ca2+ were eluted with a high salt concentration and the Ca2+ chelator EDTA and screened for DNA binding activity by EMSA. Figure 1A shows that the eluate contained an activity that bound specifically to a κB DNA sequence. This DNA binding activity could be stimulated by GTP (data not shown), a previously reported property of NF-κB (36). Thus, a nuclear protein(s) with NF-κB-like DNA binding properties binds CaM.
To directly investigate whether NF-κB/Rel can bind CaM, we expressed cDNAs for the NF-κB/Rel proteins NF-κB1 (also designated p50), RelA (p65), and c-Rel in vitro and analyzed the proteins for their ability to bind to CaM-Sepharose. While NF-κB1 did not bind (Fig. 1B), both RelA and c-Rel bound in the presence and, to a much lower extent, in the absence of Ca2+ (Fig. 1B, lanes 2 and 3, respectively). The interaction was CaM specific, since binding could be inhibited by prebinding the CaM-Sepharose with a known CaM-binding peptide from CaM kinase II (Fig. 1B, lane 4) and RelA and c-Rel did not bind to the protein A-Sepharose control (Fig. 1B, lanes 5 and 6). E. coli-produced and purified RelA also bound to the CaM-Sepharose (see below), showing that the interaction is direct and is not mediated by other proteins in the in vitro-translated protein preparation.
CaM binding to RelA and c-Rel is induced by NF-κB/Rel-activating agents.
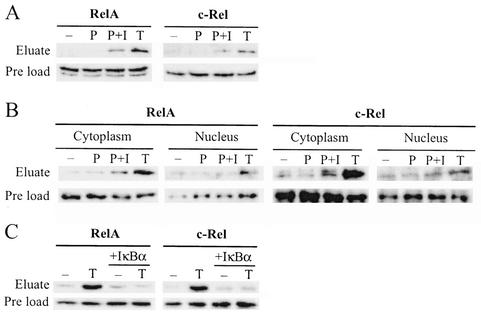
To further characterize the identified interaction between CaM and the NF-κB/Rel proteins, we analyzed the CaM binding ability of RelA and c-Rel in extracts from mammalian cells. Jurkat T cells were stimulated for 1 h with the phorbol ester mitogen phorbol 12-myristate 13-acetate (PMA), PMA plus the Ca2+ ionophore ionomycin, or the cytokine TNF-α, and whole-cell extracts were prepared. The extracts were incubated together with CaM affinity resin, and eluates were subjected to Western blot analysis with RelA- and c-Rel-specific antibodies. As shown in Fig. 2A, RelA and c-Rel from Jurkat whole-cell extracts bind to CaM affinity resin and the binding is dramatically enhanced after cell stimulation either by a combination of PMA and ionomycin or by TNF-α. Next, we investigated whether this interaction is present throughout the cell or if it is restricted to either the cytoplasmic or the nuclear compartment. Cytoplasmic and nuclear fractions of stimulated Jurkat cells were analyzed as above, revealing that the induced binding occurs predominantly in the cytoplasm (Fig. 2B). Doublets, as those seen in Fig. 2A (RelA preload) and 2B (c-Rel cytoplasm), appeared in some experiments and are probably caused by degradation or a modification of the protein.
FIG. 2.
RelA and c-Rel binding to CaM is increased by NF-κB/Rel-activating stimuli. (A) Jurkat cells were unstimulated (−) or stimulated for 1 h with PMA (P), PMA plus ionomycin (P+I), or TNF-α (T), and whole-cell extracts were prepared. Extracts were incubated with CaM affinity resin, bound proteins were eluted and separated by SDS-PAGE, and RelA and c-Rel were detected by Western blot analysis. Preload lanes contain an equal volume of each extract before incubation with the CaM affinity resin. (B) Cytoplasmic and nuclear extracts of Jurkat cells, stimulated as described for panel A, were analyzed as for panel A. (C) Whole-cell extracts of Jurkat cells, stimulated where indicated for 1 h with TNF-α (T), were incubated with CaM affinity resin in the presence or absence of recombinant IκBα. Eluates were analyzed as for panel A.
In an unstimulated cell, NF-κB/Rel proteins are retained in the cytoplasm by their interaction with IκB. Cell stimulation releases NF-κB/Rel from IκB, and this coincides with the observed dramatic increase in RelA and c-Rel interaction with CaM. We investigated whether one interaction excludes the other or if RelA and c-Rel are capable of simultaneous binding to both CaM and IκB. Jurkat whole-cell extracts were incubated with CaM affinity resin in the absence or presence of recombinant IκBα. Western blot analysis of the CaM affinity resin eluates showed that IκBα abolishes the TNF-α stimulation-dependent increase in RelA and c-Rel binding to CaM (Fig. 2C). This suggests that RelA and c-Rel can bind either CaM or IκB and that binding to CaM is dependent on the induced degradation of IκB.
Identification of the CaM binding site of RelA and c-Rel.
To identify the segment of RelA that mediates its interaction with CaM, we constructed various deletion derivatives of RelA in E. coli and assayed for the ability of the purified proteins to bind to CaM-Sepharose (Fig. 3A ). This CaM binding analysis with deletion derivatives localized the interacting region to a 37-amino-acid segment at the C-terminal end of the Rel domain, from amino acids 283 to 319. The loss of CaM binding upon deletion of the C terminus to amino acid 313 but not to amino acid 319 shows that the segment of amino acids 314 to 319 is essential for the CaM interaction. Common among many CaM targets is that they are basic and amphiphilic α-helical segments, in which basic and hydrophobic amino acids are critical for the CaM interaction (48). The C-terminal end of the identified 37-amino-acid segment of RelA, as well as the corresponding sequence of c-Rel, resembles previously described CaM targets in that it is particularly rich in basic and hydrophobic residues (see below). Furthermore, this sequence in RelA has been shown to be in an α-helical conformation when in complex with NF-κB1 and IκB (28). Mutation of either two basic residues (RelA B−) or two hydrophobic residues (RelA H−) to the uncharged polar amino acid asparagine resulted in a decrease in the ability of RelA to bind CaM (Fig. 3B), with RelA H− being particularly affected. Similarly, mutation of two hydrophobic residues (c-Rel H−) or one hydrophobic and one basic residue (c-Rel HB−) in the corresponding sequence of c-Rel resulted in severely decreased CaM binding (Fig. 3B).
We further analyzed the RelA H− and c-Rel HB− mutants by CaM pull-down experiments and coimmunoprecipitations. In the pull-down experiments, whole-cell extracts of transfected Jurkat cells, transiently expressing either wild-type or mutated proteins, were incubated together with CaM-affinity resin. Cells transfected with c-Rel constructs were stimulated with PMA, PMA plus ionomycin, or TNF-α for 1 h, and cells transfected with RelA constructs were stimulated for 20 min (Fig. 3C). Exogenous wild-type mouse RelA and c-Rel were found to behave like their endogenous human counterparts in that they bound to the CaM affinity resin in a stimulation-dependent manner (Fig. 3C). RelA H−, however, did not bind, and c-Rel HB− bound only weakly, and this weak binding was not enhanced by stimulation. Stimulation of RelA-transfected cells for 1 h gave results identical to those obtained with a 20-min stimulation, except that the longer stimulation caused up-regulation of RelA from the expression vector (data not shown). Coimmunoprecipitation analyses were performed with DG75 B cells. In this cell line, like in many other B-cell lines, the half-life of IκB is much shorter than in nonstimulated Jurkat T cells and the continuous degradation of IκB results in a high endogenous NF-κB-activity that is not enhanced either by PMA alone or by a combination of PMA and ionomycin (data not shown). As discussed above, CaM binding to the NF-κB/Rel proteins appears to be dependent on the degradation of IκB. Together this suggests that in DG75 cells, CaM and RelA or c-Rel would interact independently of stimulation. Indeed, in extracts from DG75 cells transfected with wild-type RelA or c-Rel, both proteins were found to interact with endogenous CaM (Fig. 3D). The two mutants, RelA H− and c-Rel HB−, had lost most of the binding ability compared to that of the wild-type proteins (Fig. 3D). These mutation analyses demonstrate that RelA and c-Rel interact with CaM through corresponding sequences both in vitro and in vivo and that, as with several other CaM targets, hydrophobic and basic amino acids are critical for the interaction.
Mutation of the CaM binding site does not affect DNA binding, dimerization, or IκB interaction properties of RelA or c-Rel.
The amino acid residues mutated in RelA and c-Rel are directly C terminal to the Rel domain, the critical structure for DNA binding, dimerization, and interaction with IκB. Therefore, to address whether any of these properties were affected in the CaM binding-deficient proteins, we in vitro translated the three mutants with the most severe effects on CaM binding and analyzed their DNA binding by EMSA. RelA H−, c-Rel HB−, and c-Rel H− all bound DNA as well as did their corresponding wild-type proteins (Fig. 4). This shows that the mutated residues just outside the Rel domain do not affect the DNA binding structure of the Rel domain. This also shows that the mutated proteins are still able to form dimers, since dimerization is a necessity for their DNA binding.
FIG. 4.
Mutation of the CaM binding sequences does not affect DNA binding or IκB inhibition properties of RelA or c-Rel. RelA (A) or c-Rel (B) and their respective CaM binding-deficient derivatives were in vitro translated by using a rabbit reticulocyte lysate system and analyzed for DNA binding activity by EMSA with a κB sequence as probe. Where indicated (+), proteins were preincubated with E. coli-produced and purified IκBα before addition of DNA. In the first lane of each gel, no protein was added to the binding reaction. lysate, binding reactions with rabbit reticulocyte lysate without in vitro-translated protein.
IκB binds to the Rel domain of NF-κB/Rel proteins and, in addition to preventing their nuclear import, inhibits their DNA binding. Like the wild-type proteins, the DNA binding of RelA H−, c-Rel HB−, and c-Rel H− was efficiently inhibited by IκBα (Fig. 4), showing that these mutated proteins still can interact with IκBα.
Since DNA binding, dimerization, and IκB interaction of RelA and c-Rel are unaffected in the mutated proteins, we conclude that these mutations do not result in general structure changes but specifically affect CaM binding.
The CaM binding site of c-Rel is of functional importance in vivo.
In order to address the question of whether the interactions of RelA and c-Rel with CaM are relevant for their activity in vivo, we analyzed the wild-type and CaM binding-deficient proteins in Jurkat cells. Cells were cotransfected with the indicated NF-κB/Rel expression plasmids and reporter plasmids in which the expression of the reporter gene luciferase is driven by the transcription control regions of either the IL-2 or the GM-CSF genes. The IL-2 and GM-CSF reporters were chosen as examples of cytokines that are transcriptionally regulated by both NF-κB/Rel and Ca2+ (29, 58, 59). A Ca2+ signal by itself did not activate NF-κB/Rel, since an additional signal is also needed for the degradation of IκB (data not shown). Cells were therefore stimulated with PMA plus ionomycin or PMA alone, and the effect of Ca2+was determined by calculating the ratio of the reporter activities resulting from the two different stimulations (Fig. 5). Overexpression of wild-type c-Rel was found to potentiate the Ca2+-dependent activation of transcription of both cytokine reporters. Most interestingly, the CaM binding-deficient c-Rel mutants activated better than did wild-type c-Rel under this circumstance. RelA, however, did not positively affect the Ca2+-induced reporter activities. The apparent decrease in the effect of Ca2+ on the GM-CSF reporter activity induced by RelA (Fig. 5B) is the result of a high responsiveness to PMA compared to that observed in cells transfected with c-Rel, c-Rel H−, c-Rel HB− or in vector control-transfected cells (data not shown). In contrast to the c-Rel mutants, a mutation in the CaM binding site of RelA does not alter the effect of RelA on Ca2+ activation of the GM-CSF reporter (Fig. 5B) and alters only slightly its effect on the IL-2 reporter (Fig. 5A). Western blot analysis verified that transfected wild-type and mutated proteins were expressed at the same level (data not shown).
FIG. 5.
Functional importance of the CaM binding sequences of c-Rel and RelA. c-Rel, c-Rel H−, c-Rel HB−, RelA, or RelA H− were transiently expressed in Jurkat cells together with an IL-2 (A) or GM-CSF (B) reporter plasmid. Twenty-two hours after transfection, cells were stimulated for 2 h with PMA or PMA plus ionomycin. Values shown are the ratios between the reporter gene activity in the presence of ionomycin and the reporter gene activity in the absence of ionomycin and are normalized to empty vector. Bars represent average ratios from at least three independent transfections ± standard errors of the means.
These data suggest that the CaM binding site of c-Rel is of functional importance in vivo, since the interaction between CaM and c-Rel is one way by which Ca2+ regulates the expression of the cytokines IL-2 and GM-CSF.
Ca2+- and CaM-mediated regulation of c-Rel nuclear localization.
The effects of mutating the CaM binding sites in c-Rel, demonstrated in the reporter gene assay, could possibly be explained by an altered cellular distribution of the mutated proteins compared to that observed with their wild-type counterpart. To test this hypothesis, Jurkat cells were transiently transfected with wild-type or the CaM binding-deficient derivatives of c-Rel or RelA and stimulated with PMA, PMA plus ionomycin, or TNF-α. Cytoplasmic and nuclear extracts were prepared and analyzed by Western blotting to determine the localization of the proteins. The extracts from cells overexpressing c-Rel were also subjected to Western blotting with antibodies against α-tubulin and histone H1 as a control of the fractionation method. The cytoplasmic protein α-tubulin was found exclusively in the cytoplasmic extracts, and the nuclear protein histone H1 was found only in nuclear extracts, showing that there were no significant cross-contaminations (Fig. 6A). Interestingly, both CaM binding-deficient c-Rel proteins accumulated in the nucleus to a much higher extent than the wild-type protein did (Fig. 6A). This, together with the observation of the increased ability of c-Rel H− and c-Rel HB− to activate transcription (Fig. 5), suggests that when c-Rel is unable to bind CaM, its nuclear concentration increases and thus more protein is available to activate transcription. The enhanced nuclear accumulation of CaM binding-deficient mutants of c-Rel was small upon stimulation with PMA alone, but upon combined stimulation with PMA and ionomycin, the nuclear accumulation/cytoplasmic accumulation ratio became approximately twofold higher than for the wild-type protein (Fig. 6A and B). Ionomycin alone had no effect on the nuclear localization of NF-κB/Rel, since an additional signal is also needed for the degradation of IκB (data not shown). TNF-α stimulation, which involves Ca2+ signaling (33, 39), resulted in approximately fourfold-higher nuclear/cytoplasmic accumulation ratios for the mutants than for the wild type (Fig. 6A and B). The Ca2+-dependent effects emphasize the importance of the CaM binding site for controlling the nuclear localization of c-Rel. In addition, analysis of the cellular distribution of c-Rel and c-Rel HB− in DG75 cells, in which IκB degradation is constitutively high (see above), demonstrated a much higher nuclear accumulation of CaM binding-deficient c-Rel (Fig. 6C).
In contrast to c-Rel, mutation of the CaM binding site of RelA did not increase its nuclear accumulation, since accumulation of RelA H− in the nucleus was not significantly different from that of the wild-type protein (Fig. 6D and E). This finding reveals that there is a differential regulation of these two NF-κB/Rel proteins at this level.
DISCUSSION
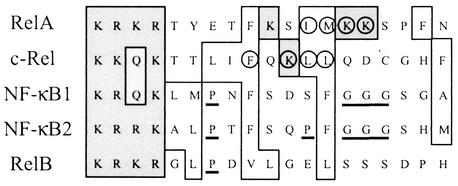
NF-κB/Rel proteins are primarily regulated by subcellular location, being sequestered by IκB in the cytoplasm and released into the nucleus upon stimulation by one of the numerous NF-κB/Rel activating signals (17, 27, 31, 32, 50). There is, however, increasing evidence that NF-κB/Rel is also regulated by other mechanisms, such as phosphorylation and interaction with other proteins (17). Here we report that some NF-κB/Rel proteins are regulated through interaction with the Ca2+ binding protein CaM, and our analysis of the function of this interaction in vivo reveals possible mechanisms by which NF-κB/Rel proteins can be differentially regulated. c-Rel and RelA, but not NF-κB1 (p50), interact Ca2+ specifically with CaM. The interaction is dramatically enhanced by NF-κB/Rel-activating agents. This enhancement occurs predominantly in the cytoplasm and can be blocked by the addition of IκBα, suggesting that c-Rel and RelA can bind to either CaM or IκB and that degradation of IκB is a prerequisite for c-Rel and RelA to interact with CaM. This conclusion was supported by analyses with purified E. coli-produced proteins, in which IκBα directly inhibits binding of RelA to CaM-Sepharose (data not shown). CaM targets are often basic and amphiphilic α-helical segments, in which basic and hydrophobic amino acids are critical for the CaM interaction (48). It has been shown, in a RelA/NF-κB1-IκBα complex, that the CaM binding sequence of RelA is an α-helical structure (28) and the CaM binding sequences of c-Rel and RelA contain basic and hydrophobic residues. The importance of these residues for the CaM interaction is illustrated in Fig. 3B to D, which show that mutation of basic or hydrophobic amino acids in these sequences severely reduced the ability of c-Rel and RelA to bind CaM both in vitro and in vivo. The corresponding sequence of NF-κB1 (Fig. 7) contains a proline residue and a stretch of three glycines, features that destabilize α-helices, which might explain why NF-κB1 does not bind CaM. NF-κB2 and RelB also contain α-helix-destabilizing residues in their corresponding sequences, and no other sequence in NF-κB2 or RelB is homologous to the CaM binding site of c-Rel or RelA, suggesting that these proteins are not CaM binding.
FIG. 7.
Comparison of the CaM binding sequences of RelA and c-Rel with the corresponding sequences of other mammalian NF-κB/Rel proteins. Sequences surrounding the identified residues important for CaM binding of mouse RelA (amino acids 301 to 319) and c-Rel (amino acids 294 to 312) were aligned with the corresponding sequences of NF-κB1 (amino acids 369 to 387), RelB (amino acids 388 to 406), and the cloned human NF-κB2 (amino acids 338 to 356) by using the ClustalW program (http://www2.ebi.ac.uk/clustalw). Hydrophobic and basic residues are indicated by open and shaded boxes, respectively. Residues of RelA and c-Rel that were mutated to disrupt CaM binding are circled, and potential α-helix-disturbing amino acids, P or rows of G, in the corresponding regions of NF-κB1, NF-κB2, and RelB are underlined.
NF-κB/Rel proteins play important roles in the regulation of the expression of many cytokines, such as IL-2 and GM-CSF (50). Mutation of the CaM binding site of c-Rel resulted in an increased ability to activate transcription from the promoters of IL-2 and GM-CSF in the presence of a Ca2+ signal. Further analysis of the transfected proteins revealed that CaM binding-deficient c-Rel proteins accumulate in the nucleus to a greater extent than does the wild-type protein upon cellular stimulation. The simplest interpretation of these observations is that when c-Rel cannot bind CaM, NF-κB-inducing signals lead to concentration of c-Rel in the nucleus, and thus, the amount of protein available to activate transcription increases. This would imply that the role of CaM is to negatively regulate c-Rel nuclear accumulation, a task that could be achieved in two ways. First, CaM may positively regulate c-Rel nuclear export. The involvement of nuclear export in the regulation of NF-κB/Rel has recently been described (1, 20, 23, 30, 54, 55, 62, 63). The emerging picture is that IκB is continuously shuttling between the cytoplasm and the nucleus. In the nucleus, IκB binds NF-κB/Rel and transports it back to the cytoplasm. In addition to being subject to IκB-mediated nuclear export, RelA has also been reported to have its own nuclear export signal (20, 55). However, c-Rel lacks a nuclear export signal, suggesting that it is not subject to direct nuclear export (63). Furthermore, our finding that the inducible interaction between c-Rel and CaM occurs mainly in the cytoplasm argues against CaM being involved in a hypothetical regulation of c-Rel nuclear export. The second alternative is that CaM may negatively regulate nuclear import of c-Rel. There are several possible mechanisms by which CaM could achieve this. (i) It has been shown that removal of C-terminal residues of c-Rel allows the protein to accumulate in the nucleus and that fusion of the C-terminal half of c-Rel to v-Rel, which is normally nuclear, sequesters v-Rel in the cytoplasm (5). This suggests that mechanisms exist to prevent c-Rel from entering the nucleus and that this is mediated through sequences in its C terminus. CaM could perhaps be part of this regulatory mechanism, and when the CaM binding site of c-Rel is mutated this negative regulation is overcome. (ii) As mentioned above, CaM targets are usually α-helical structures (48). Interestingly, the NLS of RelA is known to be part of an α-helix when in a complex with IκBα (28). However, it has been demonstrated that an NLS bound to its nuclear import receptor, importin-α, is an extended chain (9). It is conceivable that CaM, by binding to c-Rel, imposes an α-helical structure on the NLS that is incompatible with importin-α interaction and subsequently nuclear import. (iii) The NLS of c-Rel is directly adjacent to the CaM binding site that we have identified. It is therefore possible that CaM could have a negative effect on c-Rel nuclear import simply by competing for binding to the amino acids that the nuclear import receptor importin-α binds to. It has been shown that basic amino acids in the N terminus of CaM-binding sequences are electrostatically interacting with acidic residues in CaM (26, 42, 43). If the basic residues in the NLS are responsible for such interactions, it is easy to envision how CaM would directly interfere with nuclear import of c-Rel.
In contrast to what was observed with c-Rel, nuclear accumulation of RelA was not affected by a mutation of the domain that interacts with CaM. The mechanism of this differential regulation is at present not known, but it may be that nuclear transport of RelA and c-Rel is mediated by different transport receptors. If that is the case, our data would imply that CaM is one of the factors involved in the regulation of c-Rel nuclear transport whereas regulation of RelA nuclear transport is independent of CaM. Another possibility is that CaM influences a c-Rel modification that controls its nuclear accumulation and that this modification does not occur in RelA. Interestingly, endogenous RelA is transported to the nucleus much faster than c-Rel following stimulation (11, 44; data not shown). The slower kinetics of c-Rel nuclear accumulation could be a result of the differential effect of CaM binding to RelA and c-Rel. Wild-type and CaM binding-deficient RelA proteins activate transcription nearly equally well on both IL-2 and GM-CSF reporter plasmids (data not shown). However, on a luciferase reporter plasmid with only two κB-sites, the RelA mutant is 3.3-fold less efficient in activating transcription than the wild-type protein (data not shown). This would imply that RelA also is regulated by the CaM interaction, but not in the same way as c-Rel. We have not found any direct effect of CaM on the DNA binding properties of RelA, or c-Rel, in vitro (data not shown), but it is possible that the interaction with CaM is affecting the ability of RelA to activate transcription. However, further studies are required in order to address the functional consequences of this interaction.
In earlier studies, it was found that inhibitors of CaM completely block PMA-induced phosphorylation of IκBα (24), with CaM acting via CaMKII at a step prior to IKK activation (25). Taking all the information together, it is clear that CaM is an important regulatory protein for members of the NF-κB/Rel family, involved in the inactivation of their inhibitor IκBα, their nuclear accumulation, and possibly their ability to activate transcription. Defining the mechanism by which CaM acts at each of these levels and how these mechanisms differ for NF-κB/Rel family members may reveal important regulatory steps in the control of NF-κB/Rel proteins.
Acknowledgments
This work was supported by grants from the Swedish Natural Science Research Council and the Swedish Cancer Society.
REFERENCES
- 1.Arenzana Seisdedos, F., P. Turpin, M. Rodriguez, D. Thomas, R. T. Hay, J. L. Virelizier, and C. Dargemont. 1997. Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J. Cell Sci. 110:369-378. [DOI] [PubMed] [Google Scholar]
- 2.Barkett, M., and T. D. Gilmore. 1999. Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene 18:6910-6924. [DOI] [PubMed] [Google Scholar]
- 3.Beg, A. A., T. S. Finco, P. V. Nantermet, and A. S. Baldwin, Jr. 1993. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of I kappa B alpha: a mechanism for NF-kappa B activation. Mol. Cell. Biol. 13:3301-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird, T. A., K. Schooley, S. K. Dower, H. Hagen, and G. D. Virca. 1997. Activation of nuclear transcription factor NF-kappaB by interleukin-1 is accompanied by casein kinase II-mediated phosphorylation of the p65 subunit. J. Biol. Chem. 272:32606-32612. [DOI] [PubMed] [Google Scholar]
- 5.Capobianco, A. J., D. L. Simmons, and T. D. Gilmore. 1990. Cloning and expression of a chicken c-rel cDNA: unlike p59v-rel, p68c-rel is a cytoplasmic protein in chicken embryo fibroblasts. Oncogene 5:257-265. [PubMed] [Google Scholar]
- 6.Carafoli, E., P. Nicotera, and L. Santella. 1997. Calcium signalling in the cell nucleus. Cell Calcium 22:313-319. [DOI] [PubMed] [Google Scholar]
- 7.Chen, F. E., and G. Ghosh. 1999. Regulation of DNA binding by Rel/NF-kappaB transcription factors: structural views. Oncogene 18:6845-6852. [DOI] [PubMed] [Google Scholar]
- 8.Cockerill, P. N., M. F. Shannon, A. G. Bert, G. R. Ryan, and M. A. Vadas. 1993. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc. Natl. Acad. Sci. USA 90:2466-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193-204. [DOI] [PubMed] [Google Scholar]
- 10.Corneliussen, B., M. Holm, Y. Waltersson, J. Onions, B. Hallberg, A. Thornell, and T. Grundström. 1994. Calcium/calmodulin inhibition of basic-helix-loop-helix transcription factor domains. Nature 368:760-764. [DOI] [PubMed] [Google Scholar]
- 11.Doerre, S., P. Sista, S. C. Sun, D. W. Ballard, and W. C. Greene. 1993. The c-rel protooncogene product represses NF-kappa B p65-mediated transcriptional activation of the long terminal repeat of type 1 human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 90:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolmetsch, R. E., R. S. Lewis, C. C. Goodnow, and J. I. Healy. 1997. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature 386:855-858. [DOI] [PubMed] [Google Scholar]
- 13.Dolmetsch, R. E., K. Xu, and R. S. Lewis. 1998. Calcium oscillations increase the efficiency and specificity of gene expression. Nature 392:933-936. [DOI] [PubMed] [Google Scholar]
- 14.Durand, D. B., J. P. Shaw, M. R. Bush, R. E. Replogle, R. Belagaje, and G. R. Crabtree. 1988. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol. Cell. Biol. 8:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerondakis, S., M. Grossmann, Y. Nakamura, T. Pohl, and R. Grumont. 1999. Genetic approaches in mice to understand Rel/NF-kappaB and IkappaB function: transgenics and knockouts. Oncogene 18:6888-6895. [DOI] [PubMed] [Google Scholar]
- 16.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 19.Govind, S. 1999. Control of development and immunity by Rel transcription factors in Drosophila. Oncogene 18:6875-6887. [DOI] [PubMed] [Google Scholar]
- 20.Harhaj, E. W., and S. C. Sun. 1999. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Mol. Cell. Biol. 19:7088-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harley, V. R., R. Lovell Badge, P. N. Goodfellow, and P. J. Hextall. 1996. The HMG box of SRY is a calmodulin binding domain. FEBS Lett. 391:24-28. [DOI] [PubMed] [Google Scholar]
- 22.Hermann, S., J. Saarikettu, J. Onions, K. Hughes, and T. Grundstrom. 1998. Calcium regulation of basic helix-loop-helix transcription factors. Cell Calcium 23:135-142. [DOI] [PubMed] [Google Scholar]
- 23.Huang, T. T., N. Kudo, M. Yoshida, and S. Miyamoto. 2000. A nuclear export signal in the N-terminal regulatory domain of IkappaBalpha controls cytoplasmic localization of inactive NF-kappaB/IkappaBalpha complexes. Proc. Natl. Acad. Sci. USA 97:1014-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes, K., Å. Antonsson, and T. Grundström. 1998. Calmodulin dependence of NFkappaB activation. FEBS Lett. 441:132-136. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, K., S. Edin, A. Antonsson, and T. Grundstrom. 2001. Calmodulin-dependent kinase II mediates T cell receptor/CD3- and phorbol ester-induced activation of IkappaB kinase. J. Biol. Chem. 276:36008-36013. [DOI] [PubMed] [Google Scholar]
- 26.Ikura, M., G. M. Clore, A. M. Gronenborn, G. Zhu, C. B. Klee, and A. Bax. 1992. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256:632-638. [DOI] [PubMed] [Google Scholar]
- 27.Israel, A. 2000. The IKK complex: an integrator of all signals that activate NF-kappaB? Trends Cell Biol. 10:129-133. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs, M. D., and S. C. Harrison. 1998. Structure of an IkappaBalpha/NF-kappaB complex. Cell 95:749-758. [DOI] [PubMed] [Google Scholar]
- 29.Jain, J., C. Loh, and A. Rao. 1995. Transcriptional regulation of the IL-2 gene. Curr. Opin. Immunol. 7:333-342. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, C., D. Van Antwerp, and T. J. Hope. 1999. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IkappaBalpha. EMBO J. 18:6682-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karin, M. 1999. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene 18:6867-6874. [DOI] [PubMed] [Google Scholar]
- 32.Karin, M., and M. Delhase. 2000. The I kappa B kinase (IKK) and NF-kappa B: key elements of proinflammatory signalling. Semin. Immunol. 12:85-98. [DOI] [PubMed] [Google Scholar]
- 33.Kim, B. C., H. T. Kim, M. Mamura, I. S. Ambudkar, K. S. Choi, and S. J. Kim. 2002. TNF induces apoptosis in hepatoma cells by increasing Ca2+ release from the endoplasmic reticulum and suppressing Bcl-2 expression. J. Biol. Chem. 277:31381-31389. [DOI] [PubMed] [Google Scholar]
- 34.Lahdenpohja, N., T. Henttinen, and M. Hurme. 1996. Activation of the protein kinase A increases the DNA-binding and transcriptional activity of c-Rel in T cells. Scand. J. Immunol. 43:640-645. [DOI] [PubMed] [Google Scholar]
- 35.Lars, N., and S. Paschalis. 1993. The human I alpha 1 and I alpha 2 germline promoter elements: proximal positive and distal negative elements may regulate the tissue specific expression of C alpha 1 and C alpha 2 germline transcripts. Int. Immunol. 5:271-282. [DOI] [PubMed] [Google Scholar]
- 36.Lenardo, M. J., A. Kuang, A. Gifford, and D. Baltimore. 1988. NF-kappa B protein purification from bovine spleen: nucleotide stimulation and binding site specificity. Proc. Natl. Acad. Sci. USA 85:8825-8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, C. C., R. M. Dai, E. Chen, and D. L. Longo. 1994. Phosphorylation of NF-KB1-p50 is involved in NF-kappa B activation and stable DNA binding. J. Biol. Chem. 269:30089-30092. [PubMed] [Google Scholar]
- 38.Liljeholm, S., K. Hughes, T. Grundström, and P. Brodin. 1998. NF-kappaB only partially mediates Epstein-Barr virus latent membrane protein 1 activation of B cells. J. Gen. Virol. 79:2117-2125. [DOI] [PubMed] [Google Scholar]
- 39.MacEwan, D. J. 2002. TNF receptor subtype signalling: differences and cellular consequences. Cell. Signal. 14:477-492. [DOI] [PubMed] [Google Scholar]
- 40.Martin, A. G., and M. Fresno. 2000. Tumor necrosis factor-alpha activation of NF-kappa B requires the phosphorylation of Ser-471 in the transactivation domain of c-Rel. J. Biol. Chem. 275:24383-24391. [DOI] [PubMed] [Google Scholar]
- 41.May, M. J., and S. Gosh. 1999. I-kappaB kinases: kinsmen with different crafts. Science 284:271-273. [DOI] [PubMed] [Google Scholar]
- 42.Meador, W. E., A. R. Means, and F. A. Quiocho. 1993. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 262:1718-1721. [DOI] [PubMed] [Google Scholar]
- 43.Meador, W. E., A. R. Means, and F. A. Quiocho. 1992. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 257:1251-1255. [DOI] [PubMed] [Google Scholar]
- 44.Molitor, J. A., W. H. Walker, S. Doerre, D. W. Ballard, and W. C. Greene. 1990. NF-kappa B: a family of inducible and differentially expressed enhancer-binding proteins in human T cells. Proc. Natl. Acad. Sci. USA 87:10028-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosialos, G., P. Hamer, A. J. Capobianco, R. A. Laursen, and T. D. Gilmore. 1991. A protein kinase-A recognition sequence is structurally linked to transformation by p59v-rel and cytoplasmic retention of p68c-rel. Mol. Cell. Biol. 11:5867-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naumann, M., and C. Scheidereit. 1994. Activation of NF-kappa B in vivo is regulated by multiple phosphorylations. EMBO J. 13:4597-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ning, Y. M., and E. R. Sanchez. 1995. Evidence for a functional interaction between calmodulin and the glucocorticoid receptor. Biochem. Biophys. Res. Commun. 208:48-54. [DOI] [PubMed] [Google Scholar]
- 48.O'Neil, K. T., and W. F. DeGrado. 1990. How calmodulin binds its targets: sequence independent recognition of amphiphilic alpha-helices. Trends Biochem. Sci. 15:59-64. [DOI] [PubMed] [Google Scholar]
- 49.Onions, J., S. Hermann, and T. Grundström. 1997. Basic helix-loop-helix protein sequences determining differential inhibition by calmodulin and S-100 proteins. J. Biol. Chem. 272:23930-23937. [DOI] [PubMed] [Google Scholar]
- 50.Pahl, H. L. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 51.Parry, G. C., and N. Mackman. 1997. Role of cyclic AMP response element-binding protein in cyclic AMP inhibition of NF-kappaB-mediated transcription. J. Immunol. 159:5450-5456. [PubMed] [Google Scholar]
- 52.Perkins, N. D. 1997. Achieving transcriptional specificity with NF-kappa B. Int. J. Biochem. Cell Biol. 29:1433-1448. [DOI] [PubMed] [Google Scholar]
- 53.Perkins, N. D., L. K. Felzien, J. C. Betts, K. Leung, D. H. Beach, and G. J. Nabel. 1997. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science 275:523-527. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez, M. S., J. Thompson, R. T. Hay, and C. Dargemont. 1999. Nuclear retention of I-kappaBalpha protects it from signal-induced degradation and inhibits nuclear factor kappaB transcriptional activation. J. Biol. Chem. 274:9108-9115. [DOI] [PubMed] [Google Scholar]
- 55.Sachdev, S., and M. Hannink. 1998. Loss of IkappaB alpha-mediated control over nuclear import and DNA binding enables oncogenic activation of c-Rel. Mol. Cell. Biol. 18:5445-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitz, M. L., S. Bacher, and M. Kracht. 2001. I kappa B-independent control of NF-kappa B activity by modulatory phosphorylations. Trends Biochem. Sci. 26:186-190. [DOI] [PubMed] [Google Scholar]
- 57.Schmitz, M. L., M. A. dos Santos Silva, and P. A. Baeuerle. 1995. Transactivation domain 2 (TA2) of p65 NF-kappa B. Similarity to TA1 and phorbol ester-stimulated activity and phosphorylation in intact cells. J. Biol. Chem. 270:15576-15584. [DOI] [PubMed] [Google Scholar]
- 58.Serfling, E., A. Avots, and M. Neumann. 1995. The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation Biochim. Biophys. Acta 1263:181-200. [DOI] [PubMed] [Google Scholar]
- 59.Shannon, M. F., L. S. Coles, M. A. Vadas, and P. N. Cockerill. 1997. Signals for activation of the GM-CSF promoter and enhancer in T cells. Crit. Rev. Immunol. 17:301-323. [DOI] [PubMed] [Google Scholar]
- 60.Shen, C. H., and J. Stavnezer. 1998. Interaction of stat6 and NF-kappaB: direct association and synergistic activation of interleukin-4-induced transcription. Mol. Cell. Biol. 18:3395-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szymanski, D. B., B. Liao, and R. E. Zielinski. 1996. Calmodulin isoforms differentially enhance the binding of cauliflower nuclear proteins and recombinant TGA3 to a region derived from the Arabidopsis Cam-3 promoter. Plant Cell 8:1069-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tam, W. F., L. H. Lee, L. Davis, and R. Sen. 2000. Cytoplasmic sequestration of rel proteins by IkappaBalpha requires CRM1-dependent nuclear export. Mol. Cell. Biol. 20:2269-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tam, W. F., W. Wang, and R. Sen. 2001. Cell-specific association and shuttling of IkappaBalpha provides a mechanism for nuclear NF-kappaB in B lymphocytes. Mol. Cell. Biol. 21:4837-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thornell, A., B. Hallberg, and T. Grundstrom. 1988. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol. Cell. Biol. 8:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Eldik, L., and D. M. Watterson (ed.). 1998. Calmodulin and signal transduction. Academic Press, New York, N.Y.
- 66.Wang, D., and A. S. Baldwin, Jr. 1998. Activation of nuclear factor-kappaB-dependent transcription by tumor necrosis factor-alpha is mediated through phosphorylation of RelA/p65 on serine 529. J. Biol. Chem. 273:29411-29416. [DOI] [PubMed] [Google Scholar]
- 67.Zhong, H., H. SuYang, H. Erdjument Bromage, P. Tempst, and S. Ghosh. 1997. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89:413-424. [DOI] [PubMed] [Google Scholar]
- 68.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1:661-671. [DOI] [PubMed] [Google Scholar]